Significance
Our study investigates chemical damage associated with chronic inflammation and relates these macromolecular damage products to inflammatory bowel disease activity. Using mice as a model system, we show that chronic inflammatory responses that are common to mice and humans produce similar types and quantities of damage products in both species. Additional analysis of signaling molecules in the serum and tissue of diseased samples highlights the role of the innate immune response in the overall pathology of inflammatory bowel disease.
Keywords: myeloperoxidase, nitric oxide, oxidative stress, granulocyte
Abstract
Inflammatory bowel disease (IBD) arises from inappropriate activation of the mucosal immune system resulting in a state of chronic inflammation with causal links to colon cancer. Helicobacter hepaticus-infected Rag2−/− mice emulate many aspects of human IBD, and our recent work using this experimental model highlights the importance of neutrophils in the pathology of colitis. To define molecular mechanisms linking colitis to the identity of disease biomarkers, we performed a translational comparison of protein expression and protein damage products in tissues of mice and human IBD patients. Analysis in inflamed mouse colons identified the neutrophil- and macrophage-derived damage products 3-chlorotyrosine (Cl-Tyr) and 3-nitrotyrosine, both of which increased with disease duration. Analysis also revealed higher Cl-Tyr levels in colon relative to serum in patients with ulcerative colitis and Crohn disease. The DNA chlorination damage product, 5-chloro-2′-deoxycytidine, was quantified in diseased human colon samples and found to be present at levels similar to those in inflamed mouse colons. Multivariate analysis of these markers, together with serum proteins and cytokines, revealed a general signature of activated innate immunity in human IBD. Signatures in ulcerative colitis sera were strongly suggestive of neutrophil activity, and those in Crohn disease and mouse sera were suggestive of both macrophage and neutrophil activity. These data point to innate immunity as a major determinant of serum and tissue profiles and provide insight into IBD disease processes.
Inflammatory bowel disease (IBD) is a chronic and relapsing intestinal inflammatory disease that arises through unknown genetic, environmental, and bacterial origins (1, 2). Ulcerative colitis (UC) and Crohn disease (CD) are the two main forms of IBD, and their incidence is increasing in industrialized countries (3). Furthermore, IBD is a risk factor for the development of colon cancer (4). Although the specific determinants remain elusive, persistent inflammation is believed to play a significant role in colon cancer development (5).
Neutrophil recruitment and activation are key steps in the intestinal innate immune response observed in IBD (6–8), and studies with animal models of colitis highlight the relationship between neutrophil infiltration and disease severity (9–11). We recently reported results of a comprehensive analysis of histopathology, changes in gene expression, and nucleic acid damage occurring during progression of lower bowel disease in Rag2−/− mice infected with Helicobacter hepaticus (Hh) (10). This mouse model emulates many aspects of human IBD, and infected mice develop severe colitis that progress into colon carcinoma, with pronounced pathology in the cecum and proximal colon marked by infiltration of neutrophils and macrophages (12, 13).
Phagocytes produce strong oxidants and radicals that damage cellular macromolecules and promote tissue damage at sites of inflammation (14–16). Myeloperoxidase (MPO) is an abundant enzyme in neutrophils that produces hypochlorous acid (HOCl) from hydrogen peroxide (H2O2) and chloride ion (17, 18). HOCl can oxidize and chlorinate DNA, proteins, and lipids (19, 20). A prominent target of HOCl is tyrosine, which leads to the formation of the stable aromatic residue, 3-chlorotyrosine (Cl-Tyr) (21, 22). MPO also produces chlorinating species that react with DNA to form chlorinated adducts such as 5-chloro-2′-deoxycytidine (5-Cl-dC) (23), the presence of which was identified in colon tissue of H. hepaticus-infected Rag2−/− mice (10). This modification of DNA may provide a mechanistic link between neutrophil activity and colitis-associated carcinoma (10, 24, 25).
Macrophages also contribute to the array of oxidants and radicals at sites of inflammation through release of nitric oxide (NO) generated by the inducible NO synthase (iNOS) enzyme. NO reacts with superoxide anion (O2−•) at diffusion-controlled rates to yield highly reactive peroxynitrite (ONOO−) (26, 27). MPO also reacts H2O2 with nitrite (NO2−, the endpoint of cellular NO oxidation) to produce the strong nitrating agent, nitrogen dioxide radical (NO2•) (28). Both NO2• and ONOO− can react with tyrosine residues to generate the stable tyrosine nitration product, 3-nitrotyrosine (Nitro-Tyr) (29, 30).
Multiple MS methods have been applied for determination of Cl-Tyr and Nitro-Tyr levels in biological systems (10, 31–38), and both have been detected in inflamed tissues from animals and humans (11, 39). The presence of Nitro-Tyr has been demonstrated in colon tissue of IBD patients by immunohistochemistry, and levels were reported to correlate with disease activity (40, 41). We undertook the present study to test the null hypothesis that the H. hepaticus-infected mouse model of colitis and colitis-associated carcinoma represents a useful surrogate of human IBD. To examine this hypothesis, we first quantified levels of Nitro-Tyr and Cl-Tyr in proteins and 5-Cl-dC in DNA of colon tissues of IBD patients. Comparison of these data with our previous findings (10) further assessed the validity of this animal model. We then tested the hypothesis that inflammation-induced damage in the colon would be reflected in changes in serum constituents, and would therefore serve as a noninvasive measure of IBD activity. For this purpose, we determined levels of protein chlorination and nitration products, acute-phase proteins, cytokines, and chemokines in human and mouse sera. In addition, gene expression of several inflammatory signaling molecules was monitored in mice colons to determine whether colonic inflammation was directly associated with serum cytokine levels. We then used multivariate analysis to determine which systemic inflammatory markers in serum were most closely associated with disease activity and were also common to human IBD and H. hepaticus-associated colitis in Rag2−/− mice.
Results
Evaluation of Chlorination Damage in Inflamed Human and Mice Colons.
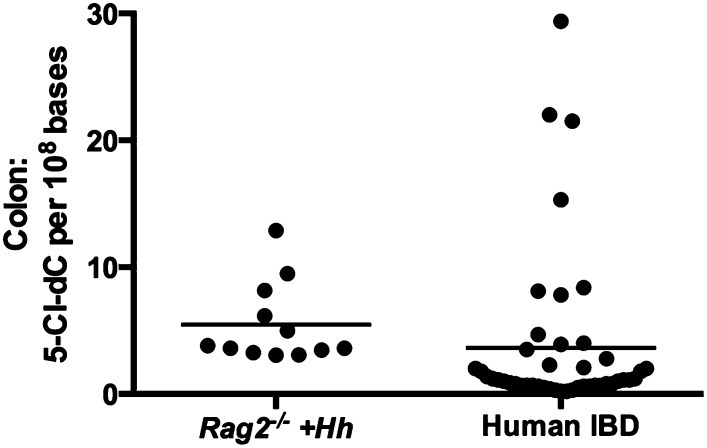
We recently reported a comprehensive characterization of nucleic acid damage products in the lower bowel of Rag2−/− mice induced by chronic H. hepaticus infection (10). Chlorinated adducts of DNA and RNA, which are generated from reaction with MPO products, were associated with persistent infiltration of neutrophils and macrophages at the site of inflammation. Furthermore, these chlorination products increased in amount as the inflammatory state lengthened. Importantly, infiltration by these innate immune cells was accompanied by cancer initiation and progression. Using the mouse as a model for colonic inflammation, we tested the hypothesis that similar chlorination events occurred during human IBD. MPO-derived chemical damage to nucleic acids and proteins has not previously been evaluated in colon tissues of IBD patients. Therefore, we obtained colon biopsy tissue samples collected during resection surgery to determine the abundance of chlorination products in protein and DNA. We first isolated DNA to quantify 5-Cl-dC for analysis by LC-MS/MS (10, 42), which showed the presence of 5-Cl-dC at levels of 0.2–29.4 (mean ± SD 3.6 ± 6.3) adducts per 108 nucleotides (Fig. 1). A similar range of 5-Cl-dC levels was found in colons of H. hepaticus-infected mice (3.1–12.9 adducts per 108 nucleotides), although mean values in mice were significantly higher (5.5 ± 3.2 adducts 108 nucleotides) (Fig. 1) (10). The similarity of adduct abundance in mouse and human genomic DNA suggests a possible link between chlorination damage to DNA and development of colitis-associated carcinoma in humans.
Fig. 1.
5-Cl-dC was analyzed in human colon tissue derived from patients with IBD and compared with previously determined levels in mice (UC, n = 18; CD, n = 22, Rag2−/− H. hepaticus-infected mice, n = 12) (11). The 5-Cl-dC levels in Rag2−/− H. hepaticus-infected mice were significantly higher than those observed in IBD colon tissues (P < 0.0002). Statistical significance between groups was assessed by the Mann–Whitney U test and the line represents the mean value in each dataset.
