Abstract
The discovery of genomic imprinting through studies of manipulated mouse embryos indicated that the paternal genome has a major influence on placental development. However, previous research has not demonstrated paternal bias in imprinted genes. We applied RNA sequencing to trophoblast tissue from reciprocal hybrids of horse and donkey, where genotypic differences allowed parent-of-origin identification of most expressed genes. Using this approach, we identified a core group of 15 ancient imprinted genes, of which 10 were paternally expressed. An additional 78 candidate imprinted genes identified by RNA sequencing also showed paternal bias. Pyrosequencing was used to confirm the imprinting status of six of the genes, including the insulin receptor (INSR), which may play a role in growth regulation with its reciprocally imprinted ligand, histone acetyltransferase-1 (HAT1), a gene involved in chromatin modification, and lymphocyte antigen 6 complex, locus G6C, a newly identified imprinted gene in the major histocompatibility complex. The 78 candidate imprinted genes displayed parent-of-origin expression bias in placenta but not fetus, and most showed less than 100% silencing of the imprinted allele. Some displayed variability in imprinting status among individuals. This variability results in a unique epigenetic signature for each placenta that contributes to variation in the intrauterine environment and thus presents the opportunity for natural selection to operate on parent-of-origin differential regulation. Taken together, these features highlight the plasticity of imprinting in mammals and the central importance of the placenta as a target tissue for genomic imprinting.
Keywords: chorionic girdle, interspecific hybrid equids, mule, hinny
Genomic imprinting is a form of epigenetic modification in which gene expression differs in an allele-specific manner depending on parent of origin (1). Imprinted genes are important in normal fetal and placental development (2, 3), and their dysregulation in humans has been implicated in developmental abnormalities (4) and cancer (5). Although genomic imprinting has been recognized for more than 25 y, only about 120 imprinted genes have been verified in humans and mice, despite strenuous efforts to identify more (6, 7).
The first unambiguous experimental evidence for a division of labor between maternal and paternal genomes in mammalian development came from studies of manipulated mouse embryos (8, 9). With the identification of imprinted genes (10, 11), in which only the maternally or paternally inherited copy of a gene is active in any given cell, the emphasis in imprinting studies switched to investigating the role of these genes in development. Most of the known imprinted genes were identified in mice and humans, and studies in other organisms primarily have confirmed the imprinting status of previously identified genes (12, 13). However, studies in domestic livestock have revealed variation in normal phenotypes associated with imprinted genes for muscle growth and insulin-like growth factor-2 (IGF2) in pigs (14), for the delta-like 1 homolog (DLK) region in sheep (15), and for production traits in dairy cattle (16). In an investigation of horse, donkey, mule, and hinny pregnancies, peak serum concentrations of the placental hormone equine chorionic gonadotropin (eCG) were markedly higher when the sire was a horse than in pregnancies in which the sire was a donkey (17). Thus, the paternal genotype appeared to have a dominant influence on eCG production, consistent with the action of paternally expressed imprinted genes in placental development demonstrated 15 y later in mice (18, 19).
In the case of reciprocal horse–donkey hybrids, the mule (donkey father) and hinny (horse father) differ physiologically and in temperament, despite sharing nuclear genomes, leading to speculation that these phenotypic differences might be attributable to the action of imprinted genes. Evidence suggests that genome-wide methylation does not undergo any organizational alteration in interspecific hybrids of placental mammals (20); therefore, the mule and hinny may provide a model for the identification of imprinted genes.
Results and Discussion
Transcriptional Profiling of Horse, Donkey, Mule, and Hinny Trophoblast.
Transcriptome sequencing of the progeny of reciprocal mouse crosses has been used to discover unbiased sets of imprinted genes (21–25). We used RNA sequencing (RNA-seq) to obtain transcriptome sequences from the trophoblast cells of the chorionic girdle from conceptuses of horse and donkey and from the reciprocal F1 hybrids, the mule and hinny (Fig. 1 A and B). Chorionic girdle can be isolated as a single cell type, and these cells give rise to the endometrial cup trophoblasts that display an imprinted phenotype of eCG secretion (17). The RNA-seq procedure produced 11.4 Gbp, and 70% of the reads were uniquely mapped to the horse RefSeq database and whole-genome assembly (SI Appendix, Table S1). Although few donkey cDNA sequences are in public databases, the high homology between horse and donkey permitted assembly and identification of sequences from both species. From the cDNA sequences, we determined ∼50,000 mRNA single-nucleotide differences between horse and donkey (SI Appendix, SI Methods). We estimated the exonic SNP density between horse and donkey to be about 4–5/kb, a figure substantially higher than the 1/kb SNP density reported within horses (26). In total, 10,937 Ensembl transcripts were covered in the four genotypes (two parents and reciprocal F1 hybrids) with reads per kilobase per million (RPKM) >1.0 (SI Appendix, SI Methods).
