For decades, biologists have discussed and experimented with bacterial viruses, called phage, as a means of treating bacterial infections (1, 2). Remarkably, our own evolution may have beaten us to it. In PNAS, Barr et al. present a compelling and unique hypothesis that animal cells use phage as weapons against bacterial pathogens (3). Animal cells ranging from cnidarian (coral) to human excrete mucus to protect themselves from their external environment. Barr et al. propose a unique function for mucus, to trap phage to intersect and destroy invading bacteria before they reach the mucus-encapsulated tissue. This hypothesis (termed BAM for bacteriophage adhering to mucus) suggests a unique component of the animal immune system governed by a cross-kingdom animal-phage mutualism.
The BAM model proposed by Barr et al. (3) is as follows: (i) Animal epithelial cells secrete mucus that is rich with mucin glycoproteins, which work like Velcro. These proteins have a unique structure that incorporates hundreds of negatively charged glycan chains, which extend nanometers into the surrounding environment (4). (ii) The other side of the Velcro is the bacteriophage capsid, which has an Ig-like protein domain that interacts with the glycoproteins. The capsid is the DNA reservoir or “head” of the phage, from which the “tail” or other structures that trigger infection extend. (iii) With the phage embedded headfirst into the mucus matrix, their protruding tails may provide protection by infecting and killing invading bacteria (Fig. 1).
Fig. 1.
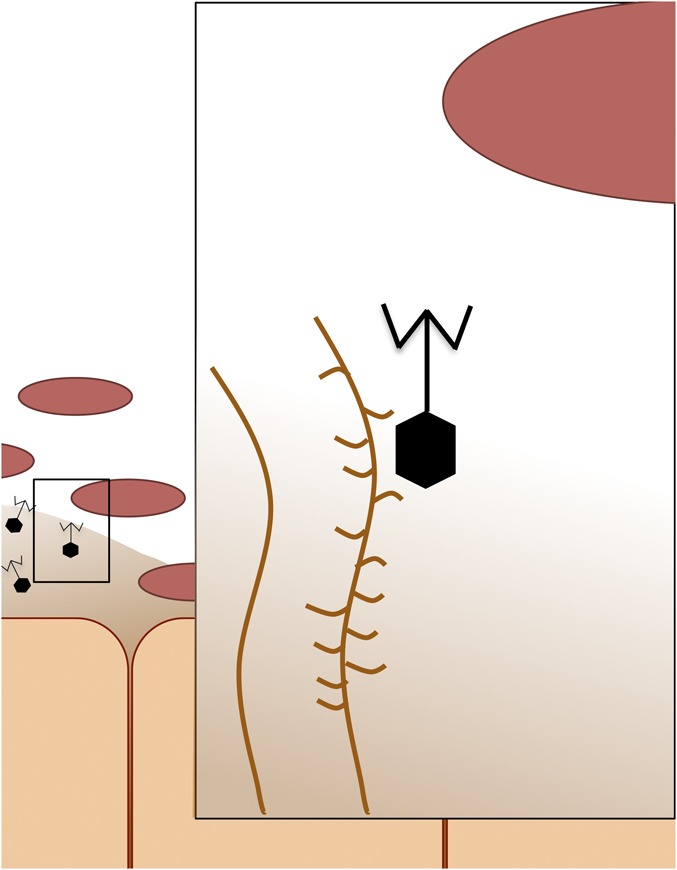
Bacteriophage adherence to mucus. Ig-like properties of phage capsids (hexagon shape) adhere to glycan residues on mucin glycoproteins (branched structure) of epithelial cell’s mucus layer. Once embedded within the mucus, phage defend animal cells against invading pathogenic bacteria.
In this interaction, animal cells benefit by gaining protection against bacterial pathogens, but the phage associating with the mucus layer may also benefit. Bacteria tend to congregate in and near mucus layers because the mucus provides a high resource habitat (5). Therefore, phage embedded in the mucus have an increased probability of encountering a host. In this model, chemical properties of mucus and the Ig-like domains of capsid proteins have coevolved to interact and reinforce this mutualistic relationship.
In support of this complex model, Barr et al. (3) identify two natural patterns consistent with BAM. First, and most importantly, mucus is enriched with phage. This finding is true for all mucus layers sampled, which ranged from invertebrates to humans. Second, phage with Ig-like domains were overrepresented in samples taken on or near mucus.
