Abstract
Recently it has been discovered that a number of eukaryotic viruses, including HIV, coopt the cellular Endosomal Sorting Complex Required for Transport (ESCRT) machinery to affect egress from infected cells. Strikingly, the ESCRT apparatus is conserved in a subset of Archaea, including members of the genus Sulfolobus where it plays a role in cytokinesis. In the current work, we reveal that the archaeal virus Sulfolobus turreted icosahedral virus isolated from Yellowstone National Park's acidic hot springs also exploits the host ESCRT machinery in its replication cycle. Moreover, perturbation of normal ESCRT function abrogates viral replication and, thus, prevents establishment of a productive Sulfolobus turreted icosahedral virus infection. We propose that the Sulfolobus ESCRT machinery is involved in viral assembly within the cytoplasm and in escape from the infected cell by using a unique lysis mechanism. Our results support an ancient origin for viruses “hijacking” ESCRT proteins to complete their replication cycle and thus identify a critical host–virus interaction conserved between two domains of life.
The formation and evolutionary history of the three domains of life (Eukarya, Bacteria, and Archaea) and their viruses remains controversial. Compared with the bacterial and eukaryal domains, little is known about the archaeal domain of life and the viruses that infect these organisms. Archaea appear to contain a combination of bacterial and eukaryotic features. The cell structure and metabolic functions of Archaea more closely resemble Bacteria, whereas information processing in Archaea, such as DNA replication, transcription, and translation, share more similarities with Eukarya.
Detailed examination of archaeal viruses is likely to provide fundamental insights into the evolutionary relationships among viruses infecting hosts in the three domains of life (1–7). The Sulfolobus turreted icosahedral virus (STIV) has emerged as a model system for examining archaeal viruses. It was originally isolated from an acidic (pH 2) hot (82 °C) spring within Yellowstone National Park (8). STIV has a double-stranded circular DNA genome and replicates in the hyperthermophile Sulfolobus solfataricus (8, 9). STIV particles possess an internal membrane surrounded by a T = 31 icosahedral capsid with turret-like projections extending from each of the virion’s fivefold axis (8, 10–14). Structural similarities of the major capsid protein (MCP), suggest an evolutionary link between archaeal viruses and viruses that replicate in bacterial and eukaryotic hosts (8, 12, 13). An examination of STIV–host interactions has revealed a novel virus-induced cell lysis mechanism (2, 7). This involves assembly of seven-sided pyramid-like structures on the surface of infected cells, which open to disrupt the cell and release progeny virus particles. Importantly, maturing STIV particles gain their internal membrane in the cytosol of the infected cell, before their movement to the cell periphery and release via the pyramid structures.
In eukaryotes, Endosomal Sorting Complex Required for Transport (ESCRT) proteins are part of a conserved pathway important for multivesicular body biogenesis, metazoan cytokinesis, macroautophagy, and the budding of viruses (15–19). Eukaryotic ESCRT systems are composed of four complexes: ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III as well as an AAA+ ATPase, Vps4. ESCRT-III proteins can be recruited to membranes by multiple pathways, and it is thought that polymerization of these proteins is involved in the constriction of cellular membranes. In eukaryotic systems, Vps4 recycles the ESCRT-III components from the membranes (20, 21). Many eukaryotic enveloped viruses [for example, HIV and hepatitis C virus (HCV)] hijack the cellular ESCRT system for assembly and for trafficking viral components to the cellular membrane to complete their replication cycle (15, 22–26). Domains within eukaryotic viral proteins, termed late domains, have been shown to recruit components of the ESCRT complex, thereby redirecting the ESCRT machinery from endosomes to sites of virion assembly and budding (23).
Sulfolobus species encode ESCRT-III homologs that are required for cell division (reviewed in ref. 27). Sulfolobus species lack homologs of ESCRT-0, ESCRT-I, and ESCRT-II complexes, and although they also lack Alix and Cep55 proteins that are involved in ESCRT-III positioning during viral budding and midcell localization in eukaryotes, they encode a protein called CdvA (SSO0911) that is responsible for recruiting ESCRT-III proteins to Sulfolobus membranes (28). In addition, is has been reported that CdvA from another archaeal species may also play a role in binding DNA (29). The S. solfataricus P2 genome encodes four ESCRT-III homologs: SSO0451, SSO0619, SSO0881, and SSO0910, and a Vps4 homolog (30). Vps4 and ESCRT-III proteins form ring-like structures, between segregated chromosomes, that appear to shrink concomitantly with membrane ingression during cell division in Sulfolobus (31). When a Walker B mutant form of the Vps4 AAA+ ATPase that is unable to hydrolyze ATP was overexpressed in Sulfolobus cells, cell division was perturbed, supporting the role of the ESCRT apparatus in archaeal cell division (32).