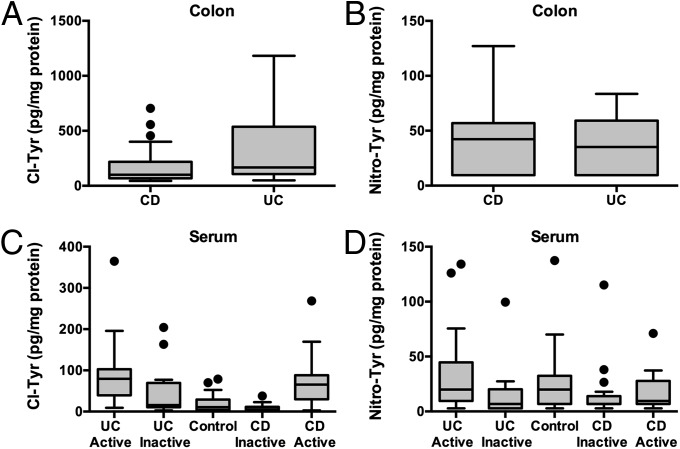
In addition to DNA, proteins are also major targets of cellular damage associated with inflammation. MPO-derived products, such as HOCl and NO2•, react with tyrosine residues in proteins to yield the stable chlorination and nitration products Cl-Tyr and Nitro-Tyr, respectively. Previous studies suggest that Nitro-Tyr is associated with human IBD severity (40, 41) and Cl-Tyr is associated with colitis in mice (11). To examine the possible association of Cl-Tyr and Nitro-Tyr with disease activity, we quantified their levels in colon biopsy samples from UC and CD patients using stable isotope dilution LC-MS/MS analysis; comparison of the two diseased tissues revealed no significant differences in levels of either Cl-Tyr or Nitro-Tyr (Fig. 2 A and B). Mean values for Cl-Tyr and Nitro-Tyr in colon tissues were 248 ± 266 and 54 ± 71 pg/mg protein, respectively. We also assessed the possible association between 5-Cl-dC and Cl-Tyr levels in the same tissue by Spearman correlation analysis (ρ), but no significant correlation was found (ρ = −0.18). This result suggests that altered rates of adduct formation or repair need to be considered when assessing macromolecular chlorination damage products.
Fig. 2.
Cl-Tyr and Nitro-Tyr were quantified in human tissues using stable isotope dilution LC-MS/MS analysis. (A and B) Quantitation of Cl-Tyr and Nitro-Tyr in IBD colon tissue (UC, n = 18; CD, n = 22). (C and D) Quantitation of Cl-Tyr and Nitro-Tyr in IBD and non-IBD serum (Control, n = 29; UC active, n = 38; UC inactive = 19; CD active, n = 42; CD inactive, n = 20). Cl-Tyr levels in both active UC (P value < 0.0001) and active CD (P value < 0.0001) serum samples were significantly higher than those observed in non-IBD control sera. Significant elevation in Cl-Tyr levels were also observed between UC active and UC inactive (P value = 0.0003), and CD active and CD inactive (P value < 0.0001) samples. A significant elevation was found in Nitro-Tyr levels between UC active and UC inactive (P value = 0.0038) samples. Statistical analysis for all panels is presented as box-and-whisker plots showing the median value (line), the interquartile range (box), and Tukey whiskers defining data within 1.5-fold of the interquartile range; all data outside the whiskers are presented as individual data points. Statistical significance between groups (two groups) was assessed by the Mann–Whitney U test.
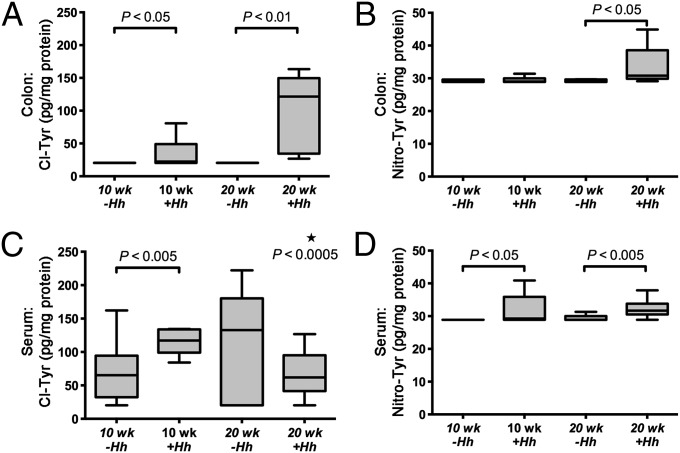
To evaluate the validity of the mouse as a model for chlorination damage in human IBD, we measured Cl-Tyr and Nitro-Tyr levels in colons from H. hepaticus-infected mice used in our previous study (10). Analyses were carried out on tissues collected from mice at 10 and 20 wk postinfection. In uninfected control animals (n = 6 at 10 wk and n = 5 at 20 wk) Cl-Tyr was not detectable (limit of detection of 2.8 pg/mg protein). However, tissues from infected animals contained Cl-Tyr at significantly elevated levels (P < 0.05 at 10 wk and P < 0.01 at 20 wk postinfection) (Fig. 3A). The mean Cl-Tyr level in colons of infected mice was 63.1 ± 53.3 pg/mg protein. In comparison with human tissues, the mean level of Cl-Tyr in inflamed mice colons was lower than the level determined in human IBD colon tissues (Figs. 2 and 3).
Fig. 3.
Analysis of modified tyrosine residues following 10 and 20 wk postinfection with H. hepaticus or from sham-infected Rag2−/− mice. (A and B) Cl-Tyr levels in Rag2−/− mouse colon significantly increased at both 10 and 20 wk postinfection, and Nitro-Tyr levels in the Rag2−/− mouse colon significantly increased at 20 wk postinfection (10 wk +Hh or −Hh, n = 6; 20 wk +Hh or −Hh, n = 5). (C and D) Cl-Tyr analysis in Rag2−/− mouse serum demonstrated a significant increase in chlorination damage at 10 wk and a significant decrease at 20 wk (*), and Nitro-Tyr levels in Rag2−/− mouse serum increased at both 10 and 20 wk postinfection (10 wk +Hh or −Hh, n = 12; 20 wk +Hh, n = 13; 20 wk −Hh, n = 11). Statistical analysis performed as described in Fig. 2.
Nitro-Tyr levels were also determined in mouse colon tissues. In animals 20 wk postinfection (but not 10 wk postinfection), Nitro-Tyr levels (31.3 ± 4.7 pg/mg protein) were significantly higher than in controls (Fig. 3B). Overall, the amounts of Nitro-Tyr were lower than those of Cl-Tyr in colon tissues of both human IBD patients and infected mice, indicating that protein damage induced by nitration chemistry is less prevalent than that caused by chlorination chemistry in vivo.
Evaluation of Cl-Tyr and Nitro-Tyr in Mouse and Human Serum.
With the objective of developing a noninvasive analytical platform with the potential to accurately reflect cellular damage occurring in colon tissue, we analyzed serum samples from IBD patients for the presence of protein chlorination and nitration products. To this end we quantified Cl-Tyr and Nitro-Tyr in serum proteins from 57 UC patients, 62 CD patients, and 29 non-IBD controls (Fig. 2 C and D). A portion of these UC and CD samples were classified further as active disease (recurrence) and inactive disease (remission) for the purpose of monitoring these protein damage products as a function of disease activity. The levels of Cl-Tyr were significantly elevated in UC active sera (87.1 ± 63.6 pg/mg protein) relative to inactive (44.3 ± 55.1 pg/mg protein, P value = 0.0003) and non-IBD controls (19.6 ± 20.8 pg/mg protein, P value < 0.0001). Cl-Tyr levels in CD active sera (69.4 ± 51.5 pg/mg protein) were also elevated relative to CD inactive sera (11.1 ± 8.2 pg/mg protein, P value < 0.0001) and non-IBD controls (P value <0.0001). Quantitation of Nitro-Tyr in IBD and non-IBD sera revealed only slight increase in the mean values of UC active sera (29.8 ± 31.5 pg/mg protein) relative to control (23.4 ± 26.6 pg/mg protein). A slight decrease in Nitro-Tyr levels were recorded between UC inactive sera relative to controls (14.7 ± 22.2 pg/mg protein, P value = 0.0309). Changes in Nitro-Tyr levels of CD active (16.7 ± 14.4 pg/mg protein) and CD inactive (15.8 ± 24.9 pg/mg protein) were not statistically different from control values.
Analysis of mouse serum revealed that infected animals at the 10-wk time point (n = 15) contained significantly higher Cl-Tyr levels than age-matched controls (128.9 ± 39.2 vs. 107.2 ± 84.8 pg/mg protein, respectively). By 20 wk, the Cl-Tyr serum levels from H. hepaticus-infected animals declined to 74.1 ± 42.9 pg/mg protein. Nitro-Tyr levels were significantly elevated from control values at both 10 and 20 wk postinfection (Fig. 3 C and D); infected animals were quantified at 34.5 ± 12.3 and 31.8 ± 2.9 pg/mg protein at 10 and 20 wk, respectively, and age-matched controls exhibited Nitro-Tyr levels of 28.9 ± 0.1 and 29.6 ± 1.6 pg/mg protein at 10 and 20 wk, respectively.
