Abstract
Diatoms are microalgae that possess so-called “complex plastids,” which evolved by secondary endosymbiosis and are surrounded by four membranes. Thus, in contrast to primary plastids, which are surrounded by only two membranes, nucleus-encoded proteins of complex plastids face additional barriers, i.e., during evolution, mechanisms had to evolve to transport preproteins across all four membranes. This study reveals that there exist glycoproteins not only in primary but also in complex plastids, making transport issues even more complicated, as most translocation machineries are not believed to be able to transport bulky proteins. We show that plastidal reporter proteins with artificial N-glycosylation sites are indeed glycosylated during transport into the complex plastid of the diatom Phaeodactylum tricornutum. Additionally, we identified five endogenous glycoproteins, which are transported into different compartments of the complex plastid. These proteins get N-glycosylated during transport across the outermost plastid membrane and thereafter are transported across the second, third, and fourth plastid membranes in the case of stromal proteins. The results of this study provide insights into the evolutionary pressure on translocation mechanisms and pose unique questions on the operating mode of well-known transport machineries like the translocons of the outer/inner chloroplast membranes (Toc/Tic).
Diatoms are a group of microalgae with great ecological relevance that evolved by secondary endosymbiosis ∼185 million years ago (1). From an evolutionary perspective these microalgae are very interesting because the endosymbiont, an ancient red alga, was completely reduced to a so-called complex or secondary plastid. This plastid has a more complex architecture than primary plastids and is surrounded by four instead of only two membranes (2). In the process of secondary endosymbiosis, the red algal nucleus was eliminated, so that nowadays the host nucleus encodes nearly all of the symbiont’s proteome. Hence, preprotein transport mechanisms had to evolve to transport nucleus-encoded preproteins into the plastid (3). It was unknown for quite a long time how protein transport across the four plastid surrounding membranes is managed, and which kind of machineries are involved. However, research especially from the last 10 y led to a basic knowledge on that issue, revealing most notably that preexisting transport mechanisms were used or even machineries from a different context were recycled and relocated during evolution to fulfill new duties in preprotein transport (4, 5).
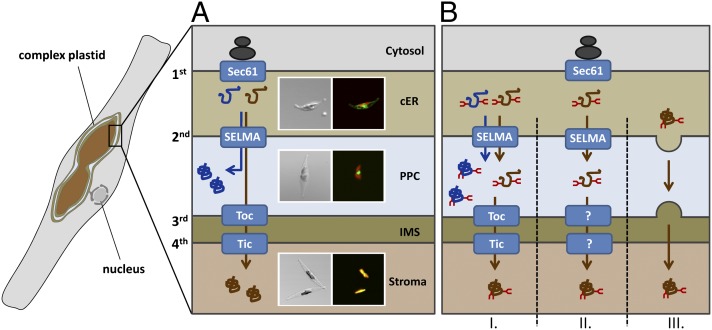
In diatoms, transport of nucleus-encoded plastid proteins starts at the endoplasmic reticulum (ER) membrane as the ER is continuous with the outermost plastid membrane (Fig. 1A). Transport across that first membrane is mediated cotranslationally via the Sec61 translocation complex and a signal peptide at the N terminus is essential for that translocation step into the chloroplast ER (cER) (6–8). To cross the remaining plastid membranes i.e., the second plastid membrane, which presumably originated from the plasma membrane of the red algal endosymbiont, and membranes three and four, a second targeting signal is necessary, which is a transit peptide-like sequence (9–11). Signal peptide and transit peptide together are called bipartite targeting signal (BTS) and represent the classical targeting signature for preproteins destined to complex plastids. Recently, it was proposed that transport across the second plastid membrane of diatoms is mediated by an ER-associated degradation (ERAD)-derived translocation machinery, which was termed symbiont-specific ERAD-like machinery (SELMA) (12, 13). Phylogenetic data together with molecular analyses suggest that during evolution, core components of the red algal ERAD-transport system were relocalized from the symbiont’s ER to the second plastid membrane to mediate preprotein translocation (12–16). After transport across the second membrane, two main protein populations have to be discriminated: proteins that remain within the space between membrane two and three—the periplastidal compartment (PPC), which corresponds to the cytosol of the red algal endosymbiont—and proteins that are destined to the plastid stroma and have to be transported across two further membranes (Fig. 1A). In the case of proteins with a function within the PPC, the transit peptide is cleaved off after transport. Stromal proteins are transported across membranes three and four, presumably mediated by a Toc/Tic (translocon of the outer/inner chloroplast membrane)-like system as it is known from primary plastids. Core components like a Toc75 homolog and several Tic factors were identified by in silico analyses and partially characterized in diatoms and related organisms (17–19).
Fig. 1.