Transcriptomic analysis of STIV-infected S. solfataricus revealed that one of the most up-regulated gene clusters during a STIV infection was an ESCRT gene cluster consisting of Vps4 (SSO0909), an ESCRT-III gene (SSO0910), and CdvA (SSO0911) (4, 33). Transcripts for the SSO0881 ESCRT-III gene were also elevated. A proteomic analysis of the host response to STIV infection showed up-regulation of CdvA and two ESCRT-III proteins, SSO0451 and SSO0881 (34). A previous proteomic study also revealed association of the SSO0881 ESCRT-III protein with purified STIV virions (35). In these analyses, an ESCRT-III protein, the STIV MCP, and other viral proteins were found in the membrane subcellular fraction. In the current work, we examine the functional relevance of this apparent interplay between STIV and the host ESCRT machinery.
Results
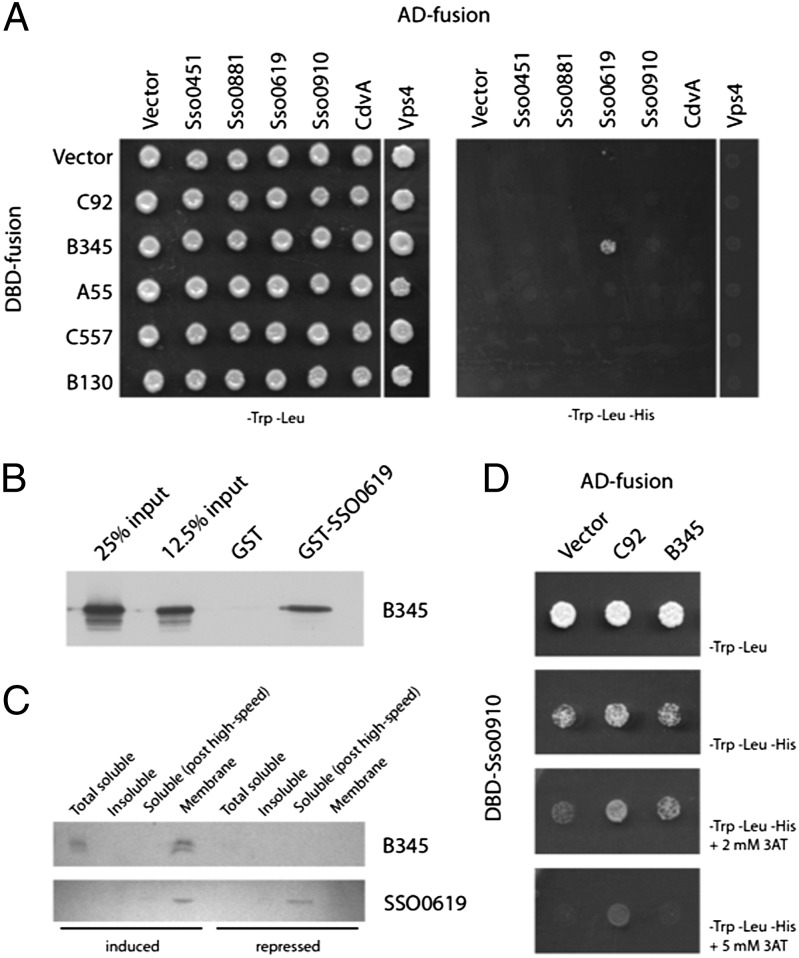
First, we tested for interactions between viral and host proteins. A targeted yeast two-hybrid screen was performed with S. solfataricus ESCRT components and all STIV proteins that were predicted to interact with membranes (Fig. 1A). An interaction was detected between the STIV MCP (B345) and SSO0619 (Fig. 1A), and this interaction was confirmed by GST pull-down assays (Fig. 1B). Furthermore, when B345 was expressed in Sulfolobus cells in the absence of other viral proteins, it enriched SSO0619 levels in the membrane fraction (Fig. 1C). When we performed the reciprocal yeast two-hybrid screen, reversing the GAL4 DNA binding domain and activation domain fusions to the ESCRT and STIV proteins, we additionally observed interactions between another ESCRT-III paralog, SSO0910, and the viral protein C92 (Fig. 1D). We did not detect any direct interactions between viral gene products and either CdvA or Vps4 (Fig. 1A). Overall, these results are consistent with our previous proteomic and microarray analysis that show increased levels of ESCRT RNA and protein levels in response to STIV infection (4, 35).