A subset of these serum samples were collected from IBD patients at the time of resection surgery to make a direct comparison between serum and colon tissue levels of Cl-Tyr and Nitro-Tyr. Similarly, mouse sera were acquired at the time of colon tissue collection. Spearman correlation analysis (ρ), revealed that neither the modified tyrosine levels in human colon (Cl-Tyr ρ = 0.33, Nitro-Tyr ρ = 0.30) nor those in mouse colon (Cl-Tyr ρ = −0.15, Nitro-Tyr ρ = 0.07) correlated with their respective serum values. When human serum proteins were fractionated into separate pools, 83% of Cl-Tyr was in the albumin fraction, about 2% in the IgG fraction, and the remaining in other serum proteins.
Characterization of Serum Acute-Phase Proteins in Infected Mice and Humans with IBD.
We quantified serum acute-phase proteins in humans and mice in an attempt to identify serum components that may reflect disease pathogenesis and progression. We first performed a serum proteomics analysis to identify changes in the abundance of serum proteins of UC active patients relative to non-IBD controls. UC active (defined by Simple Clinical Colitis Activity Index as > 4) and non-IBD control samples were pooled separately and processed for MS analysis. Peptides were labeled with iTRAQ reagent for relative quantification between groupings (43). The samples were separated by reverse-phase chromatography and analyzed by quadrupole time-of-flight (QTOF) MS. Seventy-seven serum proteins were identified: 16 proteins increased and 9 proteins decreased in serum from UC patients relative to controls (Table 1). The classic acute-phase proteins, serum amyloid A (SAA, 11.7 -fold), α-1-antichymotrypsin (SERPINA3, 4.2-fold), haptoglobin (2.6-fold), α-1-acid glycoprotein (2.5-fold), and α-1-antitrypsin (SERPINA1) (2.3-fold) showed the largest increases in abundance. Several proteins decreased in abundance in serum including apolipoproteins A-I and A-II and α-2-macroglobulin (A2M), which all decreased 0.5-fold relative to non-IBD controls (Table 1 and Table S1).
Table 1.
Untargeted serum proteomic analysis in human UC: Up- and down-regulated proteins
| Uniprot suggested name | Fold-change (increase or decrease) | UniProtKB | %Coverage | %Coverage(95%) | Peptides(95%) |
| Increase | |||||
| Serum Amyloid A | 11.7 | P02735 | 85.3 | 35.2 | 4 |
| α-1-Antichymotrypsin (SERPINA3) | 4.2 | P01011 | 63.8 | 17.3 | 8 |
| Haptoglobin | 2.6 | P00738 | 86.9 | 56.4 | 50 |
| α-1-Acid glycoprotein 1 (Orosomucoid 1) | 2.5 | P02763 | 74.1 | 36.3 | 12 |
| α-1-Antitrypsin (SERPINA1) | 2.3 | P01009 | 82.1 | 60.3 | 52 |
| Leucine-rich α-2-glycoprotein | 1.9 | P02750 | 55.0 | 8.1 | 3 |
| Peptidoglycan recognition protein 2 | 1.7 | Q96PD5 | 35.1 | 2.4 | 1 |
| Plasma protease C1 inhibitor (SERPING1) | 1.5 | P05155 | 53.0 | 14.2 | 10 |
| β-2-glycoprotein 1 | 1.5 | P02749 | 63.5 | 11.6 | 4 |
| Hemopexin | 1.4 | P02790 | 58.9 | 25.1 | 15 |
| α-1-Acid glycoprotein 2 (Orosomucoid 2) | 1.3 | P19652 | 74.6 | 22.4 | 6 |
| Complement factor H | 1.3 | P08603 | 50.7 | 9.2 | 11 |
| Vitronectin | 1.3 | P04004 | 54.4 | 13.0 | 7 |
| Heparin cofactor 2 (SERPIND1) | 1.3 | P05546 | 50.9 | 8.8 | 5 |
| Complement factor B | 1.3 | P04186 | 51.4 | 13.3 | 12 |
| α-1B-glycoprotein | 1.3 | P04217 | 53.3 | 17.2 | 7 |
| Decrease | |||||
| Transferrin | 0.7 | P02787 | 87.4 | 63.7 | 93 |
| Tissue-type plasminogen activator | 0.7 | P00750 | 46.8 | 10.4 | 7 |
| α-2-HS-glycoprotein | 0.7 | P02765 | 49.9 | 20.4 | 5 |
| Interα-trypsin inhibitor heavy chain H2 | 0.7 | P19823 | 47.7 | 4.3 | 3 |
| Retinol-binding protein 4 | 0.6 | P02753 | 59.7 | 10.5 | 3 |
| Hemoglobin subunit-β | 0.6 | P68871 | 87.1 | 43.5 | 6 |
| Apolipoprotein A-II | 0.5 | P02652 | 80.0 | 41.0 | 10 |
| Apolipoprotein A-I | 0.5 | P02647 | 94.0 | 69.7 | 46 |
| α-2-Macroglobulin | 0.5 | P01023 | 76.5 | 47.7 | 90 |
Based on our serum proteomics results, we subsequently measured acute-phase protein concentrations in individual UC and CD serum samples. Using multiplex analysis, we quantified SAA, fibrinogen, A2M, serum amyloid P (SAP), procalcitonin, ferritin, tissue plasminogen activator protein, and C-reactive protein (CRP). Quantitation of human serum proteins from individual samples identified increases in SAA (7.4-fold) and fibrinogen (1.4-fold) that were consistent with proteomics findings. SAP showed an insignificant increase (1.3-fold) and A2M was unchanged (Fig. 4 and Fig. S1). When a similar collection of proteins was assayed in mouse serum, we observed significant and infection-related increases in the abundance of haptoglobin (300-fold), SAP (7.8-fold), SAA3 (6.1-fold), α-1-acid glycoprotein (2.9-fold), CRP (2.0-fold), and fibrinogen (2.0-fold) (Fig. 4 and Fig. S1).
Fig. 4.
Acute-phase proteins were quantified in the serum of humans (A–E) and mice (F–K) by multiplex methodologies (Control, n = 29; UC, n = 20; CD, n = 20; 10 wk +Hh, n = 10; 10 wk −Hh, n = 11; 20 wk +Hh, n = 13; 20 wk −Hh, n = 12). Results were considered significant when P < 0.05. Statistical analysis performed as described in Fig. 2.
Serum Analysis of Cytokines and Chemokines.
Levels of 27 cytokines in human sera and 23 cytokines in mouse sera were measured to identify inflammatory signals that may be reflective of disease activity. In human UC sera, IL-8, G-CSF, IL-5, and eotaxin were found to be significantly increased, with a trend toward significant elevation for IP-10 (P < 0.06) (Fig. S2). No cytokines were found to increase significantly above control levels in CD sera, although IL-8 trended toward significance (P < 0.06). In mice, several serum cytokines were significantly elevated at 10 and 20 wk in response to infection (Table S1). A strong Th17-type inflammatory response [IL-17, G-CSF, Kupffer cell (KC), and IL-6] was observed in the serum of infected mice. Additionally, IL-12(p35), IL-9, and IL-13 also displayed significant serum increases relative to age-matched controls.
Quantitative PCR Array-Based Transcript Profiling of Inflammatory Stimuli in Mouse Colon.
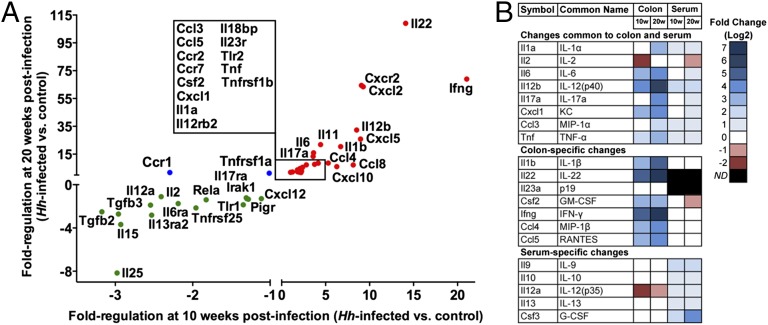
Colon tissues were collected from a subset of mice for the purpose of monitoring the gene expression of several inflammatory signaling molecules and to determine if colonic inflammation was directly associated with serum cytokine levels. Expression profiles were characterized in samples collected at 10 and 20 wk postinfection from H. hepaticus-infected and age-matched control mice. A distinct pattern of cytokine gene expression was observed in colon relative to serum cytokine protein levels (Fig. 5, Fig. S3, and Tables S2 and S3). In tissue, the IL-17 and IL-22 pathways were robustly induced (GM-CSF, IFN-γ, IL-6, IL-1β, KC, TNF-α). In the Rag2−/− mouse these expression patterns point to phagocytes, innate lymphoid cells, or epithelial cells as possible sources (44–47). Furthermore, the signals strongly connect neutrophils to the site of inflammation, confirming the previous demonstration based on histological evaluation (10). Neutrophil activation in the colon is also implicated, based on inflammation-dependent accumulation of Cl-Tyr in these tissues. An additional cytokine network observed in the colon produced IL-12(p40), mitogen-activated protein kinase (MIP)-1α, and MIP-1β. Release of these cytokines and chemokines [particularly IL-12(p40)] in the colon also suggests a role for monocytes, macrophages, and dendritic cells in the pathogenesis of disease. Pronounced expression of IL-12(p40) and RANTES coupled to the increased transcript levels of MIP-1β, TNF-α, and IFN-γ suggest the possible activation of NK cells in the colon, which may also contribute to tissue injury.