Schematic depiction on preprotein transport into the complex plastid of P. tricornutum. (A) Nucleus-encoded preproteins of the complex plastid have to be transported across multiple membranes—the two outermost membranes in the case of a periplastidal localization (marked in blue) and across all four membranes in the case of stromal proteins (marked in brown). Transport across the first membrane is mediated cotranslationally via Sec61 and thereafter proteins are transported by the SELMA system across the second plastid membrane. Transport across membranes three and four occurs via a Toc/Tic-like transport machinery. Different compartments of the complex plastid can be discriminated easily in fluorescence microscopy using GFP-labeled reporter proteins. GFP is depicted in green, plastid autofluorescence is displayed in red. (B) Our studies reveal that there exist glycoproteins in both protein populations. Periplastidal glycoproteins are most likely transported via the SELMA system into the PPC. How stromal glycoproteins are transported across membranes three and four can only be speculated. Models I–III are presented here. Model I: Stromal glycoproteins use the same route as periplastidal proteins and are thereafter transported via the Toc/Tic machinery. Model II: The Toc/Tic machinery might not be capable of transporting bulky proteins, suggesting that an alternative route exists involving as yet unknown translocation machineries within membranes three and four. Model III: Stromal glycoproteins are already discriminated within the cER and transported via a vesicle-mediated mechanism to the plastid envelope. cER, chloroplast ER; IMS, intermembrane space; PPC, periplastidal compartment; SELMA, symbiont-specific ERAD-like machinery; Tic, translocon of the inner chloroplast membrane; Toc, translocon of the outer chloroplast membrane.
In 2005 it was shown for primary plastids of higher plants that in addition to the well-investigated Toc/Tic translocation system, an alternative transport route exists to import glycoproteins, which get N-glycosylated in the ER (20). Such proteins do not carry a transit peptide but a signal peptide instead and are transported via ER and Golgi in a vesicle-mediated manner, thereby probably bypassing the Toc/Tic system (20–25). So far it is not known whether complex plastids are able to import glycoproteins as well, but as nucleus-encoded preproteins necessarily pass the ER during plastid import they might potentially get N-glycosylated en route. If so, this would inevitably raise the question of how these proteins are transported across the remaining membranes as machineries like Toc/Tic are not believed to be able to transport bulky molecules (26).
In this study, we provide first evidence for the existence of glycoprotein transport into complex plastids. We generated constructs of plastid preproteins with up to five synthetic N-glycosylation sites and analyzed the glycosylation status as well as in vivo targeting. Additionally, we identified several endogenous periplastidal and stromal glycoproteins that get N-glycosylated during the passage of the cER before being transported to their final destination across multiple membranes.
Results
N-Glycosylation in the cER and Glycoprotein Transport Across the Second Plastid Membrane.
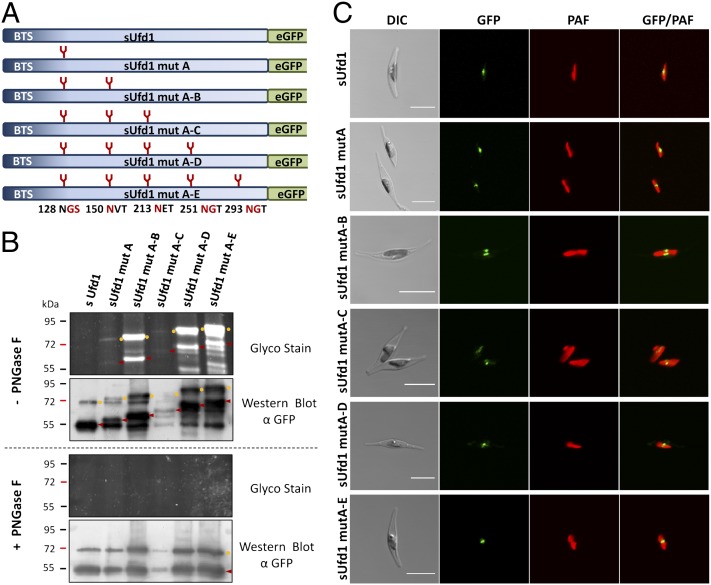
In the diatom Phaeodactylum tricornutum nucleus-encoded proteins of the periplastidal compartment are imported cotranslationally into the lumen of the cER and subsequently are transported via the SELMA complex across the second plastid membrane. To test whether (i) this special ER subcompartment—the cER—provides generally the equipment to mediate N-glycosylation and (ii) N-glycosylation of preproteins would have an effect on transport across the second plastid membrane, we generated synthetic constructs of the periplastidal preprotein sUfd1 (symbiont-specific ubiquitin fusion degradation protein 1). This protein was mutated by inserting up to five artificial N-glycosylation sites (mutA–E) predicted by the program NetNGlyc 1.0 with high confidence (Fig. 2A). Subsequently, the genuine sUfd1 protein as well as the mutated versions mutA, mutA–B, mutA–C, mutA–D, and mutA–E were expressed as GFP fusion proteins in P. tricornutum. All constructs were expressed under the inducible nitrate reductase promotor system (12) for 2 d. To check whether the sUfd1 constructs get glycosylated during the passage of the cER GFP fusion proteins of the respective P. tricornutum transfectants were immunoprecipitated, separated by gel electrophoresis, and periodate oxidized for glycoprotein detection. Thereby, we could demonstrate that indeed all mutated versions are labeled in Glyco-Stain and form a descending ladder indicating a distinct mass shift with each N-glycosylation site added (Fig. 2B, Upper). The original sUfd1, which does not contain any predicted N-glycosylation sites, is negative for Glyco-Stain. In the case of sUfd1 mutA–C, the expression level of the GFP fusion protein is rather low, hence the signal detected in the Glyco-Stain is less strong. The occurrence of a double ladder—one starting at about 55 kDa and a second at about 67 