Fig. 1.
Interactions between Sulfolobus ESCRT proteins and STIV. (A) Interactions between STIV proteins and Sulfolobus ESCRT-III homologs were identified by yeast two-hybrid assays. Growth on media lacking tryptophan, leucine, and histidine indicated interactions. AD, activation domain fusion; DBD, DNA-binding domain fusion. (B) Interaction of SSO0619 (ESCRT-III) with B345. Pull-down assays using either GST or GST-SSO0619 and B345 protein were performed as described in Materials and Methods. B345 was detected by western blot analysis with anti-B345 antiserum. (C) Association of SSO0619 with B345 in vivo. After induction of B345 from a plasmid-based system, the presence of B345 and SSO0619 in soluble, insoluble, and membrane fractions of cell extract was analyzed by western blot analysis. (D) Interaction between STIV C92 protein and the Sulfolobus ESCRT-III paralog SSO0910. A yeast two-hybrid screen reciprocal to that in A, where the GAL4 fusions to the ESCRT and STIV proteins were reversed, identified potential interactions with SSO0910. Autoactivation of the GAL4 DNA binding domain fusion to SSO0910 was repressed by the addition of 3-AT to selective media.
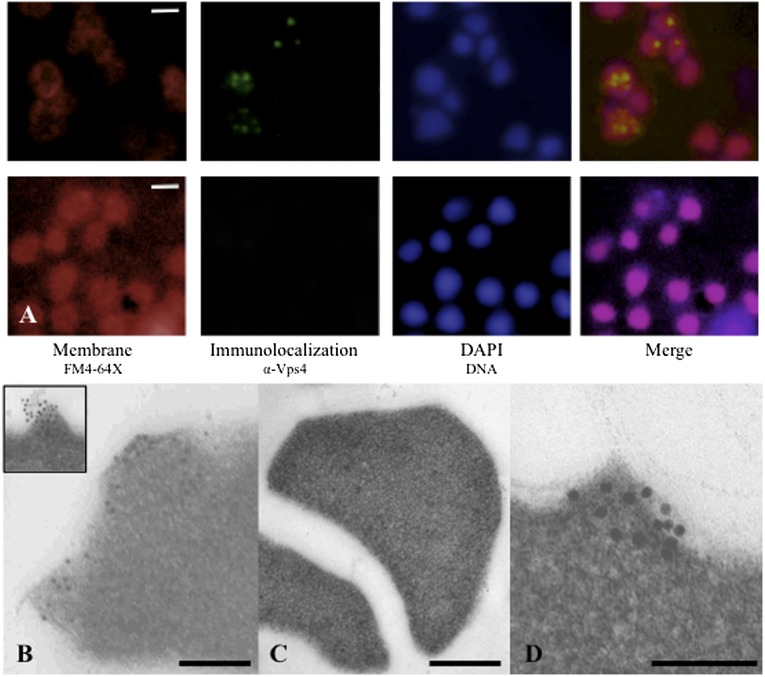
We next determined the subcellular localization of host ESCRT components during STIV infection. Sulfolobus undergoes dramatic ultrastructural changes to its membrane before viral-induced lysis (2). Before lysis, seven-sided pyramid-like structures are formed that protrude through the exterior S layer. A viral-encoded protein, C92, is solely responsible for the formation of these pyramid lysis structures (7). To determine where the cellular ESCRT components localized in STIV-infected cells, we used epifluorescence microscopy (EfM) and transmission electron microscopy (TEM) immunolocalization (Fig. 2). Immunostaining of Vps4 in S. solfataricus cells infected with STIV revealed several bright foci within cells by EfM (Fig. 2A). The uninfected cells did not show the bright centers of Vps4 protein that are obvious in the STIV-infected cells (Fig. 2A). Next, we used immunolocalization with gold-labeled secondary antibodies and found that Vps4 localized to the virus-induced pyramids that are formed before lysis of the cell (Fig. 2B). Although the Vps4 protein is constitutively expressed in cells, we do not detect any specific localization of Vps4 in noninfected asynchronous cells (Fig. 2C). Importantly, a nonspecific control antibody in conjunction with the gold-labeled secondary antibody did not localize to the pyramid lysis structures. Interestingly, it was recently shown that the Vps4 component of the eukaryotic ESCRT machinery is localized to the HIV budding site in eukaryotic cells (36).