Fig. 5.
qPCR analysis of cytokine and chemokine transcripts in H. hepaticus-infected Rag2−/− mouse colon. (A) Gene-expression data for cytokines and chemokines in the Rag2−/− mouse colon at 10 and 20 wk postinfection. All inflammatory targets in this plot exhibited significant changes (P < 0.05) at least one time point (10 wk +Hh, n = 5; 10 wk −Hh, n = 5; 20 wk +Hh, n = 7; 20 wk −Hh, n = 5). A full list of the gene-expression data can be found in Table S3. Red circles denote inflammatory targets that were up-regulated at both time points; green circles denote inflammatory targets that were down-regulated at both time points; blue circles denote inflammatory targets that were differentially regulated. (B) Comparison of colon transcript levels and serum cytokine levels to highlight common and compartment-specific changes.
Multivariate Analysis of Serum Targets.
We further analyzed systemic inflammatory markers of IBD patients using multivariate analysis to determine which serum features were most associated with disease activity in vivo. IBD patients classified as having clinically active disease were modeled against non-IBD controls. UC disease activity was measured using the Simple Clinical Colitis Activity Index (active defined as > 4), and CD disease activity was measured using the Harvey–Bradshaw Index (active defined as ≥ 5). In total, a collection of 35 serum targets (Cl-Tyr, Nitro-Tyr, cytokines, and acute-phase proteins) from 69 patients were subjected to multivariate analysis using partial least-squares (discriminant analysis). To identify features that were most relevant in classifying disease activity, the data were analyzed by variable importance on the projection (VIP), which provides an estimate of the discriminatory strength of a feature (a more detailed description can be found in Materials and Methods). We divided UC and CD into separate disease classes and modeled each independently relative to non-IBD controls.
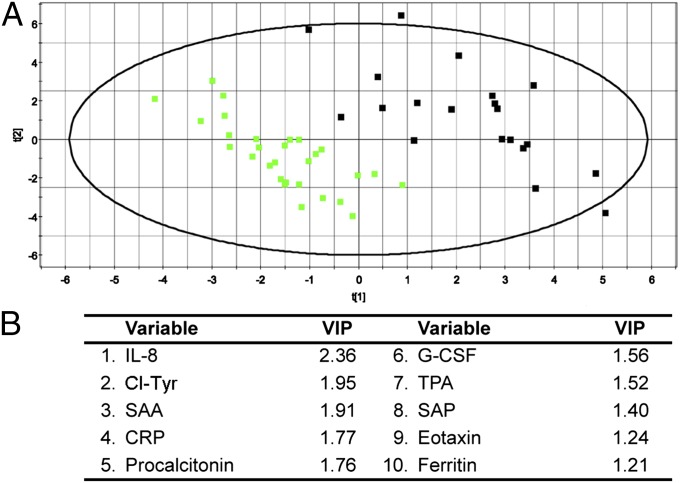
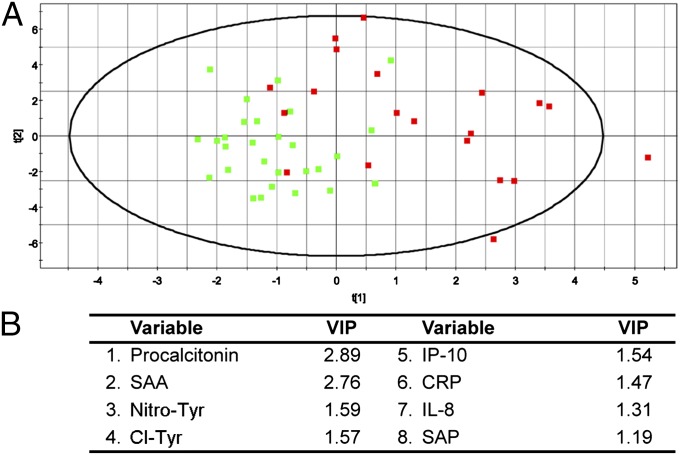
Partial least-squares analysis was more successful for UC than CD. The results can be visually interpreted by inspection of the 2D projections, and a quantitative assessment of the model performance is reflected in the R2Y and Q2(cum) outputs (assessments of the model fit and the model prediction, respectively). R2Y and Q2(cum) range from 0 to 1, with 0 indicating poor performance and 1 indicating ideal performance. The UC samples demonstrated good separation from the non-IBD controls (Fig. 6), whereas the CD samples were not well-resolved from the non-IBD controls (Fig. 7). This result is also reflected in the model outputs, which produced R2Y = 0.87 and Q2(cum) = 0.82 for UC and R2Y = 0.55 and Q2(cum) = 0.15 for CD. The UC model was then analyzed by VIP, which identified IL-8, Cl-Tyr, SAA, CRP, procalcitonin, G-CSF, and tissue plasminogen activator as the variables that were most influential for the separation (VIP > 1.5). VIP analysis of the CD model identified procalcitonin, SAA, Nitro-Tyr, Cl-Tyr, and IP-10 as the most influential variables for the separation (VIP > 1.5). These top variables from the VIP analyses are therefore the most predictive of disease activity in this model for UC and CD, respectively.
Fig. 6.
Multivariate modeling of UC serum analytes. Multivariate data analysis of serum analytes using partial least squares analysis to identify discriminatory features between UC patients (black squares, n = 20) and non-IBD controls (green squares, n = 29). (A) A 2D projection of the partial least-squares multivariate analysis, which produced separation of UC patients from non-IBD controls. The model produced R2Y = 0.87 (model fit) and Q2(cum) = 0.82 (model prediction). (B) List of the top VIPs (VIP > 1.0) in the dataset that were most influential on the data separation.
Fig. 7.
Multivariate modeling of CD serum analytes. Multivariate data analysis of serum analytes to identify discriminatory features between CD patients (red squares, n = 20) and non-IBD controls (green squares, n = 29). (A) A 2D projection of the partial least-squares multivariate analysis, which shows an overlapping distribution of CD patients and non-IBD controls. The model produced R2Y = 0.55 (model fit) and Q2(cum) = 0.15 (model prediction). (B) List of the top VIPs (VIP > 1.0) in the dataset that were most influential on the data separation.
We applied the same multivariate analysis to the H. hepaticus-infected mice. Mouse sera were classified into two groups based on H. hepaticus-infected (10 and 20 wk postinfection) and noninfected controls (10 and 20 wk). In total, a collection of 30 serum targets (cytokines and acute-phase proteins) from 33 mice were analyzed and modeled using partial least-squares analysis. The model outputs for the mouse analysis exceeded those attained from the UC modeling and produced an R2Y = 0.93 and Q2(cum) = 0.91 (Fig. 8). When VIP analysis was applied to the model, haptoglobin, G-CSF, IL-12(p40), SAP, and α-1-acid glycoprotein were identified as the most influential variables on the data separation indicating that they are predictive of colitis in this model (Fig. 8).
Fig. 8.
Multivariate data analysis of serum analytes to identify discriminatory features between Rag2−/− H. hepaticus-infected mice and the uninfected controls. All infected (10 wk +Hh, black squares, n = 8; 20 wk +Hh, green squares, n = 10) and control mice (10 wk −Hh, red squares, n = 6; 20 wk −Hh, blue squares, n = 9) were pooled into two separate groups in this model. (A) A 2D projection of the partial least squares multivariate analysis, which produced clear separation of infected mice and uninfected controls. The model produced R2Y = 0.93 (model fit) and Q2(cum) = 0.91 (model prediction). (B) List of the top VIPs (VIP > 1.0) in the dataset that were most influential on the data separation.
Discussion
Determinants of inflammation-associated cancer development in H. hepaticus-infected Rag2−/− mice were recently described (10). The contributing factors to the onset of colon carcinoma were infiltration of phagocytes and subsequent macromolecular damage to DNA and RNA (principally chlorination) mediated by neutrophil activity. The present study was designed to quantify levels of neutrophil-derived damage products in tissues of IBD patients and in infected Rag2−/− mice tissues. Cl-Tyr and Nitro-Tyr were quantified in human colon and serum from UC and CD patients, and Cl-Tyr levels were found to be higher in colon than in serum. In H. hepaticus-infected mice, both Cl-Tyr and Nitro-Tyr were present and increased in amounts as the disease progressed over time. Additionally, 5-Cl-dC was found to be present in colon tissue of IBD patients at levels similar to those in colons of H. hepaticus-infected mice. We also measured serum levels of other protein products of inflammatory cells, including cytokines, chemokines, and acute-phase proteins. A general signature of activated innate immune response was observed in human sera; the signature in sera from UC patients was strongly suggestive of neutrophil activity, but that in sera of CD patients was suggestive of macrophage and neutrophil activity. A similar spectrum of serum inflammatory proteins was elevated in sera of infected mice. Collectively, these data suggest that phagocytes contribute to the development of IBD in both humans and infected mice, and that Cl-Tyr may prove to be a useful marker of disease activity in human IBD.