kDa—is due to the fact that not all proteins are completely processed within the periplastidal compartment and partially still carry the transit peptide at the N terminus. This is an observation, which is seen not only in diatoms, but also in Apicomplexa (27) and might be a result of overexpression. However, all processing variants of the protein are glycosylated (Fig. 2B, Upper) and complete processing already provided a first hint that the glycosylated protein is completely imported into the PPC where the targeting signal is cleaved off. Intermediate signals are a result of partial glycosylation with not all inserted N-glycosylation sites uniformly covered. This is confirmed by treatment with Peptide-N-Glycosidase F (PNGase F) of the samples, which leads to complete deglycosylation of all sUfd1 variants. The signals in the Western blot collapse to the wild-type pattern of the endogenous sUfd1 protein with no signal being detected in the Glyco-Stain (Fig. 2B, Lower). In the following, in vivo localization studies were performed demonstrating that not only the genuine sUfd1 protein but also all glycosylated versions are completely imported into the periplastidal compartment. GFP fusion proteins of all constructs accumulate in the classical blob-like structure (Fig. 2C), which is typical for a localization within the periplastidal compartment (9, 12, 28, 29). Altogether, these results demonstrate that even very bulky, highly N-glycosylated proteins can be transported across the second plastid membrane into the PPC.
Fig. 2.
Analyses on the N-glycosylation status and in vivo localization of synthetic sUfd1 constructs. (A) Schematic depiction on sUfd1 constructs with up to five synthetic N-glycosylation sites. Mutated amino acids and their positions are marked in red. (B) Glyco-Stain of the isolated GFP fusion proteins reveals that all inserted N-glycosylation sites get glycosylated in vivo, resulting in a descending ladder from sUfd1 mutA to sUfd1 mutA–E. Not all proteins are completely processed, however, and partially still carry the N-terminal targeting signal. Mature proteins are marked with red arrowheads; versions still carrying the targeting signal are labeled with yellow circles. Intermediate signals are a result of partial glycosylation with not all inserted N-glycosylation sites being uniformly covered. Specificity of N-gylcosylation was additionally confirmed by PNGase F treatment resulting in complete deglycosylation. In the case of all variants, Western blot signals collapse to the wild-type pattern of endogenous sUfd1. (C) In vivo localization studies in P. tricornutum demonstrate that the endogenous sUfd1-GFP fusion protein as well as the glycosylated versions localize to the periplastidal compartment visible as a blob-like structure close to the plastid. (Scale bar, 10 µm.) BTS, bipartite targeting signal; DIC, differential interference contrast; PAF, plastid autofluorescence.
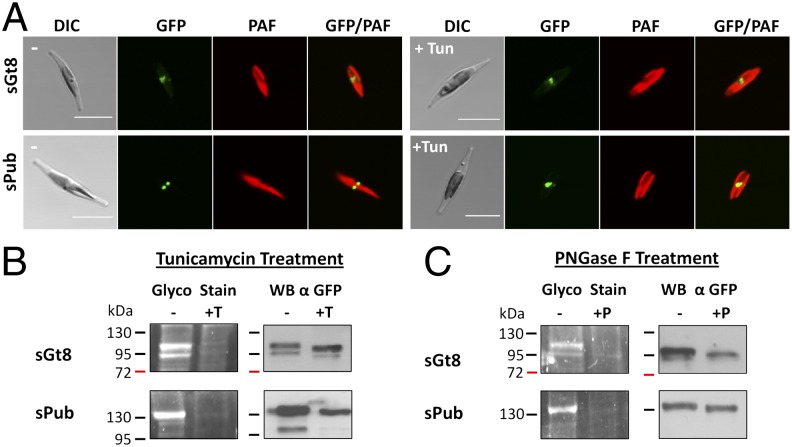
Besides such synthetic constructs, we screened all known endogenous PPC proteins (9, 10, 13, 16, 29–31) for potential N-glycosylation sites using the program NetNGlyc 1.0. Interestingly, 23% of the 52 proteins possess potential N-glycosylation sites with high confidence predictions (++) within the mature, soluble protein (Table S1). For further analyses, we selected two proteins, that were analyzed in previous studies and have good N-glycosylation predictions (Fig. S1). The protein sPub (symbiont-specific PNGase/UBA or UBX) is a potential adapter protein of the AAA-ATPase Cdc48 containing an additional thioredoxin domain and was previously shown to be localized within the PPC (29). The protein sGt8 (symbiont-specific glycosyltransferase family 8) shows homology to glycosyltransferases of the GT8 family. Both proteins were expressed in fusion with GFP and in vivo localization was investigated after 2 d revealing a classical periplastidal localization (Fig. 3A). Subsequently, GFP fusion proteins were purified by immunoprecipitation and the glycosylation status was analyzed demonstrating that both proteins indeed are glycosylated in vivo (Fig. 3B). To test whether N-glycosylation plays a role for preprotein targeting, all experiments were carried out in double with and without the N-glycosylation inhibitor tunicamycin in the culture medium. Tunicamycin treatment had no effect on in vivo expression and targeting of the GFP fusion proteins, which still accumulate within the PPC (Fig. 3A). The Glyco-Stain, however, confirms that N-glycosylation is efficiently blocked by tunicamycin treatment in the case of both proteins and Western blot analyses reveal a slight mass shift due to the lack of glycan moieties (Fig. 3B). In the case of sPub, this mass shift is less obvious as the separation in the range of 130 kDa is rather poor. Specificity of N-glycosylation was additionally confirmed by PNGase F treatment of the purified proteins resulting in complete deglycosylation (Fig. 3C). Altogether, one can conclude from these assays that the periplastidal proteins sGt8 and sPub get N-glycosylated during the passage of the cER before being transported across the second plastid membrane into the PPC.