Fig. 2.
Localization of SSO0909 (Vps4) in S. solfataricus P23 and PH1-16 cells. (A) Epifluorescence localization of Vps4 in STIV-infected P23 cells (Upper) and uninfected P23 cells (Lower); samples were collected 20 h post infection (hpi); images show the FM4-64 staining for membranes (red), antibody-labeling of Vps4 (green), DAPI staining for DNA (blue), and merged. (B) Immunogold localization of SSO0909 (Vps4) in STIV-infected P23 cells collected at 32 hpi. (B Inset) Close-up view of Vps4 localized to virus-induced pyramid lysis structure. (C) Immunogold localization of Vps4 in uninfected P23 control cells collected at 32 hpi. (D) Immunolocalization of SSO0909 in PH1-16 cells overexpressing C92 (induced with 0.4% arabinose) collected at 24 hpi. See Fig. S3 for control of immunolocalization of Vps4 in PH1-16 cells repressing C92 (repressed with 0.4% galactose). (Scale bars: A, 1 μm; B and C, 250 nm; and D, 100 nm.)
We have shown that expression of C92 in S. solfataricus resulted in pyramid structures on the cell surface (7). Previous immunogold labeling of Vps4 in noninfected S. solfataricus cells revealed that Vps4 is distributed throughout the cell and can be membrane associated (37). Strikingly, Vps4 localizes to the pyramid structures formed at the cellular membrane during C92 expression (Fig. 2D). As a control, we did not detect any specific localization of Vps4 in C92-repressed cultures (Fig. S1). Our interaction data did not detect any direct interactions between C92 and Vps4; however, they reveal that C92 interacts with the ESCRT-III paralog, SSO0910 (Fig. 1B). Significantly, this protein is the sole ESCRT-III paralog in Sulfolobus that possesses a MIM2 motif that is necessary and sufficient for interaction with Vps4 (32).
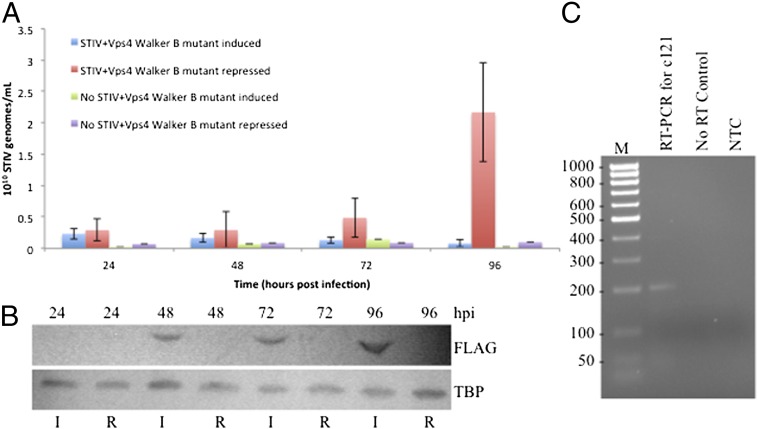
Our data establish a linkage between the ESCRT apparatus and STIV biology. To determine whether normal functioning of the ESCRT machinery was required for productive infection by STIV, we expressed a trans-dominant negative Vps4 (E206Q) Walker B mutant via an arabinose-inducible system (32) in STIV-infected S. solfataricus PH1-16. STIV replication was monitored by quantitative PCR of the viral MCP gene, and expression of the Walker B Vps4 protein was monitored by western blot analysis (Fig. 3). The results reveal that cells expressing the mutant Vps4 fail to support STIV replication, whereas control cultures are able to sustain a viral infection (Fig. 3 A and B). We excluded the possibility that arabinose induction alone may have a negative effect on STIV entry and/or attachment by demonstrating that STIV infects PH1-16 cells in the presence of arabinose (Fig. S2). Furthermore, we tested the hypothesis that ESCRT proteins may be required in the attachment and/or entry of STIV. However, analysis of an early viral gene transcript revealed the viral DNA is being transcribed in PH1-16 cells expressing the trans-dominant Vps4 Walker B mutant (Fig. 3C), indicating that the STIV genome is successfully entering the cells and being transcribed.