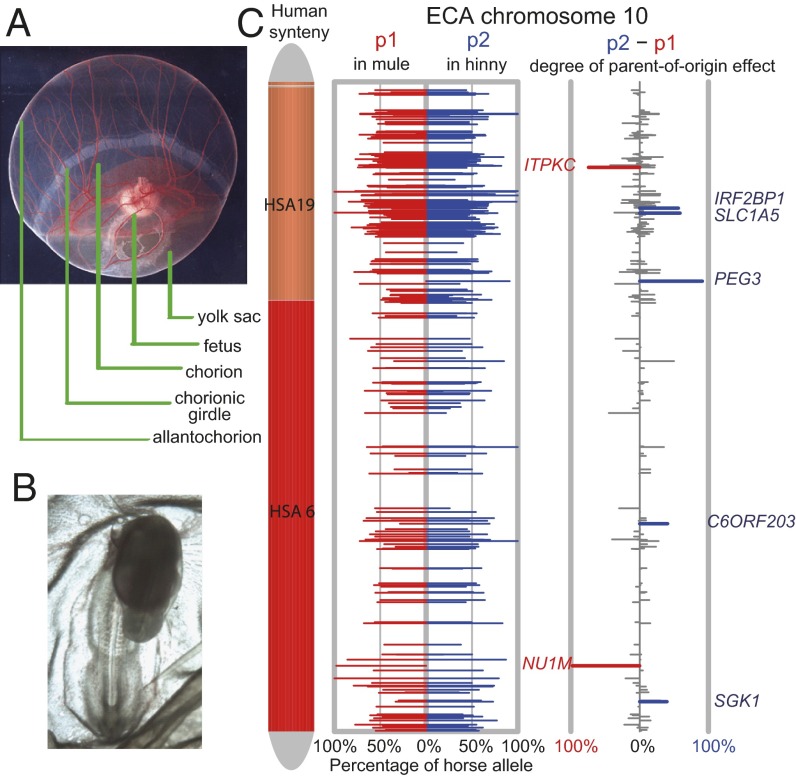
Fig. 1.
The mule–hinny chorionic girdle and fetal imprinting assay system. (A) Day 33 horse conceptus, showing the principal fetal and placental tissues, including the invasive trophoblast of the chorionic girdle. (B) Day 19 mule embryo. (C) Distribution of allelic expression ratio and parent-of-origin effect along equine chromosome 10 for all transcribed genes. (Left) Horse chromosome 10, color-coded with human chromosome synteny, showing two large syntenic blocks of human chromosomes 19 and 6. (Center) Plots of allelic expression bias for genes on chromosome 10 for both mule and hinny chorionic girdle samples. The x-axis shows the percentage of allelic expression from the horse allele (0–100%) in the two reciprocal F1 hybrids. The red bar depicts the percentage of horse allelic expression (p1) in the mule (horse allele is maternal in mules), and the blue bar represents the proportion of horse allelic expression (p2) in the hinny (paternal allele). (Right) Plot of the degree of parent-of-origin bias on chromosome 10. Red indicates statistically significant overexpression of the maternal allele, and blue indicates statistically significant overexpression of the paternal allele with a cutoff of q value < 0.01. Gray represents nonsignificant genes. The height of each bar is the degree of parent-of-origin effect, which is computed as (p2 − p1). Gene names of significant candidates are labeled (q-value < 0.05, Storer–Kim test).
Quantification of Allele-Specific Expression and Detection of Parent-of-Origin Effects in Mule and Hinny Placental Tissue.
Transcriptome-wide allele-specific expression was quantified in hybrid tissues using nucleotide sites that were nonpolymorphic in both horse and donkey but were different between the species (SI Appendix, SI Methods). Allele-specific expression ratios were obtained by counting the number of such fixed nucleotide differences originating from each parent (Fig. 1C). The uniform distribution of such differences enabled robust determination of paternal and maternal gene-expression levels for nearly 7,000 genes. Among the 48,125 SNPs with high coverage, 2,388 showed significant difference of allelic imbalance (q value <0.01), and 753 showed a parent-of-origin effect. In 93 candidate genes, there were reciprocal biased expression ratios of 65:35 or greater for a mule:hinny pair (SI Appendix, Table S2).
With this high level of ascertainment of parent-of-origin of transcripts, we expected to identify a large number of known imprinted genes in the hybrids. Although we detected 40 genes reported to be imprinted in human and/or mouse placenta, only 15 were imprinted in the mule and hinny. Twenty of the remaining 25 genes had informative SNPs but displayed clear biallelic expression in the equid trophoblast tissue (SI Appendix, Table S3). It is likely that many of these genes are imprinted in mice and/or humans and may also be imprinted in other equid tissues not tested in this study.
We mapped the 93 candidate imprinted genes to the horse genome (SI Appendix, Fig. S1); most of the previously identified genes were located in known clusters, whereas the 78 newly identified candidate imprinted genes were distributed across the equine genome without significant clustering, and no newly identified members of known imprinted clusters were found. We could discern no metabolic or functional gene ontology patterns common to the newly identified imprinted genes.
Newly Identified Imprinted Genes Are Trophoblast Specific and Show Interindividual Variability.
We confirmed the parent-of-origin expression bias of the 15 previously known imprinted genes and six newly identified imprinted genes using quantitative allele-specific pyrosequencing (Tables 1 and 2 and SI Appendix, Figs. S2–S19 and Table S4). These 21 genes also were tested in fetal samples from the same conceptuses. Most of the known imprinted genes displayed identical allelic expression bias in fetal tissue (Table 1), but the newly identified genes did not (Table 2 and SI Appendix, Table S2). The 21 confirmed genes were tested on additional samples of day 33–34 mule or hinny chorionic girdle trophoblasts, and all were found to be imprinted. However, although the previously known genes were imprinted in all individuals tested, two of the newly identified genes, insulin receptor (INSR) and stonin 1 (STON1), showed variability among individuals (SI Appendix, Text S1, Table S5, and Figs. S17 and S19).
Table 1.