Fig. 3.
Identification of endogenous glycoproteins of the periplastidal compartment. The proteins sGt8 and sPub were expressed as GFP fusion proteins under standard conditions as well as in the presence of the N-glycosylation inhibitor tunicamycin (+Tun). (A) In vivo localization studies demonstrate that both proteins localize to the periplastidal compartment classically visualized as a so-called blob-like structure. Tunicamycin treatment has no effect on the localization (+Tun). (B) Purified GFP fusion proteins were analyzed by Glyco-Staining as well as Western blot, demonstrating that both proteins get glycosylated in vivo (−). Tunicamycin treatment of the cultures efficiently blocks N-glycosylation in vivo and no signal is observed in the Glyco-Stain (+T). Western blot analyses confirm the presence of the purified protein and reveal a slight mass shift due to the lack of glycan moieties (+T). (C) In an additional control, purified proteins were treated with the enzyme PNGase F, which specifically cleaves off N-glycosylation residues. PNGase F treatment resulted in complete deglycosylation of both proteins (+P), confirming once more that the signal detected in the Glyco-Staining is due to N-glycosylation. (Scale bar, 10 µm.) DIC, differential interference contrast; PAF, plastid autofluorescence; Tun, tunicamycin; WB, Western blot.
Transport of N-Glycosylated Proteins Across the Third and Fourth Plastid Membrane.
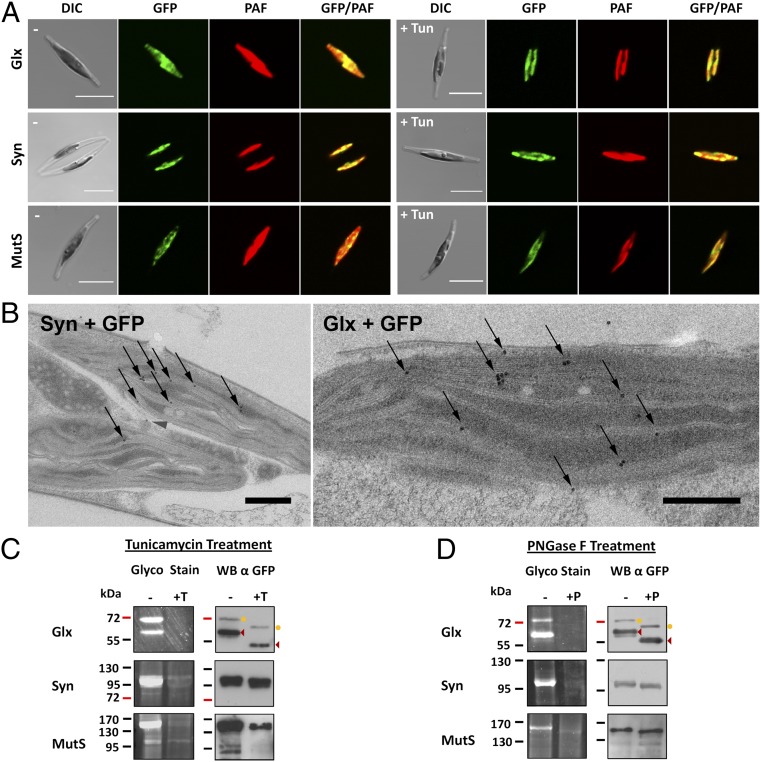
Our results on nucleus-encoded periplastidal proteins demonstrate that preproteins can get N-glycosylated in the cER and subsequently are transported across the second plastid membrane. Transport across membranes three and four of the complex plastid (corresponding to the plastid envelope of primary plastids) is mediated like in land plants by a Toc/Tic-like translocation machinery, which is so far not believed to be able to transport bulky proteins (26, 32). We screened the genome of P. tricornutum for potential stroma-specific glycoproteins with high confidence N-glycosylation predictions (>0.7) to elucidate whether glycosylated proteins can also be transported across the two innermost plastid membranes three and four or not. Three candidates with a classical bipartite stromal targeting signal that have putative functions within the plastid, and show very good predictions for N-glycosylation were picked for further analyses: The protein Glx, which contains a C terminal glyoxalase domain, the tRNA synthetase Syn and the putative mismatch repair protein MutS (Mutator S). Each of these proteins contains one or two predicted N-glycosylation sites in the mature protein sequence (Fig. S1). In vivo localization studies with GFP fusion constructs demonstrate that all three proteins localize to the plastid stroma (Fig. 4A). Immunoelectron microscopic analyses using an antibody against GFP confirm this observation, demonstrating that the GFP fusion proteins of Glx, Syn, and MutS accumulate within the plastid stroma exclusively (Fig. 4B and Fig. S2). Once again, cultures were grown in the presence of as well as without the N-glycosylation inhibitor tunicamycin showing no difference in localization (Fig. 4A). Subsequently, GFP fusion proteins were isolated, separated by gel electrophoresis, and analyzed for N-glycosylation. Indeed, all three proteins show strong signals in the Glyco-Stain and are nonglycosylated when cultures were treated with tunicamycin (Fig. 4C). In the case of the protein Glx different processing stadia are observed as proteins partially still carry the transit peptide. Both variants are