Fig. 3.
STIV infection in S. solfataricus PH1-16 expressing the trans-dominant negative Vps4 Walker B mutant. (A) qPCR measurements of STIV B345 gene abundance during the course of STIV infections and mock infections in S. solfataricus PH1-16 cells repressing or expressing a Walker B mutant form of Vps4. (B) Western blot analysis of episomal Vps4 Walker B mutant expression. Episomal Vps4 was detected with the anti-FLAG antibody (Upper) and the TATA Binding Protein (TBP) served as a loading control (Lower). Cultures were induced (I) with arabinose or repressed (R) with galactose 24 h after STIV infection. (C) RT-PCR of RNA extracted from S. solfataricus PH1-16 cultures infected with STIV shows transcription of an early viral gene product (C121). Samples were collected at 24 hpi, and the extracted RNA was subjected to a one-step RT-PCR. The Taq-only control was performed without reverse transcription to test for contaminating DNA. NTC, no template control; size standard is in base pairs.
Discussion
It has been reported that infection of Sulfolobus cells with STIV modulates cellular levels of the host ESCRT-III and Vps4 proteins. We observe physical interactions between an ESCRT-III paralog and the MCP of the virus and between another ESCRT-III paralog and the pyramid-forming C92. We observe an association of Vps4 with viral exit structures and find that expression of a trans-dominant negative allele of Vps4 abrogates viral replication. In light of these observations, we propose that the archaeal ESCRT machinery is intimately involved in at least two stages of the life cycle of STIV. We suggest that the ESCRT machinery may be used in the maturation of the internal membrane component of the assembling STIV virions via the MCP–SSO0619 interaction. Second, we hypothesize that the ESCRT machinery may play a role in the opening of pyramid exit structures. We observe a strong association between the C92 pyramid structures and Vps4. This interaction is unlikely to be involved in the assembly of these structures, because studies of the orthologous P98 of Sulfolobus islandicus rudivirus 2 (SIRV2) have revealed that the viral protein forms pyramid structures in the inner membrane of Escherichia coli when heterologously expressed (6). However, it is enticing to note that the pyramids formed in E. coli never opened, leading us to speculate that in the native host, the ESCRT machinery may play a role in the final maturation of the pyramid structures to form open exit structures (Fig. S3).
Our data reveal that in addition to playing roles in vesicle biogenesis and cell division, the archaeal ESCRT apparatus is also coopted by viruses. The parallels with the more complex eukaryal ESCRT system are striking and strongly support the proposal that the cellular ESCRT system is ancient and likely to have evolved before the separation of Archaea and Eukarya. Interestingly, as has been reviewed elsewhere, ESCRT proteins are not ubiquitous throughout the archaeal domain of life (38). The uneven phyletic distribution of the ESCRT machinery has been interpreted as suggesting extensive lineage-specific loss of this apparatus, further implying the existence of a Last Archaeal Common Ancestor that contained ESCRT machineries. We speculate that the involvement of the ESCRT apparatus with an archaeal virus could suggest an alterative explanation. Just as many present day viruses encode their own replication machineries and may even have contributed to the evolution of cellular replication machineries (39, 40), perhaps there exist, or have existed, archaeal viruses that encode their own ESCRT-related proteins to facilitate viral biology. Acquisition of these virus-derived genes could account for the strikingly disperse phyletic distributions of the archaeal ESCRT proteins.
Materials and Methods
Yeast Two-Hybrid Assays.