Chlorination Damage Products in Humans and Mice with IBD.
Cl-Tyr and Nitro-Tyr were quantified in 148 human serum samples derived from IBD patients or non-IBD controls. A subset of the IBD samples was paired with colon biopsy samples, which allowed us to assess levels of chlorination and nitration damage in inflamed tissues and compare them to systemic serum levels of the same subjects. Forty paired samples were analyzed for Cl-Tyr and Nitro-Tyr (22 from CD and 18 from UC); both adducts were found to be present at higher average levels in colon tissue than in serum. However, no clear correlation was observed between serum and colon levels of either Nitro-Tyr or Cl-Tyr, suggesting that these modified tyrosine residues in serum did not originate from the colon. Because the liver is directly connected to the intestine via the hepatic portal system, it is a likely first target of inflammatory factors arising in the colon. Thus, it is possible that the increase in serum chlorination and nitration products may be attributed at least partially to enhanced innate cell activity in the liver following immune stimulation from the colon. In Citrobacter rodentium-infected mice, liver inflammation can be observed 3 d following colonic inflammation (48). In humans, the time course required for inflammation in the colon to activation of the liver is unknown. Primary sclerosing cholangitis is one of several liver pathologies of significant incidence in IBD patients (49), indicating the importance of identifying extraintestinal factors that may impact disease outcomes and potentially confound serum profiles in IBD patients.
The association of these innate cell-mediated protein damage products with IBD is noteworthy. Both Cl-Tyr and Nitro-Tyr are stable, covalent modifications of tyrosine residues that form as products of reactive chlorinating and nitrating species, respectively (21, 29, 30). Prior studies of UC and CD investigated the role of MPO in IBD development by immunohistochemical staining or enzyme activity assays (16, 50). A limitation of such studies is that analysis of MPO by immunohistochemistry provides information regarding MPO abundance, but not activity. Activity assays require analysis of tissues in which activated neutrophils are present (colon biopsy, fecal fluid, and so forth). Cl-Tyr represents a direct product of systemic MPO activity that can be identified in a less-invasive manner in serum. When Cl-Tyr levels were determined in UC and CD patients during remission and active disease, the inactive samples appeared more similar to non-IBD controls than to active disease patients. The abundance of Cl-Tyr in the protein pool will be reflective of the half-lives of the proteins in which the modification is found. In serum, the major chlorinated species is albumin, which is also the most abundant serum protein, so the albumin signal in serum is reflective of chlorination events amortized over the half-life of albumin, which in humans is ∼20 d. Albumin is also synthesized and released in the liver, further suggesting that the liver may be a possible site for innate cell activation in IBD patients, and therefore a possible source of the chlorination and nitration products observed in serum.
We performed the same analyses of Cl-Tyr and Nitro-Tyr in colon tissue and sera of Rag2−/− mice infected with H. hepaticus. This well-defined and highly reproducible model of IBD involves a bacterial infection that induces chronic inflammation in the lower bowel (12, 13). Furthermore, H. hepaticus is also associated with a clearly defined liver pathology resulting in a predictable incidence of hepatitis in susceptible mice, including the Rag2−/− strain (51–53). Cl-Tyr and Nitro-Tyr were found to increase in colon tissue in a time- and disease-dependent manner upon infection with H. hepaticus (Fig. 3). Cl-Tyr and Nitro-Tyr levels in the colons of uninfected mice were near or below the level of LC-MS/MS quantitation, but significant increases in both modifications were observed by 20 wk postinfection. These increases were accompanied by a persistent mucosal infiltration of neutrophils and macrophages over the course of infection (10).
Previously, we reported an increase in chlorination damage to nucleosides in mouse colon at 20 wk postinfection (10). Over the same time course in the present study we observed the accumulation of Cl-Tyr and Nitro-Tyr in proteins of mouse colon. Collectively, these data suggest that chlorination damage products (both to protein and nucleic acids) arising from neutrophil activity at the site of inflammation increase over the course of disease progression. We extended these observations into humans to assess levels of 5-Cl-dC in colon samples from IBD patients. The DNA damage product 5-Cl-dC was found to be present at similar levels in human colon tissue and in inflamed colonic tissues of mice. This represents a unique finding that associates nucleoside chlorination damage products with inflammation in colon tissue of IBD patients. Although the average values in mouse colons were higher than those found in human tissues, the range of data were overlapping (Fig. 1), which suggests that 5-Cl-dC may be a contributing factor in the initiation of colon carcinogenesis through either mutational or gene silencing mechanisms (54–56).
Serum Cytokine and Acute-Phase Protein Analysis.
An array of cytokines and acute-phase proteins was analyzed in the sera of IBD patients and mice for the purpose of identifying differences between control and diseased subjects. These serum factors were then subjected to multivariate analysis (partial least-squares) to identify important features discriminatory for disease activity. The principal observation from the data analysis in IBD patients was a systemic increase in factors associated with innate immune cell recruitment and activation. Among the 27 cytokines measured in human serum, significant changes were observed almost exclusively in molecules that activate and recruit innate cells (Fig. S2). Based on multivariate analysis of data from UC patients, IL-8, G-CSF, and Cl-Tyr were all found to associate with disease activity; these are strongly associated with neutrophil activation and recruitment. Multivariate analysis of CD patients showed Nitro-Tyr, Cl-Tyr, IP-10, and IL-8, neutrophil- and macrophage-associated factors, to be discriminatory for disease activity. In the Rag2−/− mouse, G-CSF was the main neutrophil-associated factor identified in serum to be discriminatory for infection status; IL-12(p40) was also a discriminating factor in mouse serum and it is important for macrophage regulation. Correspondingly, we observed modest increases in Nitro-Tyr levels in mouse serum samples. The increase in macrophage recruitment, activation, and activity markers is consistent with prior observations on the role of iNOS in the development of colon cancer in the Rag2−/− mouse (9) and their accumulation in the inflamed colon (10).
In the Rag2−/− mouse, distinct but overlapping cytokine signatures were identified in serum compared with gene transcript levels observed in colon. Both compartments displayed an increase in IL-17–associated cytokines, consistent with previous studies of mouse models of colitis (45, 57). In the colon, although we observed a strong increase in IL-12(p40) expression, there was a notable lack of induction of IL-23 (assessed by p19 expression). IL-23 is considered to be a driver of intestinal pathology in the Rag2−/− mouse (45, 58), so the absence of this transcript is unexplained. However, several signals downstream of IL-23 were present (IL-17A, IL-22), suggesting that this pathway was indeed activated. It is possible that IL-1β may have produced a similar response to that of IL-23 in these tissues (59). Recent studies suggest that host genetic factors and alterations in the microbiome impact the immune response in the gut (60, 61). Furthermore, we cannot rule out the possibility of protein stability relative to transcript stability as a confounding factor in our analysis (62).
Our study identified several acute-phase proteins that are commonly associated with IBD (63–65). Proteolytic products of these acute-phase proteins may also be useful in assessing IBD activity (66). Two proteins of potential interest were identified from the proteomics study: orosomucoid 2 (α-1-acid glycoprotein 2) and peptidylglycan recognition protein 2 (PGRP2, also named N-acetylmuramoyl-l-alanine amidase). Association of orosmucoid 2 with UC disease activity is not established, but it has been associated with development of colorectal cancer (67). PGRP2 is a serum protein that hydrolyses peptidoglycan and transgenic knockouts of PGRP2 exhibit enhanced colitis in murine models of IBD (68); its role in human IBD is unknown.
Multivariate Models.
The partial least-squares analysis demonstrated that UC was more amenable to modeling than was CD. The reasons for the poor model behavior for CD are unclear, but two possibilities are likely. First, there are reports that the Harvey–Bradshaw Index is not completely reflective of tissue pathology. In a recent study of CD, more than one-third of enrolled patients with moderate disease activity were found to show no evidence of mucosal ulcerations during endoscopic evaluation (69). We have relied on clinical indices of disease activity, not endoscopy, and UC inflammatory activity may be more accurately reflected in the Simple Clinical Colitis Activity Index than the mucosal inflammation of CD as measured by the Harvey–Bradshaw Index. Second, the disease heterogeneity of CD may contribute to multiple subpopulations that may have subtle or significant changes in the cytokine profiles. As a corollary, the extent of disease involvement is not quantified in any clinical index, but may be critical in explaining discordance between clinical indices and serum markers (i.e., proctitis may cause an increase in symptoms compared with inflammation in a large segment of small bowel, which could be largely clinically silent). When we restricted both CD and UC to smaller subpopulations (disease location, medication, and so forth) no significant observations were made, which is likely because these groupings represent a small sample size that lack predictive power. Genetic variations in affected patients were not investigated in this study.