glycosylated, however, and the complete processing—which occurs in the plastid stroma—indicates also on a molecular level that the glycosylated protein is completely imported. PNGase F treatment of the purified proteins resulted in the case of all three proteins in complete deglycosylation, confirming once again that the signal in the Glyco-Stain is indeed a product of N-glycosylation (Fig. 4D). Western blot analyses attest to a mass shift, when proteins are deglycosylated. Depending on the molecular weight of the protein and the number of predicted N-glycosylation residues, this mass shift is more or less distinct (Fig. 4 C and D). As the nonglycosylated version of the GFP fusion proteins was never detected in the untreated Western blot samples (best seen in the case of Glx) one can conclude that the majority of the GFP fusion protein gets glycosylated in vivo. Control experiments, demonstrating that the Glyco-Stain is indeed selective for periodate labile glycans, are provided for all endogenous proteins in comparison with the stromal nonglycosylated protein FbaC2 (fructose-1,6-bisphosphate aldolase C2) (Fig. S3). Altogether, these studies with GFP fusion proteins show by means of fluorescence and electron microscopy as well as three different techniques of glycoprotein detection that plastidal glycoproteins exist that are transported across all membranes into the stroma of the complex plastid. In an additional assay the lectin concanavalinA (ConA) was used for immunogold labeling studies on electron microscopic sections of wild-type P. tricornutum cells. As expected, ConA binding was observed throughout the cell especially in the secretory system where the majority of glycoproteins would be expected. Interestingly, however, high binding affinity was also observed within the plastid stroma, suggesting in a more general approach that glycoproteins might commonly exist in the complex plastid of P. tricornutum (Fig. S4). This observation is supported by in silico analyses performed with homologs of a set of 48 verified heterokont stromal proteins (33) revealing that 10.4% have high confidence predictions for N-glycosylation (Table S1).
Fig. 4.
Identification of endogenous glycoproteins of the plastid stroma. The proteins Glx, Syn, and MutS were expressed as GFP fusion proteins in P. tricornutum. (A) In vivo localization studies demonstrate that all three proteins localize to the plastid stroma with GFP fluorescence overlapping with the autofluorescence of the plastid. Incubation with the N-glycosylation inhibitor tunicamycin has no influence on protein targeting (+Tun). (Scale bar, 10 µm.) (B) Immunoelectron microscopic analyses using an αGFP antibody confirm the fluorescence microscopic data for all three proteins. Thin sections of Glx + GFP and Syn + GFP expressing cells are shown exemplarily. Additional data are provided in Fig. S2. Gold particles (10 nm) are highlighted with arrows. (Scale bar, 500 nm.) (C) Glyco-Staining as well as Western blot assays on the purified GFP fusion proteins confirm that the proteins get N-glycosylated in vivo (−). In tunicamycin-treated cultures, no glycosylation is observed (+T). In the case of the protein Glx, different processing stadia are observed. The mature protein is marked with arrowheads; the variant still carrying the targeting signal is labeled with a circle. (D) In a second assay the purified proteins were treated with PNGase F, resulting in complete deglycosylation in the case of all three proteins (+P). DIC, differential interference contrast; PAF, plastid autofluorescence; Tun, tunicamycin; WB, Western blot.