The yeast two-hybrid assays were performed as described (41). Briefly, Saccharomyces cerevisiae AH109 cells were cotransformed with plasmids encoding either STIV genes or cellular ESCRT genes fused to the GAL4 DNA-binding domain (pGBKT7) or the GAL4 activation domain (pGADT7) (Clontech Matchmaker System). Transformants were selected by growth on SC-Leu-Trp agar for 2 d at 30 °C. The positives were spotted on both SC-Leu-Trp (as a control) and on SC-Leu-Trp-His agar and allowed to grow 3 d. Growth on plates lacking histidine was considered a positive result. To reduce autoactivation of DBD-SSO0910, 3-amino-1,2,4-triazole (3-AT) was added at 2 and 5 mM.
GST Pull-Downs.
GST pull-downs were performed as described (32). Briefly, pull-downs were performed in 100 µL of 10 mM Tris at pH 8, 150 mM NaCl, 0.1% (vol/vol) Tween-20, and 5 mM MgCl2 (TBSTM). Reactions contained ∼20 µL of bead volume of glutathione sepharose with 10 µg of either GST or GST-SSO0619. One microgram of B345 was added per reaction. Binding was for 30 min at room temperature with agitation, and reactions were spun and washed four times with 500 µL of TBSTM. Pellets were resuspended in 100 µL of 1× SDS/PAGE buffer, run on an 11.25% gel, and subjected to western blotting with detection by anti-B345 serum.
Strains and Growth Conditions.
S. solfataricus P23 was grown in Media 182 (www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium182.pdf) as described (4). Briefly, S. solfataricus P23 was revived from glycerol stocks in 25 mL of Media 182 (pH 3) at 80 °C for ∼24 h. The “starter” culture was then transferred to a larger volume (250 mL) of Media 182 (pH 2.5) at 80 °C for STIV infection. S. solfataricus strain PH1-16 (42) used for genetic studies and protein expression was grown in Brock’s medium (43) (pH 3.5) at 75 °C with 10 μg/mL uracil before transformation and no uracil afterward. All S. solfataricus plating was on Brock’s medium at pH 3.5 + 0.2% (wt/vol) tryptone solidified with 0.7% (wt/vol) Phytagel (Sigma). S. cerevisiae AH109 was grown in yeast extract peptone dextrose (YEPD) medium (44) and plated on solid selective media, as indicated.
Construction of B345+pSeSD1 and SSO0909+pSeSD1.
STIV major capsid protein (B345) was amplified from STIV viral DNA by using B345-ClaI-F and B345-SalI-R primers (Table S1) and Phusion High-fidelity DNA polymerase (NEB) under standard conditions. Purified PCR products were digested with ClaI and SalI (NEB) and ligated into ClaI/SalI-digested pSeSD1 (7). The N-terminal FLAG tag was added to the B345 gene cloned into pSeSD1 via inverse PCR (Table S1). Likewise, SSO0909 and SSO0909 E206Q were amplified from pRYS1 (32) by using Phusion DNA polymerase and standard conditions with SSO0909-NdeI-F and SSO0909+FLAG-SalI-R (Table S1). The purified PCR products were digested with NdeI and SalI (NEB) and ligated to NdeI/SalI-digested pSeSD1. All constructs were verified via sequencing.
STIV Infection of S. solfataricus P23 and PH1-16 Cultures.
Cultures of S. solfataricus P23 were grown in Media 182 at pH 2.5 until midlog (OD650 ∼ 0.2) growth phase. The culture was infected with purified STIV particles at a multiplicity of infection (MOI) of 1.7–2. A single cycle of virus infection was ∼48 h. Samples were collected at various time points after infection. Cultures of S. solfataricus PH1-16 were grown in Brock’s medium at pH 3.5 until midlog (OD650 ∼ 0.2) growth phase. The culture was infected with purified STIV particles at a MOI of 50. Lysis of cells was evident at 96 h after infection. Samples were collected at various time points after infection. To measure virus infection, quantitative PCR (qPCR) was performed on culture collected during virus infection. Standard procedures were followed by using EvaGreen Sso Fast qPCR SuperMix (Bio-Rad) and STIV 5f and STIV midr primers (7) (Table S1). The data were exported and analyzed in Microsoft Excel.