In all cases of disease modeling, the mouse models were more predictive than the human models. In fact, when the data were restricted to only the 20-wk time point, the best model was attained (Fig. S4). The mouse model of H. hepaticus-induced IBD provides several opportunities to control variables compared with the human studies; minimization of these variables likely contributed to the improved performance. All Rag2−/− mice are genetically equivalent but in human populations, the genetic heterogeneity may result in substantial variability. To initiate colitis in the mouse, animals are infected with a mouse pathogen, H. hepaticus. The initiating events that trigger IBD in human populations probably vary within each disease, although a response to a commensal or pathogenic bacterium is a likely initiating event (70). The disease duration in the mouse was experimentally controlled, whereas the human studies include differences in the age of onset and the duration of the illness. Finally, in the animal studies, there was no therapeutic intervention to alter the immunological response, whereas in human studies patients were frequently being treated with aggressive therapies, such as steroids, TNF-α inhibitors, and other immunosuppressors to attenuate disease symptoms.
Collectively, these data have evaluated the H. hepaticus-infected Rag2−/− mouse as a model for human IBD. Strong similarities were observed between the mouse and human through the accumulation of nitration and chlorination damage products in diseased colon tissue. In the mouse model of colitis, these changes were dependent on disease severity and duration. In human tissues, the serum accumulation of protein damage products was significantly increased over non-IBD controls. We also report unique detection and quantification of 5-Cl-dC in colon tissue of IBD patients, and found its levels to fall within the same range as those present in inflamed colons of H. hepaticus-infected Rag2−/− mice. Inflammatory signals in mouse colons were indicative of a Th17-type response, and were consistent with the recruitment of neutrophils and macrophages to the site of inflammation. The corresponding serum analysis suggested overlapping and distinct patterns that implicated both Th17-like and innate cell activity. Human serum markers were strongly associated with neutrophil factors in UC and both neutrophil and macrophage factors in CD. The cytokine and modified tyrosine targets in serum and tissue collectively support a role for the innate immune system in IBD disease processes, which is also reflected in the Rag2−/− H. hepaticus-induced colitis model.
Materials and Methods
Human Serum Sample Collection.
Two cohorts of IBD samples were used in this investigation. One cohort was obtained from the Cooperative Human Tissue Network (Western Division at Vanderbilt University Medical Center), funded by the National Cancer Institute, and all samples were obtained with informed written consent and were anonymized before receipt at the Massachusetts Institute of Technology (MIT). Cooperative Human Tissue Network patient cohort information is available in Table S4. All work performed at Vanderbilt University was approved by the Vanderbilt University Institutional Review Board. These samples were comprised of colon biopsy tissues paired to serum. A second cohort of serum samples was collected by the Massachusetts General Hospital (MGH) Crohn’s and Colitis Center with written informed consent obtained, and with approval by the MGH Institutional Review Board. MGH patient cohort information is available in Table S5. A set of non-IBD healthy control (n = 29) sera was also included in this cohort. All work from these two sample cohorts was performed at MIT with approval by the MIT Committee on the Use of Humans as Experimental Subjects Committee.
Animal Husbandry, H. hepaticus Infection, and Tissue Collection.
All experiments were performed in accordance with the protocols approved by the MIT Committee on Animal Care and with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, as described previously (10).
Analysis of Chlorination and Nitration Damage Products.
We analyzed 5-Cl-dC as previously described (10, 42). Cl-Tyr and Nitro-Tyr in serum and colon tissue were quantified by LC-MS/MS as previously described with modifications (34, 36). Artifactual production of Cl-Tyr and Nitro-Tyr was monitored during method development by addition of [13C6]-Tyr to tissue samples; no artifactual formation was found. Serum or colon tissue samples were thawed just before processing. Briefly, colon tissue was homogenized with lysis buffer (150 mM NaCl, 5 mM EDTA, 25 mM bicine, 0.5% CHAPS, aprotinin, phenylmethylsulfonyl fluoride, in 1x PBS), centrifuged, and the supernatant was collected. Homogenized colon protein or serum (2 mg) was spiked with internal standards (IS) (l-3-chloro-[13C9,15N]-tyrosine and l-3-nitro-[13C9,15N]-tyrosine). The pronase E digestion was performed as previously described (11) and followed by HPLC purification. The collected HPLC fractions were dried by SpeedVac before LC-MS analysis. All samples were analyzed using Agilent 1200 Series stack (degasser, capillary pump, and autosampler) and an Agilent 6430 triple quandrupole mass spectrometer. Serum samples were applied to an Xbridge Phenyl column (3.5 µm, 1.0 × 100 mm; Waters) and eluted isocratically with 25% methanol containing 0.1% formic acid. Cl-Tyr and IS were analyzed by multiple reaction monitoring (MRM) as follows: m/z 216→170 (Cl-Tyr) and m/z 226→179 (Cl-Tyr IS). Nitro-Tyr and IS were analyzed by MRM with in-source fragmentation (MS3) as follows: m/z 227→181→117 (Nitro-Tyr) and m/z 237→190→125 (Nitro-Tyr IS). Because of inefficient separation on the Phenyl column, colon samples were applied to a Cogent Diamond Hydride column (4 µm, 2.1 × 150 mm; Cogent) and eluted isocratically with 65% acetonitrile containing 0.1% formic acid. Cl-Tyr and IS transitions were monitored as above. Nitro-Tyr and IS were monitored by MRM as follows: m/z 227→181 (Nitro-Tyr) and m/z 190→125 (Nitro-Tyr IS). The Diamond Hydride column was washed with 50% methanol containing 0.1% formic acid for 15 min after every three injections to ensure optimum performance. Quantitation of Cl-Tyr and Nitro-Tyr was based on six-point calibration curves (6.7–267 nM for Cl-Tyr and 8.7–267 nM for Nitro-Tyr) in reference to the internal standard. All analyses were carried out in triplicate. To identify the major serum proteins containing Cl-Tyr, serum was fractionated into a pool containing albumin and IgG and pool containing the remaining (depleted) proteins using Vivapure Anti-HAS/IgG Kit (Sartorius-Stedim). Subsequently, the albumin was separated from the IgG using a Protein G HP SpinTrap column (GE Healthcare). Each of these fractions were analyzed separately as outlined above for the presence of Cl-Tyr.
Serum Proteomics Analysis.
Serum samples were first depleted of albumin and IgG using the Sartorius-Stedim Vivapure Anti-HSA/IgG Kit (Sartorius-Stedim) as suggested by the manufacturer. UC samples were pooled into two separate, randomly selected fractions of 10 samples for both active disease and inactive disease sera. Pooled samples were processed as outlined by the manufacturer for 8-plex iTRAQ Reagent (ABSciex) labeling (43). Pooled and labeled peptides were desalted by solid phase extraction (SPE) using Strata-X, 60 mg, polymeric reversed phase resin (Phenomenex). SPE eluted peptides were dried in a SpeedVac and redissolved in Offgel rehydration buffer (Agilent Technologies), applied to a 24-well gel strip (pH 3–10; GE Healthcare), and separated using an Offgel Fractionator as outlined by the manufacturer (Agilent Technologies). Following separation, fractions were removed and stored at −20 °C until LC-MS/MS analysis.
Sample injection and separation were performed using an Agilent 1100 series capillary pump and microwell plate autosampler, and an Agilent 1200 series nanopump, in line with a Cheminert 10 port dual, position nanovolume, switching valve (Vici). Samples were applied to a custom-packed capillary column (50-μm ID capillary tubing) using 5 μm, YMC ODS-AQ C18 resin (YMC-Gel). The following gradient was applied using the nanopump (mobile phase A was 0.4% acetic acid in water, and mobile phase B was 0.4% acetic acid in acetonitrile): isocratic hold of 2% B for 15 min, linear increase from 2% B to 55% B over 60 min, linear increase to 95% B in 15 min, isocratic hold at 95% B for 10 min, linear decrease to 2% B in 15 min, followed by an isocratic hold for 30 min (re-equilibration). Peptide analysis was performed using an AB Sciex QSTAR Elite QTOF MS equipped with a nanospray source (Proxeon Biosystems) using an 8-μm silica PicoTip Emitter (New Objective). The electrospray voltage was typically 1,300–1,500 V and applied at a conductive union just downstream of the capillary column. Information-dependent MS/MS analysis was performed on the four most abundant peaks in each TOF-MS spectrum, which selected only multiply charged ions (+2 to +5). The following instrument parameters were used: accumulation time 2 s, total cycle time of 8.99 s, mass tolerance 50 ppm. Data were analyzed using Spectrum Mill software (Agilent Technologies) for protein identification, and using Protein Pilot (Sciex) for protein identification and relative quantitation. Proteins were quantified by determining the ratio of the iTRAQ marker ions for each combination of inactive and active samples, and then averaging the response to determine fold-change.
Multiplex Detection of Serum Cytokines, Chemokines, and Acute-Phase Proteins.