RNA-seq allele-specific read counts and significance tests for known imprinted genes in mule and hinny placenta
| Rank* | Gene name | No. of significant SNPs | Known in mouse and/or human | Exp. allele | p1†, % | p2§, % | q-value | Pyro ratio¶ | Imprinted in fetus | Pyro ratio fetus |
| 1 | H19 | 10 | Human, mouse | M | 99.76 | 0.89 | 0 | 100:0 | Yes | 100:0 |
| 2 | IGF2 | 18 | Human, mouse | P | 0.67 | 99.95 | 0 | 0:100 | Yes | 0:100 |
| 3 | INS | 17 | Human, mouse | P | 0.68 | 99.95 | 0 | 0:100 | Yes | — |
| 4 | PHLDA2 | 8 | Human, mouse | M | 99.92 | 1.05 | 0 | 100:0 | Yes | 60:15 |
| 7 | IGF2R | 43 | Mouse | M | 73.59 | 24.08 | 0 | 60:20 | Yes | 70:15 |
| 8 | PEG10 | 17 | Human, mouse | P | 0.52 | 100.00 | 0 | 0:100 | Yes | 0:100 |
| 9 | MEST | 6 | Human, mouse | P | 2.69 | 100.00 | 0 | 5:100 | Yes | 10:90 |
| 11 | PEG3 | 17 | Human, mouse | P | 0.00 | 91.15 | 1.69E-205 | 0:85 | Yes | 5:100 |
| 21 | NAP1L4 | 10 | Mouse | M | 72.70 | 17.92 | 1.46E-47 | 75:20 | No | 45:40 |
| 24 | SNRPN | 2 | Human, mouse | P | 0.00 | 100.00 | 5.03E-12 | 0:100 | Yes | 10:100 |
| 36 | DLK1 | 1 | Human, mouse | P | 0.00 | 100.00 | 6.97E-09 | 0:100 | Yes | 0:100 |
| 37 | NDN | 2 | Human, mouse | P | 0.00 | 100.00 | 1.68E-08 | 20:100 | Yes | 10:100 |
| 46 | PAR-SN | 1 | Human | P | 0.00 | 100.00 | 1.61E-06 | 0:100 | Yes | 0:100 |
| 58 | MEG3 | 1 | Human, mouse | M | 100.00 | 0.00 | 7.39E-05 | 95:5 | Yes | 95:0 |
| 70 | SGCE | 1 | Human, mouse | P | 0.00 | 100.00 | 4.79E-04 | 0:90 | Yes | 15:85 |
| Fetus 7 | NNAT | 8 | Human, mouse | P | — | — | — | — | Yes | 0:100 |
| Fetus 25 | MAGEL2 | 2 | Human, mouse | P | — | — | — | — | Yes | 0:100 |
| Fetus 56 | DIRAS3 | 1 | Human | P | — | — | — | — | Yes | 0:100 |
The q-value ranking for the candidate imprinted genes in chorionic girdle. For genes only detected in fetus, the ranking is in the fetus candidates.
The allelic expression (percent horse allele) estimated from RNA-seq data in mule chorionic girdle.
The allelic expression (percent horse allele) estimated from RNA-seq data in hinny chorionic girdle.
The allelic expression (percent horse allele) quantified by allele-specific pyrosequencing (horse allele % in mule: horse allele % in hinny).
Table 2.
RNA-seq results for confirmed newly identified imprinted genes in mule and hinny placenta
| Rank* | Gene name | No. of significant SNPs | Exp. allele | p1†, % | p2§, % | q-value | Pyro ratio¶ | Variable? | Status in fetus |
| 13 | HAT1 | 3 | P | 2.8 | 100.0 | 5.24E-111 | 10:100 | No | Not imprinted |
| 14 | INSR | 17 | P | 15.7 | 100.0 | 3.96E-146 | 15:100 | Yes | Not imprinted |
| 25 | LY6G6C | 3 | M | 68.4 | 9.0 | 4.64E-19 | 85:10 | No | Not covered |
| 27 | D7ERTD715E | 2 | P | 0.0 | 100.0 | 2.42E-18 | 0:100 | No | Imprinted |
| 32 | STON1 | 1 | P | 4.9 | 100.0 | 3.43E-10 | 20:90 | Yes | Not imprinted |
| 54 | CFH | 1 | M | 100.0 | 0.0 | 3.88E-05 | 100:0 | No | Not covered |
The q-value ranking in the candidate imprinted genes in chorionic girdle. For genes only detected in fetus, the ranking is in the fetus candidates.
The allelic expression (percent horse allele) estimated from RNA-seq data in mule chorionic girdle.
The allelic expression (percent horse allele) estimated from RNA-seq data in hinny chorionic girdle.
The allelic expression (percent horse allele) quantified by allele-specific pyrosequencing (percent horse allele % in mule: percent horse allele in hinny).
Limited transcriptome sequencing of mule fetal tissue resulted in the identification of three additional known imprinted genes (Table 1). None of the 78 newly identified candidate imprinted genes displayed a parent-of-origin bias in fetal tissue (SI Appendix, Text S2). Thus, the parent-of-origin expression bias of the genes described here seems to be restricted to extraembryonic tissue.
Genomic Imprinting and Epigenetic Reprogramming Are Properly Regulated In Equid Hybrids.
Although we recognize that genomic imprinting might be abnormal in interspecific hybrids, our data do not support this possibility. For all the 15 equid imprinted genes identified in this study whose orthologs also are imprinted in mouse and human, the imprinting direction is the same in mule and hinny as in mouse/human (Table 1). Insulin-like growth factor 2 receptor (IGF2R) and nucleosome assembly protein 1-like 4 (NAP1L4) are preferentially expressed from the maternal allele in mouse, and they also are maternally expressed with paternal leakage in mule and hinny (Table 1). The conservation of direction and degree of imprinting for known imprinted genes suggests that genomic imprinting is not dysregulated in equid hybrids.