Discussion
In 2005 it was demonstrated by Villarejo et al. that in addition to the known Toc/Tic transport machinery of primary plastids, an alternative pathway exists for the import of plastid glycoproteins (20). These proteins are most likely transported via vesicles from the ER to the Golgi and further to the plastid envelope (23). However, detailed mechanisms on glycoprotein transport in primary plastids are still unknown. The study presented here reveals that not only in primary but also in complex plastids, which evolved by secondary endosymbiosis and are surrounded by additional membranes, plastidal glycoproteins do exist. These proteins get N-glycosylated in the cER after transport across the first plastid membrane, demonstrating that this subcompartment basically possesses the equipment to provide N-glycosylation, which was so far only studied for the ER in P. tricornutum (34). Subsequently, plastidal glycoproteins are transported across the periplastidal membrane in the case of PPC-specific proteins and pass two further membranes in the case of stroma-specific proteins. As for most cellular translocation machineries transport of bulky glycoproteins is not an essential task, because glycosylation of proteins is restricted to ER and Golgi, this issue appears as a new challenge for the evolution of preprotein import into complex plastids that was not discussed hitherto.
One machinery that has the capability to transport glycosylated proteins is the ERAD-system transporting misfolded proteins from the ER back into the cytosol (35, 36). With that information in mind, one can predict that SELMA, the ERAD-derived preprotein transport system of the second membrane of many complex plastids, transports glycosylated proteins as well. During evolution the ERAD system of the red algal symbiont was relocalized from the ER membrane to the periplastidal membrane and thus recycled to fulfill new functions in preprotein transport (14). The results of this study cast a unique light on the evolutionary scenario as the ability to transport glycoproteins might even have served as selective pressure leading to the establishment of SELMA, as nucleus-encoded plastid proteins have to cross the ER with its glycosylation machinery.
Besides periplastidal glycoproteins, stromal glycoproteins were also identified, which are transported further across the two innermost plastid membranes. These proteins carry a typical bipartite targeting signal for a stromal localization at the N terminus and contain one or two N-glycosylation sites that get loaded during the passage of the cER. In P. tricornutum preprotein transport across these membranes is presumably mediated by a Toc/Tic-like machinery similar to primary plastids. A Toc75 homolog, the protein ptOmp85 (P. tricornutum outer membrane protein 85), was recently identified and characterized by Bullmann and colleagues (17, 37). However, Toc75 as well as the Tic complex within the inner plastid membrane are generally not believed to be able to transport bulky proteins (26) and it has been shown that plastid preproteins necessarily have to be transported in an unfolded conformation (32). Whereas the pore diameter of Toc75 measures about 14–26 Å (38) and the translocons of the inner plastid membrane Tic110 and Tic20 are in a similar range with 15–31 Å and 7.8–14.1 Å, respectively (39, 40), typical N-glycosylation residues comprising 12 glycans have a size of ∼10 × 10 × 30 Å (41, 42), and hence might be too big for translocation. However, as in primary plastids, there is no need for Toc75 to transport glycoproteins, because the addition of glycans occurs exclusively in the ER and Golgi and stromal glycoproteins necessarily use a vesicle-mediated route (20, 22–25), analyses on that issue are not available so far. The pore diameter of the Toc75 homolog in P. tricornutum measures about 15 Å (17), thus it is not bigger than Toc75 in primary plastids and might be too narrow to allow the transport of N-glycosylated proteins.
At this point, it can only be speculated how plastid glycoproteins are transported across membranes three and four in P. tricornutum. Models I–III are discussed (Fig. 1B) here. Model I: Stromal glycoproteins get N-glycosylated in the cER, are subsequently transported across the second membrane using the SELMA system (like periplastidal proteins and nonglycosylated stromal proteins), and finally reach the plastid stroma via the general Toc/Tic pathway. In that case, glycan residues might be sterically less problematic than expected or the pore-forming units Toc75 and Tic110/Tic20 might be flexible enough or can actively stretch to allow transport. Model II: Of course there might also exist hitherto unknown translocation systems that are capable of transporting bulky glycoproteins across membranes three and four but are not yet identified. In that case, one has to postulate that glycosylation residues in combination with the transit peptide serve as a targeting signal. Model III: A third possibility would be that there exists an alternative vesicle-mediated pathway between the second and third plastid membranes, which was postulated in a different context (not for glycoproteins) previously (11). Also in that model the transit peptide together with the N-glycans should serve as a recognition signal to discriminate these proteins from other secretory glycoproteins. However, as no SNAREs or other classic components of vesicle-mediated transport pathway were identified so far within the periplastidal compartment (29), this scenario might be rather unlikely or other components should be involved.
In our experiments, we studied the influence of tunicamycin on protein targeting, demonstrating that tunicamycin treatment hinders N-glycosylation, but does not interfere with the final destination of the expressed proteins. Thus, glycosylation per se is not essential for the process of protein import into the plastid. Nevertheless, this finding does not necessarily exclude models II and III, as stromal glycoproteins possess typical bipartite targeting signals and hence might use the classic Toc/Tic mediated route when N-glycosylation is inhibited.