Human serum was analyzed using the human 27-plex cytokine panel (Bio-Rad) as specified by the manufacturer and contained the following targets: IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12(p70), IL-13, IL-15, IL-17, Basic FGF, Eotaxin, G-CSF, GM-CSF, IFN-γ, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF, RANTES, TNF-α, and VEGF. Mouse serum was analyzed using the mouse 23-plex cytokine panel (Bio-Rad) as specified by the manufacturer and contained the following targets: IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-17, Eotaxin, G-CSF, GM-CSF, IFN-γ, KC, MCP-1, MIP-1α, MIP-1β, RANTES, and TNF-α. For human acute-phase protein analysis, serum was analyzed using a human acute-phase panel (Bio-Rad) as specified by the manufacturer and contained the following targets: A2M, CRP, ferritin, fibrinogen, haptoglobin, procalcitonin, SAA, SAP, and tissue plasminogen activator. Mouse acute-phase proteins in serum were analyzed using the separate platforms designed by Millipore as specified by the manufacturer and contained the following targets: Fibrinogen, SAA3, adipsin, α-1-acid glycoprotein, A2M, CRP, haptoglobin, and SAP. The following three human analytes did not fall within the range of the standard curve for the majority of samples and were excluded from further analysis: PDGF, RANTES, and haptoglobin.
Gene Expression.
Analyses were performed using customized quantitative PCR (qPCR) arrays (Qiagen) on an ABI 7900HT qPCR system (384-well block; Life Technologies). All steps were performed according to the manufacturers’ instructions with the following specifications: RNA quality and quantity was verified using a RNA 6000 Nano LabChip on an Agilent 2100 Bioanalyzer (Agilent). RNA integrity numbers were consistently >7, indicating high-quality RNA. Reverse transcription was performed with 400 ng of total RNA. Genomic DNA and reverse transcription controls were included in each PCR run to check for genomic DNA contaminations and impurities in RNA samples that may affect reverse transcription, respectively. All experiments met quality control standards as recommended by the manufacturer. Ct values were normalized to four different control genes (Actb, Gapdh, Hprt1, and 18SrRNA) for comparison of control and infected samples. Data were analyzed using the PCR Array Data Analysis Web Portal (www.sabiosciences.com/pcrarraydataanalysis.php) according to the ΔΔCt method.
Multivariate and Statistical Data Analysis.
All multivariate analysis was carried out using SIMCA-P+ v11.5 software (Umetrics). The data were log10-transformed before multivariate analysis. Disease classification was assessed clinically and used as the discriminant variable (dependent, Y-variable) in the partial least-squares discriminant analysis. Serum analytes were used as the independent variable (X-variable). Assessment of the model was carried out by inspecting the R2Y and Q2(cum), which are outputs that describe the model’s performance. R2Y is the fraction of the total Y variation that can be explained by the model (model fit), and Q2(cum) is the total variation of Y that can be predicted by the model (model prediction). An R2Y or Q2(cum) of 1.0 indicates an ideal model fit and a 100% relationship between all X and Y variables. VIP analysis was used to estimate the importance that a particular variable has on the classification accuracy. The average VIP equals 1; therefore, VIPs >1 provide greater predictive value than VIPs <1. A VIP value significantly less than 1 indicates that the feature is not predictive. All statistical analyses were performed using GraphPad Prism software, v6.0a for Mac (GraphPad Software). Statistical significance in serum targets between patients groups (two groups) were assessed by the Mann–Whitney U test; P values < 0.05 were considered significant. The nonparametric Spearman’s rank correlation coefficient (ρ) was determined to identify if statistical dependence existed between measured variables.
Supplementary Material
Acknowledgments
We thank Agilent Technologies for providing access to the triple quadrupole mass spectrometer; M. Kay Washington and Kerry R. Wiles (Vanderbilt University Medical Center) for helpful discussions; and Laura J. Trudel for assistance with the manuscript preparation. Tissue samples were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. Other investigators may have received specimens from the same subjects. This work was supported by National Institutes of Health Grant CA26731 and Massachusetts Institute of Technology Center for Environmental Health Sciences Grant ES002109; a Massachusetts Institute of Technology-Merck Fellowship (to C.G.K.); and a fellowship of the German Academic Exchange Service (to A.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222669110/-/DCSupplemental.
References
- 1.Danese S, Fiocchi C. Etiopathogenesis of inflammatory bowel diseases. World J Gastroenterol. 2006;12(30):4807–4812. doi: 10.3748/wjg.v12.i30.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347(6):417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 3.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142(1):46–54.e42; quiz e30. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Feagins LA, Souza RF, Spechler SJ. Carcinogenesis in IBD: Potential targets for the prevention of colorectal cancer. Nat Rev Gastroenterol Hepatol. 2009;6(5):297–305. doi: 10.1038/nrgastro.2009.44. [DOI] [PubMed] [Google Scholar]
- 5.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korzenik JR, Dieckgraefe BK, Valentine JF, Hausman DF, Gilbert MJ. Sargramostim in Crohn’s Disease Study Group Sargramostim for active Crohn’s disease. N Engl J Med. 2005;352(21):2193–2201. doi: 10.1056/NEJMoa041109. [DOI] [PubMed] [Google Scholar]
- 7.Reaves TA, et al. Neutrophil transepithelial migration: Regulation at the apical epithelial surface by Fc-mediated events. Am J Physiol Gastrointest Liver Physiol. 2001;280(4):G746–G754. doi: 10.1152/ajpgi.2001.280.4.G746. [DOI] [PubMed] [Google Scholar]
- 8.Kucharzik T, Walsh SV, Chen J, Parkos CA, Nusrat A. Neutrophil transmigration in inflammatory bowel disease is associated with differential expression of epithelial intercellular junction proteins. Am J Pathol. 2001;159(6):2001–2009. doi: 10.1016/S0002-9440(10)63051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdman SE, et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci USA. 2009;106(4):1027–1032. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangerich A, et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109(27):E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBee ME, et al. Multivariate modeling identifies neutrophil- and Th17-related factors as differential serum biomarkers of chronic murine colitis. PLoS ONE. 2010;5(10):e13277. doi: 10.1371/journal.pone.0013277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdman SE, et al. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003;162(2):691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maloy KJ, et al. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med. 2003;197(1):111–119. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurst JK, Barrette WCJ., Jr Leukocytic oxygen activation and microbicidal oxidative toxins. Crit Rev Biochem Mol Biol. 1989;24(4):271–328. doi: 10.3109/10409238909082555. [DOI] [PubMed] [Google Scholar]
- 15.Weiss SJ. Tissue destruction by neutrophils. N Engl J Med. 1989;320(6):365–376. doi: 10.1056/NEJM198902093200606. [DOI] [PubMed] [Google Scholar]
- 16.Naito Y, Takagi T, Yoshikawa T. Neutrophil-dependent oxidative stress in ulcerative colitis. J Clin Biochem Nutr. 2007;41(1):18–26. doi: 10.3164/jcbn.2007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klebanoff SJ, Belding ME. Virucidal activity of H2O2-generating bacteria: Requirement for peroxidase and a halide. J Infect Dis. 1974;129(3):345–348. doi: 10.1093/infdis/129.3.345. [DOI] [PubMed] [Google Scholar]
- 18.Harrison JE, Schultz J. Studies on the chlorinating activity of myeloperoxidase. J Biol Chem. 1976;251(5):1371–1374. [PubMed] [Google Scholar]
- 19.Pattison DI, Davies MJ. Reactions of myeloperoxidase-derived oxidants with biological substrates: Gaining chemical insight into human inflammatory diseases. Curr Med Chem. 2006;13(27):3271–3290. doi: 10.2174/092986706778773095. [DOI] [PubMed] [Google Scholar]
- 20.Winterbourn CC. Biological reactivity and biomarkers of the neutrophil oxidant, hypochlorous acid. Toxicology. 2002;181-182:223–227. doi: 10.1016/s0300-483x(02)00286-x. [DOI] [PubMed] [Google Scholar]
- 21.Domigan NM, Charlton TS, Duncan MW, Winterbourn CC, Kettle AJ. Chlorination of tyrosyl residues in peptides by myeloperoxidase and human neutrophils. J Biol Chem. 1995;270(28):16542–16548. doi: 10.1074/jbc.270.28.16542. [DOI] [PubMed] [Google Scholar]
- 22.Gaut JP, et al. Neutrophils employ the myeloperoxidase system to generate antimicrobial brominating and chlorinating oxidants during sepsis. Proc Natl Acad Sci USA. 2001;98(21):11961–11966. doi: 10.1073/pnas.211190298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson JP, Byun J, Heinecke JW. Molecular chlorine generated by the myeloperoxidase-hydrogen peroxide-chloride system of phagocytes produces 5-chlorocytosine in bacterial RNA. J Biol Chem. 1999;274(47):33440–33448. doi: 10.1074/jbc.274.47.33440. [DOI] [PubMed] [Google Scholar]
- 24.Stanley NR, Pattison DI, Hawkins CL. Ability of hypochlorous acid and N-chloramines to chlorinate DNA and its constituents. Chem Res Toxicol. 2010;23(7):1293–1302. doi: 10.1021/tx100188b. [DOI] [PubMed] [Google Scholar]