Mule chorionic girdle trophoblast cells and fetal fibroblasts demonstrated robust maintenance of imprinting status for IGF2, H19, paternally expressed gene 3 (PEG3), and histone acetyltransferase-1 (HAT1) after 30 d in continuous in vitro culture (SI Appendix, SI Methods). Methylation profiling of the differentially methylated regions (DMRs) for several previously known and newly identified imprinted genes revealed differential methylation in the tissues where the genes are imprinted (SI Appendix, Text S3). Treatment of cultured mule cells with a demethylating reagent or an inhibitor of histone deacetylase resulted in biallelic expression of H19 and IGF2, respectively, as has been shown in mice (Fig. 2 and SI Appendix, SI Methods) (27), providing further evidence that imprinting in horse × donkey hybrid tissue is not disrupted.
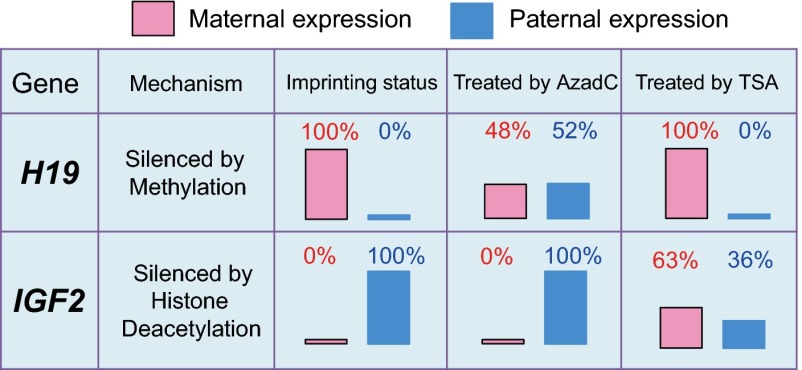
Fig. 2.
Change of imprinting status of H19 and IGF2 under 5-aza-2′-deoxycytidine (AzadC) and trichostatin A (TSA) treatments. The effect of methylation and deacetylase inhibitor on imprinting status for the H19 and IGF2 genes in mule placenta was checked in the chorionic girdle cells from a day 34 mule conceptus cultured for 33 d. In vitro treatment with the DNA methyltransferase inhibitor AzadC abolished the H19 imprinting status, but IGF2 was unaffected. When the cells were treated with the histone deacetylase inhibitor TSA, we observed biallelic expression for IGF2, but H19 imprinting status was not affected. The results in cultured mule cells are consistent with the report in mouse fibroblast cells.
One of the mechanisms for genomic imprinting is allele-specific differential DNA methylation (1). To investigate whether selected equid imprinted genes are regulated by DMRs, we checked one known paternal and one maternal imprinted gene with bisulfite sequencing to ascertain allele-specific differential methylation (Fig. 3 and SI Appendix, Text S3) (28). For H19, horse sperm was fully methylated through the relevant CpG island, and an identical methylation profile was found in the horse, donkey, mule, and hinny chorionic girdle, consistent with paternal silencing (Fig. 3A). In the CpG island of H19, we discovered a fixed single-nucleotide difference between horse and donkey which allowed identification of the parent-of-origin of methylated alleles in the mule and hinny (Fig. 3A). The paternally expressed gene PEG3 exhibited a methylation pattern opposite that of H19, in which sperm DNA was unmethylated in the CpG island. Another informative fixed nucleotide difference in the CpG island permitted unambiguous identification of the paternal and maternal DNA strands in mule and hinny (Fig. 3B).
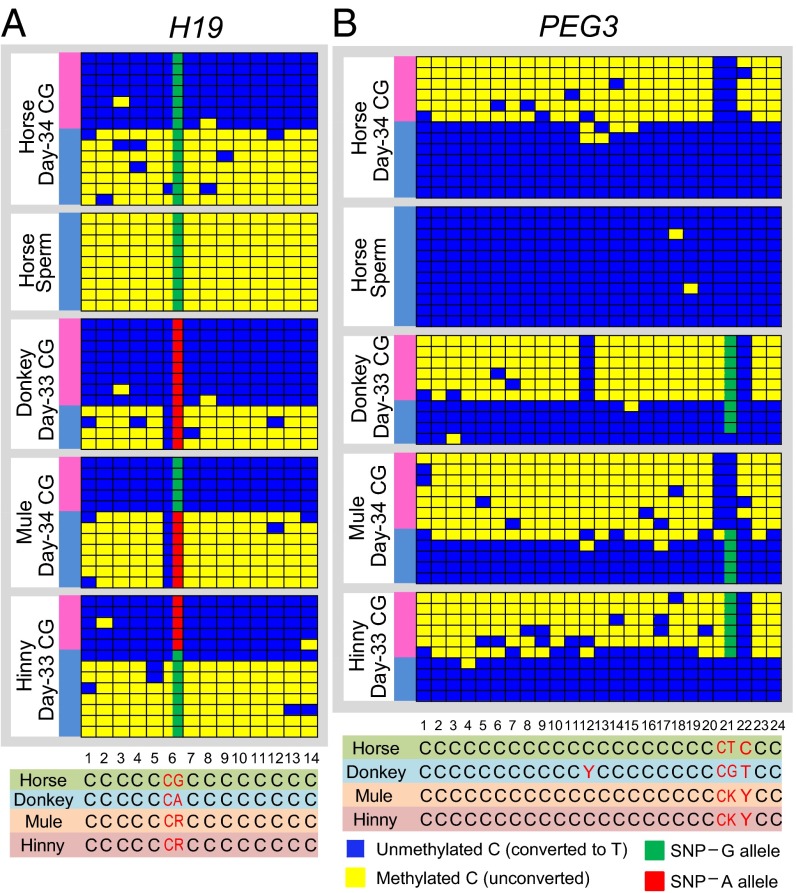
Fig. 3.