Altogether, one can conclude that not only in primary plastids but also in complex plastids glycoproteins do exist. These proteins get N-glycosylated during the passage of the cER and thereafter are transported across up to three membranes into the plastid stroma. Whereas periplastidal proteins use a transport system adapted for glycosylated proteins, stromal glycoproteins are either transported via the preexisting translocons in the plastid envelope that might have been adapted for that purpose or these proteins use so far unidentified systems for the entry into the stroma. In future, import studies with isolated plastids might give a first idea of whether the Toc/Tic machinery is in general able to transport N-glycosylated proteins or whether alternative so far uncharacterized machineries or vesicle-mediated pathways do exist. In any case, it will be very exciting to elucidate detailed mechanisms on glycoprotein transport into complex plastids.
Materials and Methods
Plasmid Construction.
All P. tricornutum sequences used in this study can be retrieved from database PhatrDBv2.0 (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html). Sequences of sUfd1 (ID Phatr2: 49319), Glx (ID Phatr2: 48863), and Syn (ID Phatr2: 43097) were amplified by standard PCR using genomic P. tricornutum DNA as a template. MutS (ID Phatr2: 47730), sPub (Phatr2: 37661), sGt8 (ID Phatr2: 40314), and FbaC2 (ID Phatr2: 22993) were amplified by RT-PCR, as EST models were either incomplete or sequences contained introns. RNA was isolated using standard phenol-chloroform extraction protocols and purified with the RNeasy Plus Mini kit (Qiagen). Reverse transcription was carried out with SuperScript II (Invitrogen) following manufacturer’s instructions. Synthetic constructs of sUfd1 were generated by multiple mutation reactions (MMRs) as described in ref. 43 using the following oligonucleotides: sUfd1 5′ GAATTCATGGCTGTTCGACGTC, sUfd1 3′ CCATGGCCTCCCTATCCTGTTCAC, sUfd1 mutA CCGGCAAACGGTTCCCAAGCCATTCAG, sUfd1 mutB GAATTGACAACGTTACTGGTGAACGG, sUfd1 mutC GTGGAATTGAATGAGACAGTACCTGC, sUfd1 mutD CATTACAGCAATGGAACGCAAGGCTCG, and sUfd1 mutE GGATTGCGATAATGGCACGGATTTTCTG, respectively. For in vivo studies on localization and glycosylation, all sequences were cloned in front of eGFP into the vector pPha-NR (GenBank: JN180663), which is a derivate of pPhaT1 with endogenous nitrate reductase promoter/terminator flanking the multiple cloning site. P. tricornutum transfection was carried out as described in ref. 44.
Cell Culture.
P. tricornutum was cultivated in f/2 medium under constant illumination (80 µmol photons⋅m−2⋅s−1) at 22 °C as described previously (44). Liquid cultures were grown with agitation (150 rpm) in a volume of 300 mL to a density of approximately OD600 = 0.8 with 1.5 mM NH4Cl as sole nitrogen source. To induce expression of recombinant proteins, cells were harvested and transferred to fresh medium containing 0.9 mM NaNO3 for 2 d. Subsequently, in vivo localization and the N-glycosylation status of GFP fusion proteins was checked. When analyzing in vivo effects of N-glycosylation inhibition, cultures were split with one-half being incubated with 0.5 µg/mL tunicamycin in the induction media and one-half being treated regularly without the inhibitor.
Glycoprotein Detection.
Cells were harvested by centrifugation (5 min, 1,500 × g), resuspended in 3 mL immunoprecipitation (IP) buffer (50 mM Tris⋅HCl, 200 mM KOAc, 1 mM EDTA, 10% (vol/vol) glycerol, 1% Nonidet P-40, protease inhibitor mixture, pH 7.5) and disrupted with a French press (20,000 psi cell pressure, two repeats). Cell debris was removed by centrifugation (30 min, 20,000 × g at 4 °C) and GFP fusion proteins from the supernatant were immunoprecipitated using the µMACS Epitope Tag Protein Isolation kit (Miltenyi Biotec) following manufacturer’s instructions. A total of 90% of the purified GFP fusion proteins was separated by SDS/PAGE and checked for glycosylation using the Staining Pro-Q Emerald 300 Glycoprotein Gel kit (Invitrogen). A total of 10% of the IP eluate was analyzed by Western blot with an anti-GFP antibody to confirm specificity of the precipitated GFP fusion proteins. To verify the binding of N-glycans to the proteins, one-half of the IP eluate was incubated with 5 units of the N-deglycosylation enzyme PNGase F from Elizabethkingia miricola (Sigma-Aldrich) following manufacturer’s instructions and analyzed as described before.
Fluorescence and Electron Microscopy.