Immunofluorescence Microscopy.
Immunofluorescence microscopy was performed as described (32). Slides were viewed with a Nikon A1RSi, and images were analyzed and merged with NIS Elements software.
TEM Localization Microscopy.
TEM immunogold localization was performed as described (7). Briefly, samples were collected from STIV-infected cultures (and C92 expressing cultures) at various times during the infection cycle (and after induction/repression). After collection, cells were centrifuged and fixed in 0.5% (vol/vol) gluteraldehyde (Ted Pella) and 2% (vol/vol) paraformaldehyde in cacodylate buffer (pH 7.2) at 4 °C. After fixation, cells were pelleted and mixed with a drop of warm 2% (wt/vol) agar. After solidification, the pellets were cut into small pieces, rinsed with buffer, and dehydrated in ethanol. Dehydration was followed by infiltration with LR White (Ted Pella) embedding medium. Pieces were placed in Beem capsules, covered with Parafilm, and baked at 70 °C overnight. Sections were cut to ∼90 nm with an OmU2 Reichert ultramicrotome and placed on extra thin bar 200-mesh nickel grids (Ted Pella). Grids containing these sections were placed on drops of PBS + 1% (vol/vol) Tween-20 (PBST) and blocked with PBST + 1% (wt/vol) BSA. Sections were incubated with primary antibody (anti-Vps4 at a 1:100 dilution) for 3–4 h, washed with blocking solution, incubated for 2 h with gold-conjugated anti-rabbit secondary antibody (Ted Pella; 1:20 dilution), washed again, and fixed with 1% (vol/vol) gluteraldehyde followed by staining with uranyl acetate and Reynold’s lead citrate. Sections were viewed with a Leo 912 AB (Zeiss) TEM.
RNA Purification and RT-PCR.
At 24 h after STIV infection, 10 mL of S. solfataricus PH1-16 infected with STIV was removed from the growth flask. The culture was immediately centrifuged at 4,388 × g for 5 min to pellet the cells. To aid in removal of unattached STIV virus particles, the cell pellets were washed three times with Brock’s media at pH 3.5. The cell pellets were resuspended in RNAlater (Qiagen) and stored at −80 °C until RNA extraction. Total RNA was extracted from washed cell pellets by using the mirVana miRNA Isolation Kit (Ambion) following the manufacturer’s instructions. DNA was removed by treatment with DNaseI (Promega), and the RNA was further purified with the DNA-Free RNA Kit (Zymo Research). A one-step reverse transcriptase reaction was performed with 100 ng of purified total RNA by using C121-F and C121-R primers (Table S1) and SuperScript-III Reverse Transcriptase/Platinum Taq enzyme mix (Invitrogen). Controls were prepared by only including Platinum Taq in the reaction to check for contaminating DNA.
Supplementary Material
Acknowledgments
We thank Dr. Frank Roberto for technical assistance with epifluorescence microscopy and Keshia Kerchner for general technical assistance. This research is supported by National Science Foundation Grants DEB-0936178 and EF-080220 (to M.J.Y.), National Aeronautics and Space Administration Grant NNA-08CN85A (to M.J.Y.), and the Wellcome Trust and The College of Arts and Sciences, Indiana University (S.D.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301605110/-/DCSupplemental.