Strand-specific methylation of CpG islands of equine imprinted genes. Bisulfite sequencing of the DMRs in H19 and PEG3 shows allele-specific differential methylation corresponding to the allelic expression bias. Parental allele-specific methylation in mule and hinny is inferred from differences in single-nucleotide sequences in horse and donkey. Yellow boxes depict methylated CpGs, and blue boxes depict unmethylated CpGs. Each panel shows multiple CpG sites at the DMR for the corresponding gene. (A) H19 is 100% methylated in sperm, consistent with paternal silencing. The H19 DMR is differentially methylated in day 33–34 chorionic girdle (CG) samples from horse, donkey, mule, and hinny, with paternal methylation. Note the concordance between the SNP allelic state and the methylation state in mule and hinny. (B) PEG3 is unmethylated in sperm, consistent with paternal expression. The PEG3 DMR is differentially methylated in day 33–34 chorionic girdle samples from horse, donkey, mule, and hinny, with maternal methylation.
Finally, we confirmed the imprinting status in at least one parental species for three previously known (IGF2, PEG3, and H19) and two newly identified (HAT1 and INSR) imprinted genes (SI Appendix, Text S4 and Figs. S20–S22). This result indicates that the epigenetic mechanisms governing imprinted genes that operate within species are retained within interspecific hybrids of horse and donkey and is not surprising, given the close evolutionary relationship between horse and donkey (29) and our own observations that the coding sequences of many genes were identical between horse and donkey.
HAT1: a Placenta-Specific Imprinted Gene Identified in Equids.
HAT1 is a newly identified imprinted gene that is directly involved in epigenetic modifications. HAT1 can acetylate soluble histone H4 in the cytoplasm at the Lys-5 and Lys-12 positions (30). Previous studies suggested that HAT1 functions only in the cytoplasm, but more recent work has shown that it has nuclear function as a histone chaperone and can assist chromatin assembly (31–33). The imprinting status of HAT1 exhibited a high degree of tissue specificity, with virtually 100% paternal expression in day 33 mule and hinny chorionic girdle trophoblasts (Fig. 4 A–C). It is possible that the imprinting of some genes arises as a byproduct of an evolutionary pressure to alter total expression levels, and we note that the total level of HAT1 message was elevated when the horse was the sire, consistent with the direction of imprinting (Fig. 4 D and E). The imprinting status of HAT1 was consistent in trophoblast tissue in all hybrid individuals tested, but paired fetal samples showed perfect biallelic expression, as did samples from adult mule and hinny liver and lymphocytes (Fig. 4F). This finding suggests that HAT1 is a placental tissue-specific imprinted gene. The promoter CpG island of HAT1 was differentially methylated in chorionic girdle samples of all four species/hybrids and was 100% unmethylated in horse sperm (Fig. 4G). We found maternal-only DNA methylation at the DMR in mule and hinny, using a fixed nucleotide difference between donkey and horse in the HAT1 DMR, which enables unequivocal identification of paternal and maternal allelic methylation status. The DMR was unmethylated in fetal and adult lymphocyte samples, confirming the tissue-restricted imprinting profile revealed by transcriptome sequencing (Fig. 4G). These results demonstrate that the tissue-specific imprinting status of HAT1 is consistent with the allele-specific methylation of this gene.
Fig. 4.
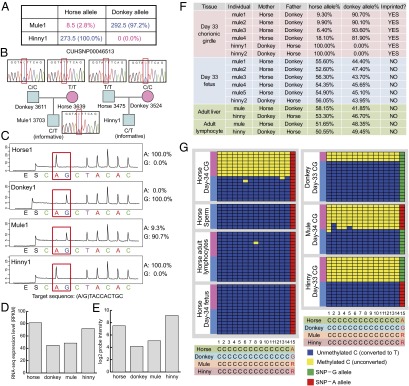
HAT1, a newly identified imprinted gene in the equine placenta. (A) Allele counts from RNA-seq data of HAT1 in chorionic girdle of mule and hinny showing the strong paternally biased allelic expression. (B) SNP genotyping in mule and hinny and their parents by Sanger sequencing, showing that HAT1 exonic SNP CUHSNP00046513 is informative. (C) Allele-specific pyrosequencing confirmation of the paternal allele overexpression of HAT1 in mule and hinny day 33 chorionic girdle samples. The target sequence is on the opposite strand. (D) Total expression levels expressed as RPKM in RNA-seq data indicating that total HAT1 expression tracks with the paternal parent. (E) Total expression levels expressed as log2 probe intensity in Agilent 4 × 44K Horse Gene Expression Microarray of the same individual samples showing that total HAT1 expression tracks with the paternal parent, as is consistent with the results shown in D. (F) Allele-specific pyrosequencing verification of HAT1 imprinting in six different mule/hinny individuals in day 33 chorionic girdle and fetus samples and in adult liver and lymphocytes in one mule and one hinny, demonstrating that imprinting is limited to the placental tissue and that fetal and adult tissues are biallelically expressed. (G) The HAT1 DMR is differentially methylated in horse, donkey, mule, and hinny day 33–34 chorionic girdle samples with maternal-only methylation, consistent with paternal allelic expression. In the horse day 34 fetus and adult lymphocytes, the HAT1 DMR was 100% unmethylated, consistent with biallelic expression.