To provide direct causal evidence for BAM, Barr et al. (3) conducted experiments that incorporated techniques from many biological disciplines. The first set used a laboratory-based model for the three-species interaction, with human tissue cultures, the bacterial pathogen Escherichia coli, and the bacteriophage T4. Barr et al. began by showing that T4 adheres to mucus and that these phage particles remain viable and able to reproduce despite being tangled in a mucus web. Next, T4 were shown to efficiently kill invading E. coli. Most importantly, phage bound to mucus protected tissue culture cells from mortality caused by E. coli. Barr et al. also ran experiments to diagnose the molecular mechanisms that facilitate the phage-mucus interaction. As predicted, adherence of the phage to the mucus layer is a result of the interaction between glycans and Ig-like domains on the capsids.
Taken together, the experiments show that the physical mechanics proposed by BAM work under controlled laboratory settings. Combined with the natural patterns, the work strongly suggests that phage enrichment in mucus provides defense from bacterial pathogens.
The next big challenge will be finding evidence that BAM adds an additional layer to the animal immune system under natural conditions. In a more natural environment, animal cells encounter tens or hundreds of potentially pathogenic strains of bacteria. On average, phage are extremely strain specific (6, 7); thus, how a mucus layer creates a reservoir of phage extensive enough to protect against a myriad of bacterial diseases remains to be seen.
An additional limitation of BAM for providing immunity is that many phage are not lethal and can donate beneficial genes to their hosts (8). One example that challenges the effectiveness of BAM are pathogenic enteric bacteria from the genera Escherichia and Shigella. Many benign E. coli strains are made pathogenic after receiving toxin genes from phage of the λ family (9, 10), which also possess Ig-like domains on their capsids (11). Therefore, under some rare conditions, BAM may increase the incidence of disease by improving the probability of benign E. coli encountering a λ phage. Animal cells could avoid this consequence by selectively binding with other phage. It will be fascinating to know whether mucus is selective and if epithelial cells can reap the benefits of BAM, but avoid this negative consequence.
Another promising line of research to explore will be the role of BAM in the context of the animal microbiome. Recent work has revealed that a healthy microbiome helps maintain host fitness (12, 13). One way animal cells maintain a healthy microbiome is by producing compounds that favor the proliferation of commensal bacteria in mucus (14, 15). Perhaps BAM has a similar effect; phage may inhibit the growth of certain microbes, clearing space and resources for commensals to flourish. Unraveling the direct benefits of phage's ability to destroy invading pathogens, and other indirect benefits moderated through the microbiome will likely reveal new therapeutic possibilities for phage.
Overall, I believe BAM is a compelling hypothesis that has likely revealed a hidden, but ubiquitous and beneficial interaction between metazoans and phage. I look forward to seeing the important work this report will undoubtedly instigate.
Footnotes
The author declares no conflict of interest.
See companion article on page 10771.
References
- 1.Levin BR, Bull JJ. Population and evolutionary dynamics of phage therapy. Nat Rev Microbiol. 2004;2(2):166–173. doi: 10.1038/nrmicro822. [DOI] [PubMed] [Google Scholar]
- 2.Pirnay J-P, et al. Introducing yesterday’s phage therapy to today’s medicine. Future Virol. 2012;7(4):379–390. [Google Scholar]
- 3.Barr JJ, et al. Bacteriophage adhering to mucus provide a non–host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cone RA. Barrier properties of mucus. Adv Drug Deliv Rev. 2009;61(2):75–85. doi: 10.1016/j.addr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. Statistical structure of host-phage interactions. Proc Natl Acad Sci USA. 2011;108(28):E288–E297. doi: 10.1073/pnas.1101595108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores CO, Valverde S, Weitz JS. Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phage. ISME J. 2013;7(3):520–532. doi: 10.1038/ismej.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daubin V, Lerat E, Perrière G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4(9):R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barondess JJ, Beckwith J. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature. 1990;346(6287):871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- 10.Cheetham BF, Katz ME. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Molec Microbiol. 1995;18(2):201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 11.Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microb and Molec Biol Rev. 2011;75(3):423–433. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaser M, et al. The microbiome explored: Recent insights and future challenges. Nat Rev Microbiol. 2013;11(3):213–217. doi: 10.1038/nrmicro2973. [DOI] [PubMed] [Google Scholar]
- 13.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 14.Vaishnava S, et al. The antibacterial lectin RegIII γ promotes the spatial segregation of microbiota and host in the Intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10(11):e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]