References
- 1.Bize A, et al. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci USA. 2009;106(27):11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumfield SK, et al. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J Virol. 2009;83(12):5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence CM, et al. Structural and functional studies of archaeal viruses. J Biol Chem. 2009;284(19):12599–12603. doi: 10.1074/jbc.R800078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortmann AC, et al. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008;82(10):4874–4883. doi: 10.1128/JVI.02583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quax TE, Krupovic M, Lucas S, Forterre P, Prangishvili D. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology. 2010;404(1):1–4. doi: 10.1016/j.virol.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Quax TEF, et al. Simple and elegant design of a virion egress structure in Archaea. Proc Natl Acad Sci USA. 2011;108(8):3354–3359. doi: 10.1073/pnas.1018052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder JC, Brumfield SK, Peng N, She Q, Young MJ. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J Virol. 2011;85(13):6287–6292. doi: 10.1128/JVI.00379-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice G, et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci USA. 2004;101(20):7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice G, et al. Viruses from extreme thermal environments. Proc Natl Acad Sci USA. 2001;98(23):13341–13345. doi: 10.1073/pnas.231170198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu CY, Johnson JE. Structure and cell biology of archaeal virus STIV. Curr Opin Virol. 2012;2(2):122–127. doi: 10.1016/j.coviro.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu CY, et al. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure. 2010;18(12):1579–1586. doi: 10.1016/j.str.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khayat R, Fu CY, Ortmann AC, Young MJ, Johnson JE. The architecture and chemical stability of the archaeal Sulfolobus turreted icosahedral virus. J Virol. 2010;84(18):9575–9583. doi: 10.1128/JVI.00708-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khayat R, et al. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci USA. 2005;102(52):18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veesler D, et al. Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography. Proc Natl Acad Sci USA. 2013;110(14):5504–5509. doi: 10.1073/pnas.1300601110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlton JG, Martin-Serrano J. The ESCRT machinery: New functions in viral and cellular biology. Biochem Soc Trans. 2009;37(Pt 1):195–199. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- 16.Ghazi-Tabatabai S, et al. Evolution and assembly of ESCRTs. Biochem Soc Trans. 2009;37(Pt 1):151–155. doi: 10.1042/BST0370151. [DOI] [PubMed] [Google Scholar]
- 17.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20(1):4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 19.Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8(5):355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Blissard GW. Cellular VPS4 is required for efficient entry and egress of budded virions of Autographa californica multiple nucleopolyhedrovirus. J Virol. 2012;86(1):459–472. doi: 10.1128/JVI.06049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuchell-Brereton MD, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449(7163):740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 22.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316(5833):1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 23.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: Can some viruses do without an ESCRT? Virology. 2008;372(2):221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corless L, Crump CM, Griffin SDC, Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. 2010;91(Pt 2):362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silvestri LS, et al. Involvement of vacuolar protein sorting pathway in Ebola virus release independent of TSG101 interaction. J Infect Dis. 2007;196(Suppl 2):S264–S270. doi: 10.1086/520610. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe T, et al. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci USA. 2007;104(24):10205–10210. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samson RY, Bell SD. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 2009;17(11):507–513. doi: 10.1016/j.tim.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Samson RY, et al. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41(2):186–196. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriscot C, et al. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS ONE. 2011;6(7):e21921. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.She Q, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98(14):7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samson RY, Bell SD. Cell cycles and cell division in the archaea. Curr Opin Microbiol. 2011;14(3):350–356. doi: 10.1016/j.mib.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322(5908):1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder JC, Young MJ. Potential role of cellular ESCRT proteins in the STIV life cycle. Biochem Soc Trans. 2011;39(1):107–110. doi: 10.1042/BST0390107. [DOI] [PubMed] [Google Scholar]
- 34.Maaty WS, et al. Proteomic analysis of Sulfolobus solfataricus during Sulfolobus Turreted Icosahedral Virus infection. J Proteome Res. 2012;11(2):1420–1432. doi: 10.1021/pr201087v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maaty WS, et al. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol. 2006;80(15):7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumgärtel V, et al. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13(4):469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 37.Ellen AF, et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles. 2009;13(1):67–79. doi: 10.1007/s00792-008-0199-x. [DOI] [PubMed] [Google Scholar]
- 38.Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8(10):731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forterre P, Krupovic M. The origin of virions and virocells: The Escape Hypothesis Revisited. In: Witzany G, editor. Viruses: Essential Agents of Life. Amsterdam: Springer; 2012. pp. 43–60. [Google Scholar]
- 40.McGeoch AT, Bell SD. Extra-chromosomal elements and the evolution of cellular DNA replication machineries. Nat Rev Mol Cell Biol. 2008;9(7):569–574. doi: 10.1038/nrm2426. [DOI] [PubMed] [Google Scholar]
- 41.Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Cell. 2003;11(1):275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 42.Martusewitsch E, Sensen CW, Schleper C. High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J Bacteriol. 2000;182(9):2574–2581. doi: 10.1128/jb.182.9.2574-2581.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brock TD, Brock KM, Belly RT, Weiss RL. Sulfolobus: A new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84(1):54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.