Newly Identified Receptor–Ligand Pair of Imprinted Genes.
IGF2 and IGF2R form a classic imprinted ligand–receptor gene pair with a pronounced effect on fetal growth. In many mammals the growth-promoting IGF2 is paternally expressed, whereas the growth attenuating IGF2R is maternally expressed (34–36). Both genes were imprinted in equids: IGF2 with 100% paternal expression and IGF2R with preferential maternal expression. In our study we discovered that INSR, the insulin receptor, which binds to insulin to stimulate glucose uptake, was imprinted in the hybrid trophoblast. This newly identified imprinting status of INSR was verified in additional mule and hinny samples and in the horse (SI Appendix, Text S4). Thus, we describe a second imprinted ligand–receptor pair, INS and INSR, but in this case both display paternal expression. Consistent imprinting status of receptor–ligand pairs might be expected if there are tight stoichiometric constraints on the respective biological process.
Paternally Expressed Imprinted Genes Predominate in Placenta.
The discovery of imprinting through the construction of androgenetic and gynogenetic mice strongly suggested that the paternal genome has a major influence on placental development (8, 37). Similar conclusions can be drawn from tissue development in human molar pregnancies (38, 39). Surprisingly, previous studies have reported an excess number of maternally expressed genes in the placenta (7) or approximately equal numbers of paternally and maternally biased genes (22). From the 93 significant candidates reported here, we found more paternally biased than maternally biased genes (53:40). If we exclude the genes that are maternally biased because of mtDNA insertion and genes on potential X-linked contigs, there are 1.7 times more paternally biased candidates (53:31). For the 15 previously known and six newly identified verified imprinted genes, 67% (14/21) were paternally expressed (Tables 1 and 2). It is likely that there may be paternal expression bias in the placentae of other species. The high degree of purity of equid trophoblast preparations compared with placental isolations from mice may have reduced false-positive assignments caused by contaminating maternal tissues and facilitated the detection of paternal-expression bias in this extraembryonic lineage.
This paternal-expression bias of imprinted genes is consistent with the early experiments in mouse embryo manipulation (8, 37). Furthermore, our data provide a list of candidate genes that could produce the pattern of eCG levels in maternal serum described long ago for pregnant equids carrying intra- and interspecies pregnancies (17). The eCG genes themselves are not imprinted; the imprinting phenotype is a result of differential cell division that determines the number of eCG-producing cells: high numbers in horse and hinny (horse father) conceptuses, and low numbers in donkey and mule (donkey father). The eCG system represents an example of fetal signaling to the mother during pregnancy that is determined by the paternal genome and thus is consistent with the paternal–maternal conflict theory (40).
Conclusions: a Spectrum of Imprinted Gene Expression in the Placenta.
The 15 genes detected in equid placenta that undergo genomic imprinting (Table 1) all displayed patterns of paternal or maternal expression and degree of gene silencing that are shared across several mammalian species (human, mouse, cow, pig, and sheep; see www.geneimprint.com). Many of these genes also are imprinted in marsupials (41), suggesting an ancient origin. In contrast, the six confirmed newly identified imprinted genes (Table 2) and the 72 additional candidates (SI Appendix, Table S2) showed several different expression patterns, including less than 100% silencing of the imprinted gene, a high degree of tissue specificity, and variation in imprinting among individuals. The imprinting status of these genes may be of more recent origin and perhaps may be restricted to equids. The predominance of paternally expressed imprinted genes in equid trophoblast supports the evidence from earlier embryologic studies that the paternal genome has a major influence on placental development (8).
The placenta arose late in vertebrate evolution as a requirement for viviparity and at the gross and microscopic level long has been regarded as the most structurally diverse organ of mammals (42). Similarly, there is rapid turnover of proteins involved in providing nutrition and waste exchange for the fetus and in protecting the fetus from damaging maternal immune responses (43). Layered over this diversity in gene expression is the epigenetic control determined by genomic imprinting, a phenomenon that has puzzled biologists since its discovery more than 25 y ago (18, 19). Our identification of 78 candidate imprinted genes in equids suggests that different species express distinct complements of imprinted genes, reflecting a possible role of epigenetic modifications in producing the diversity of placental types. The evolutionary forces that generate new patterns of imprinted gene expression remain to be defined. Often genome rearrangements that move novel genes into proximity with imprinted gene clusters can be invoked to explain some of the evolutionary fluidity of imprinting status. Within the genus Equus such mechanisms that result in rapid diversification of karyotypes are likely to be involved (44).
We find support for the idea that genomic imprinting in the placenta may be an adaptive mechanism permitting plasticity of function in response to changing environmental conditions during gestation (45). The tissue specificity, incomplete silencing of the imprinted allele, and the interindividual variation in imprinting status that we have documented for many of the newly described imprinted genes may be other manifestations of flexibility in the design and construction of the mammalian placenta.
Methods
Tissue Dissection and Illumina mRNA-Seq.