- 25.Whiteman M, Jenner A, Halliwell B. Hypochlorous acid-induced base modifications in isolated calf thymus DNA. Chem Res Toxicol. 1997;10(11):1240–1246. doi: 10.1021/tx970086i. [DOI] [PubMed] [Google Scholar]
- 26.Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: Biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6(8):662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 27.Lim CH, Dedon PC, Deen WM. Kinetic analysis of intracellular concentrations of reactive nitrogen species. Chem Res Toxicol. 2008;21(11):2134–2147. doi: 10.1021/tx800213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klebanoff SJ. Reactive nitrogen intermediates and antimicrobial activity: Role of nitrite. Free Radic Biol Med. 1993;14(4):351–360. doi: 10.1016/0891-5849(93)90084-8. [DOI] [PubMed] [Google Scholar]
- 29.Eiserich JP, et al. Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature. 1998;391(6665):393–397. doi: 10.1038/34923. [DOI] [PubMed] [Google Scholar]
- 30.Gaut JP, et al. Myeloperoxidase produces nitrating oxidants in vivo. J Clin Invest. 2002;109(10):1311–1319. doi: 10.1172/JCI15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaut JP, Byun J, Tran HD, Heinecke JW. Artifact-free quantification of free 3-chlorotyrosine, 3-bromotyrosine, and 3-nitrotyrosine in human plasma by electron capture-negative chemical ionization gas chromatography mass spectrometry and liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Biochem. 2002;300(2):252–259. doi: 10.1006/abio.2001.5469. [DOI] [PubMed] [Google Scholar]
- 32.Hazen SL, Hsu FF, Mueller DM, Crowley JR, Heinecke JW. Human neutrophils employ chlorine gas as an oxidant during phagocytosis. J Clin Invest. 1996;98(6):1283–1289. doi: 10.1172/JCI118914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeuwenburgh C, et al. Reactive nitrogen intermediates promote low density lipoprotein oxidation in human atherosclerotic intima. J Biol Chem. 1997;272(3):1433–1436. doi: 10.1074/jbc.272.3.1433. [DOI] [PubMed] [Google Scholar]
- 34.Brennan ML, et al. A tale of two controversies: Defining both the role of peroxidases in nitrotyrosine formation in vivo using eosinophil peroxidase and myeloperoxidase-deficient mice, and the nature of peroxidase-generated reactive nitrogen species. J Biol Chem. 2002;277(20):17415–17427. doi: 10.1074/jbc.M112400200. [DOI] [PubMed] [Google Scholar]
- 35.Kang JIJ, Jr, Neidigh JW. Hypochlorous acid damages histone proteins forming 3-chlorotyrosine and 3,5-dichlorotyrosine. Chem Res Toxicol. 2008;21(5):1028–1038. doi: 10.1021/tx7003486. [DOI] [PubMed] [Google Scholar]
- 36.Marvin LF, et al. Quantification of o,o’-dityrosine, o-nitrotyrosine, and o-tyrosine in cat urine samples by LC/ electrospray ionization-MS/MS using isotope dilution. Anal Chem. 2003;75(2):261–267. doi: 10.1021/ac020309w. [DOI] [PubMed] [Google Scholar]
- 37.Frost MT, Halliwell B, Moore KP. Analysis of free and protein-bound nitrotyrosine in human plasma by a gas chromatography/mass spectrometry method that avoids nitration artifacts. Biochem J. 2000;345(Pt 3):453–458. [PMC free article] [PubMed] [Google Scholar]
- 38.Chen HJ, Row SW, Hong CL. Detection and quantification of 5-chlorocytosine in DNA by stable isotope dilution and gas chromatography/negative ion chemical ionization/mass spectrometry. Chem Res Toxicol. 2002;15(2):262–268. doi: 10.1021/tx015578g. [DOI] [PubMed] [Google Scholar]
- 39.Bergt C, et al. The myeloperoxidase product hypochlorous acid oxidizes HDL in the human artery wall and impairs ABCA1-dependent cholesterol transport. Proc Natl Acad Sci USA. 2004;101(35):13032–13037. doi: 10.1073/pnas.0405292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer II, et al. Expression of inducible nitric oxide synthase and nitrotyrosine in colonic epithelium in inflammatory bowel disease. Gastroenterology. 1996;111(4):871–885. doi: 10.1016/s0016-5085(96)70055-0. [DOI] [PubMed] [Google Scholar]
- 41.Keshavarzian A, et al. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut. 2003;52(5):720–728. doi: 10.1136/gut.52.5.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taghizadeh K, et al. Quantification of DNA damage products resulting from deamination, oxidation and reaction with products of lipid peroxidation by liquid chromatography isotope dilution tandem mass spectrometry. Nat Protoc. 2008;3(8):1287–1298. doi: 10.1038/nprot.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57(7):1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 44.Boulard O, Kirchberger S, Royston DJ, Maloy KJ, Powrie FM. Identification of a genetic locus controlling bacteria-driven colitis and associated cancer through effects on innate inflammation. J Exp Med. 2012;209(7):1309–1324. doi: 10.1084/jem.20120239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hue S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203(11):2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cua DJ, Tato CM. Innate IL-17-producing cells: The sentinels of the immune system. Nat Rev Immunol. 2010;10(7):479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 47.Lau KS, et al. Multi-scale in vivo systems analysis reveals the influence of immune cells on TNF-α-induced apoptosis in the intestinal epithelium. PLoS Biol. 2012;10(9):e1001393. doi: 10.1371/journal.pbio.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raczynski AR, et al. Enteric infection with Citrobacter rodentium induces coagulative liver necrosis and hepatic inflammation prior to peak infection and colonic disease. PLoS ONE. 2012;7(3):e33099. doi: 10.1371/journal.pone.0033099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uko V, Thangada S, Radhakrishnan K. Liver disorders in inflammatory bowel disease. Gastroenterol Res Pract. 2012;2012:642923. doi: 10.1155/2012/642923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kayo S, et al. Close association between activated platelets and neutrophils in the active phase of ulcerative colitis in humans. Inflamm Bowel Dis. 2006;12(8):727–735. doi: 10.1097/00054725-200608000-00009. [DOI] [PubMed] [Google Scholar]
- 51.Fox JG, et al. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: A model of helicobacter-induced carcinogenesis. Infect Immun. 1996;64(5):1548–1558. doi: 10.1128/iai.64.5.1548-1558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers AB, et al. Progression of chronic hepatitis and preneoplasia in Helicobacter hepaticus-infected A/JCr mice. Toxicol Pathol. 2004;32(6):668–677. doi: 10.1080/01926230490524247. [DOI] [PubMed] [Google Scholar]
- 53.Ward JM, et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst. 1994;86(16):1222–1227. doi: 10.1093/jnci/86.16.1222. [DOI] [PubMed] [Google Scholar]
- 54.Kang JIJ, Jr, Sowers LC. Examination of hypochlorous acid-induced damage to cytosine residues in a CpG dinucleotide in DNA. Chem Res Toxicol. 2008;21(6):1211–1218. doi: 10.1021/tx800037h. [DOI] [PubMed] [Google Scholar]
- 55.Lao VV, et al. Incorporation of 5-chlorocytosine into mammalian DNA results in heritable gene silencing and altered cytosine methylation patterns. Carcinogenesis. 2009;30(5):886–893. doi: 10.1093/carcin/bgp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valinluck V, Sowers LC. Inflammation-mediated cytosine damage: A mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 2007;67(12):5583–5586. doi: 10.1158/0008-5472.CAN-07-0846. [DOI] [PubMed] [Google Scholar]
- 57.Alex P, et al. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis. 2009;15(3):341–352. doi: 10.1002/ibd.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464(7293):1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sutton CE, et al. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31(2):331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 60.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qian GS, et al. A follow-up study of urinary markers of aflatoxin exposure and liver cancer risk in Shanghai, People’s Republic of China. Cancer Epidemiol Biomarkers Prev. 1994;3(1):3–10. [PubMed] [Google Scholar]
- 63.Weeke B, Jarnum S. Serum concentration of 19 serum proteins in Crohn’s disease and ulcerative colitis. Gut. 1971;12(4):297–302. doi: 10.1136/gut.12.4.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meuwis MA, et al. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem Pharmacol. 2007;73(9):1422–1433. doi: 10.1016/j.bcp.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 65.Vermeire S, Van Assche G, Rutgeerts P. Laboratory markers in IBD: Useful, magic, or unnecessary toys? Gut. 2006;55(3):426–431. doi: 10.1136/gut.2005.069476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu K, Knutson CG, Wishnok JS, Fox JG, Tannenbaum SR. Serum metabolomics in a Helicobacter hepaticus mouse model of inflammatory bowel disease reveal important changes in the microbiome, serum peptides, and intermediary metabolism. J Proteome Res. 2012;11(10):4916–4926. doi: 10.1021/pr300429x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, et al. The potential role of ORM2 in the development of colorectal cancer. PLoS ONE. 2012;7(2):e31868. doi: 10.1371/journal.pone.0031868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saha S, et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe. 2010;8(2):147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colombel JF, et al. SONIC Study Group Infliximab, azathioprine, or combination therapy for Crohn’s disease. N Engl J Med. 2010;362(15):1383–1395. doi: 10.1056/NEJMoa0904492. [DOI] [PubMed] [Google Scholar]
- 70.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.