Equine conceptuses were collected on days 33–35 post ovulation (46) and were microdissected into distinct tissues (SI Appendix, SI Methods). Animal care and experiments were performed in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee of Cornell University under protocol 1986-0216. Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen). RNA quality was assayed on an Agilent BioAnalyzer 2100 (Agilent). mRNA-seq was performed on RNA from horse, donkey, mule, and hinny chorionic girdle on an Illumina Genome Analyzer with 3 µg total RNA using the mRNA Seq-8 Sample Prep Kit (Illumina). We performed one Illumina GAIIx lane each for two mule fetus samples and one hinny fetus sample, with 6 µg of starting total RNA.
Bioinformatic Analysis.
The RNA-seq reads were aligned to the horse genome using Burrows–Wheeler Aligner (BWA) with a maximum of five mismatches (47). Alignment counts were normalized by transcript length and total coverage to compute RPKM. We performed de novo SNP calling from uniquely mapped reads using both Maq and SAMtools software (48). In mule and hinny, we counted reads with the horse and the donkey allele at each SNP position (22). Allelic expression ratios were calculated on a per-gene basis by summarizing all informative SNP positions.
Detection of Significant Parent-of-Origin Effects.
We defined p1 as the expression percentage from the horse allele in mule and p2 as the horse percentage in hinny. For a nonimprinted gene with 50:50% expression ratio p1 = p2 = 0.5. For an imprinted gene with strictly paternal expression, we expect p1 = 0 and p2 = 1. We use p2 − p1 to measure the parent-of-origin effect, ranging from −1 (100% maternal expression) to 0 (nonimprinted genes), to +1 (100% paternal expression). The Storer–Kim test (49) was used to test the null hypothesis that (p2 − p1) = 0. To include the significant partially imprinted candidates, we used an arbitrary cutoff of p1 >0.65 and p2 <0.35 for maternally expressed candidates and p1 <0.35 and p2 >0.65 for paternally expressed ones.
Verification of Candidate Imprinted Genes.
Pyrosequencing primers were designed with PyroMark Assay Design Software 2.0.1.15 (Qiagen). PCR amplification of genomic DNA and cDNA was carried out in 40-µL volumes using Ampli-Taq Gold polymerase (Life Technologies). PCR products were prepared for pyrosequencing on the PSQ 96MA Pyrosequencer (Qiagen) with the PyroMark Gold Reagents (Qiagen) using the Allele Quantification (AQ) method.
Analysis of Methylation Status.
CpG islands were identified using CpG Island Explorer 2.0 (50) and MethPrimer (51). Bisulfite conversion was carried out with Qiagen EpiTect Bisulfite Kit (Qiagen) or the MethylCode Bisulfite Conversion Kit (Invitrogen). PCR primers were designed using Methyl Primer Express software v1.0 (Life Technologies). Amplification of target regions was carried out using AmpliTaq Gold DNA Polymerase (Life Technologies) with 40 cycles of three-step PCR. Products then were purified and cloned into the pGEM T-Easy vector (Promega). Positive clones were sequenced at the Cornell University DNA Sequencing Facility (Ithaca, NY).
Demethylation and Deacetylase Inhibition Assays.
Fetal fibroblasts from three mule conceptuses and chorionic girdle cells revived from cultures were plated in six-well gelatin-coated plates. When cells reached 90% confluency, culture medium was removed, and cells were treated with 1.0 µM 5-Aza-2′-deoxycytidine, 0.3 µM trichostatin A, or culture medium. Treatment medium was removed from wells after 24 h, followed by cell harvesting and DNA and RNA isolation from separate aliquots.
Supplementary Material
Acknowledgments
We thank Amanda Manfredo and Li (Grace) Chi for assistance with Illumina sequencing and pyrosequencing experiments; Jim Hardy, Scott Hoffay, and Emily Silvela for management of the Baker Institute equid herd; and Anne Ferguson-Smith and Andrew Pask for helpful discussions. This work was funded by the Cornell Center for Vertebrate Genomics, the Zweig Memorial Fund, and the Morris Animal Foundation. A.G.C. is the recipient of a Meinig Family Investigator Award. D.F.A. is an investigator of the Dorothy Russell Havemeyer Foundation, Inc.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE30243).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308998110/-/DCSupplemental.
References
- 1.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011 doi: 10.1101/cshperspect.a002592. 3(7):a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fowden AL, Coan PM, Angiolini E, Burton GJ, Constancia M. Imprinted genes and the epigenetic regulation of placental phenotype. Prog Biophys Mol Biol. 2011;106(1):281–288. doi: 10.1016/j.pbiomolbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6(7):e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eggermann T, Eggermann K, Schönherr N. Growth retardation versus overgrowth: Silver-Russell syndrome is genetically opposite to Beckwith-Wiedemann syndrome. Trends Genet. 2008;24(4):195–204. doi: 10.1016/j.tig.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol. 2004;14(6):427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21(8):457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 2001;29(1):275–276. doi: 10.1093/nar/29.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311(5984):374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- 9.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV, Surani MA. Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature. 1991;351(6328):667–670. doi: 10.1038/351667a0. [DOI] [PubMed] [Google Scholar]
- 11.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 12.Bischoff SR, et al. Characterization of conserved and nonconserved imprinted genes in swine. Biol Reprod. 2009;81(5):906–920. doi: 10.1095/biolreprod.109.078139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaitoun I, Khatib H. Assessment of genomic imprinting of SLC38A4, NNAT, NAP1L5, and H19 in cattle. BMC Genet. 2006;7:49. doi: 10.1186/1471-2156-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markljung E, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7(12):e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda H, et al. The callipyge mutation enhances bidirectional long-range DLK1-GTL2 intergenic transcription in cis. Proc Natl Acad Sci USA. 2006;103(21):8119–8124. doi: 10.1073/pnas.0602844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magee DA, et al. Single nucleotide polymorphisms within the bovine DLK1-DIO3 imprinted domain are associated with economically important production traits in cattle. J Hered. 2011;102(1):94–101. doi: 10.1093/jhered/esq097. [DOI] [PubMed] [Google Scholar]
- 17.Allen WR. Factors influencing pregnant mare serum gonadotrophin production. Nature. 1969;223(5201):64–65. doi: 10.1038/223064a0. [DOI] [PubMed] [Google Scholar]
- 18.McGrath J, Solter D. Nucleocytoplasmic interactions in the mouse embryo. J Embryol Exp Morphol. 1986;97(Suppl):277–289. [PubMed] [Google Scholar]
- 19.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308(5959):548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 20.Roemer I, et al. Genome evolution. Global methylation in eutherian hybrids. Nature. 1999;401(6749):131–132. doi: 10.1038/43607. [DOI] [PubMed] [Google Scholar]
- 21.Gregg C, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329(5992):643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, et al. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE. 2008;3(12):e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Soloway PD, Clark AG. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics. 2011;189(1):109–122. doi: 10.1534/genetics.111.130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeVeale B, van der Kooy D, Babak T. Critical evaluation of imprinted gene expression by RNA-Seq: A new perspective. PLoS Genet. 2012;8(3):e1002600. doi: 10.1371/journal.pgen.1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okae H, et al. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum Mol Genet. 2012;21(3):548–558. doi: 10.1093/hmg/ddr488. [DOI] [PubMed] [Google Scholar]
- 26.Wade CM, et al. Broad Institute Genome Sequencing Platform Broad Institute Whole Genome Assembly Team Genome sequence, comparative analysis, and population genetics of the domestic horse. Science. 2009;326(5954):865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Kharroubi A, Piras G, Stewart CL. DNA demethylation reactivates a subset of imprinted genes in uniparental mouse embryonic fibroblasts. J Biol Chem. 2001;276(12):8674–8680. doi: 10.1074/jbc.M009392200. [DOI] [PubMed] [Google Scholar]
- 28.Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17(8):4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlando L, et al. Revising the recent evolutionary history of equids using ancient DNA. Proc Natl Acad Sci USA. 2009;106(51):21754–21759. doi: 10.1073/pnas.0903672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8(2):96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 31.Parthun MR. Histone acetyltransferase 1: More than just an enzyme? Biochim Biophys Acta. 2012;1819(3-4):256–263. doi: 10.1016/j.bbagrm.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge Z, Wang H, Parthun MR. Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J Biol Chem. 2011;286(19):16790–16799. doi: 10.1074/jbc.M110.216846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ejlassi-Lassallette A, Mocquard E, Arnaud MC, Thiriet C. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol Biol Cell. 2011;22(2):245–255. doi: 10.1091/mbc.E10-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349(6304):84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 35.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64(4):849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 36.Killian JK, et al. M6P/IGF2R imprinting evolution in mammals. Mol Cell. 2000;5(4):707–716. doi: 10.1016/s1097-2765(00)80249-x. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi N, Barton SC, Kaneda M, Hajkova P, Surani MA. The continuing quest to comprehend genomic imprinting. Cytogenet Genome Res. 2006;113(1-4):6–11. doi: 10.1159/000090808. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs PA, Wilson CM, Sprenkle JA, Rosenshein NB, Migeon BR. Mechanism of origin of complete hydatidiform moles. Nature. 1980;286(5774):714–716. doi: 10.1038/286714a0. [DOI] [PubMed] [Google Scholar]
- 39.Wake N, Arima T, Matsuda T. Involvement of IGF2 and H19 imprinting in choriocarcinoma development. Int J Gynaecol Obstet. 1998;60(Suppl 1):S1–S8. [PubMed] [Google Scholar]
- 40.Antczak DF, de Mestre AM, Wilsher S, Allen WR. The equine endometrial cup reaction: A fetomaternal signal of significance. Annual Review of Animal Biosciences. 2013;1(1):419–442. doi: 10.1146/annurev-animal-031412-103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renfree MB, Suzuki S, Kaneko-Ishino T. The origin and evolution of genomic imprinting and viviparity in mammals. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20120151. doi: 10.1098/rstb.2012.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mess A, Carter AM. Evolution of the placenta during the early radiation of placental mammals. Comp Biochem Physiol A Mol Integr Physiol. 2007;148(4):769–779. doi: 10.1016/j.cbpa.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 43.Wildman DE. Review: Toward an integrated evolutionary understanding of the mammalian placenta. Placenta. 2011;32(Suppl 2):S142–S145. doi: 10.1016/j.placenta.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raudsepp T, Mariat D, Guérin G, Chowdhary BP. Comparative FISH mapping of 32 loci reveals new homologous regions between donkey and horse karyotypes. Cytogenet Cell Genet. 2001;94(3-4):180–185. doi: 10.1159/000048812. [DOI] [PubMed] [Google Scholar]
- 45.Radford EJ, Ferrón SR, Ferguson-Smith AC. Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett. 2011;585(13):2059–2066. doi: 10.1016/j.febslet.2011.05.063. [DOI] [PubMed] [Google Scholar]
- 46.Antczak DF, et al. Differentiation molecules of the equine trophoblast. J Reprod Fertil Suppl. 1987;35:371–378. [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storer BE, Kim C. Exact Properties of Some Exact Test Statistics for Comparing 2 Binomial Proportions. J Am Stat Assoc. 1990;85(409):146–155. [Google Scholar]
- 50.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20(7):1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 51.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.