For fluorescence microscopy, cells were fixed with 4% (wt/vol) paraformaldehyde (PFA) and 0.0075% glutaraldehyde (GA) for 20 min. Subsequently, in vivo localization of GFP fusion proteins was analyzed with a Leica TCS SP2 confocal laser scanning microscope using a PL APO 63×/1.32–0.6 oil Ph3 CS objective. GFP and chlorophyll fluorescence was excited with an argon 65 mW laser at 488 nm and detected at a bandwidth of 500–520 nm and 625–720 nm, respectively. For electron microscopic studies, cells were centrifuged for 5 min at 1,500 × g, high-pressure frozen with a Wohlwend HPF Compact 02 (M. Wohlwend, Engineering Office, Sennwald, Switzerland) and transferred to the automatic AFS2 freeze substitution unit (Leica Microsystems). To substitute the water and enhance the contrast of the samples, a mixture consisting of 0.2% OsO4, 0.25% uranyl acetate, and 5% (vol/vol) H2O in acetone was used. The substitution program, the following washing steps, Epon embedding, ultrathin sectioning, and immunogold labeling were performed as described previously (45). Labeling of the GFP fusion proteins was performed with a primary antibody against GFP (goat-αGFP; Rockland, diluted 1:1,000) and a secondary 10-nm gold-coupled rabbit-α-goat IgG (dilution 1:20) with subsequent poststaining as described previously (46). For ConA labeling, wild-type cells were fixed for 4 h with 0.02% GA and 4% (wt/vol) PFA in f/2 medium. Cells were washed in Sorensen’s buffer, dehydrated in a graded ethanol series, and embedded in Lowicryl resin. ConA biotin conjugate (2 µg/mL) in combination with streptavidin coupled to 20 nm gold were used. Transmission electron microscopy was performed on a JEOL JEM 2100, operating at 120 kV, and equipped with a fast-scan 2 k × 2 k CCD camera F214 (TVIPS).
Supplementary Material
Acknowledgments
We thank Marion Debus and Marianne Johannsen for technical assistance in cell preparation for electron microscopy. This work was supported by the German Research Foundation Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 593) and the LOEWE program of the state of Hessen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301945110/-/DCSupplemental.
References
- 1.Kooistra WH, Medlin LK. Evolution of the diatoms (Bacillariophyta). IV. A reconstruction of their age from small subunit rRNA coding regions and the fossil record. Mol Phylogenet Evol. 1996;6(3):391–407. doi: 10.1006/mpev.1996.0088. [DOI] [PubMed] [Google Scholar]
- 2. Gould SB, Waller RF, McFadden GI (2008) Plastid evolution. Annu Rev Plant Biol 59:491–517. [DOI] [PubMed]
- 3. Cavalier-Smith T (2003) Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Philos Trans R Soc Lond B Biol Sci 358(1429):109–133; discussion 133–134. [DOI] [PMC free article] [PubMed]
- 4.Bolte K, et al. Protein targeting into secondary plastids. J Eukaryot Microbiol. 2009;56(1):9–15. doi: 10.1111/j.1550-7408.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- 5.Hempel F, et al. Transport of nuclear-encoded proteins into secondarily evolved plastids. Biol Chem. 2007;388(9):899–906. doi: 10.1515/BC.2007.119. [DOI] [PubMed] [Google Scholar]
- 6.Apt KE, et al. In vivo characterization of diatom multipartite plastid targeting signals. J Cell Sci. 2002;115(Pt 21):4061–4069. doi: 10.1242/jcs.00092. [DOI] [PubMed] [Google Scholar]
- 7.Bhaya D, Grossman A. Targeting proteins to diatom plastids involves transport through an endoplasmic reticulum. Mol Gen Genet. 1991;229(3):400–404. doi: 10.1007/BF00267462. [DOI] [PubMed] [Google Scholar]
- 8.Lang M, Apt KE, Kroth PG. Protein transport into “complex” diatom plastids utilizes two different targeting signals. J Biol Chem. 1998;273(47):30973–30978. doi: 10.1074/jbc.273.47.30973. [DOI] [PubMed] [Google Scholar]
- 9.Gould SB, et al. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol Biol Evol. 2006;23(12):2413–2422. doi: 10.1093/molbev/msl113. [DOI] [PubMed] [Google Scholar]
- 10.Gruber A, et al. Protein targeting into complex diatom plastids: Functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64(5):519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- 11.Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005;41(2):175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- 12.Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol Biol Evol. 2009;26(8):1781–1790. doi: 10.1093/molbev/msp079. [DOI] [PubMed] [Google Scholar]
- 13.Sommer MS, et al. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24(4):918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- 14.Bolte K, et al. Making new out of old: Recycling and modification of an ancient protein translocation system during eukaryotic evolution. Mechanistic comparison and phylogenetic analysis of ERAD, SELMA and the peroxisomal importomer. Bioessays. 2011;33(5):368–376. doi: 10.1002/bies.201100007. [DOI] [PubMed] [Google Scholar]
- 15. Felsner G, et al. (2011) ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane. Genome Biol Evol 3:140–150. [DOI] [PMC free article] [PubMed]
- 16.Hempel F, Felsner G, Maier UG. New mechanistic insights into pre-protein transport across the second outermost plastid membrane of diatoms. Mol Microbiol. 2010;76(3):793–801. doi: 10.1111/j.1365-2958.2010.07142.x. [DOI] [PubMed] [Google Scholar]
- 17.Bullmann L, et al. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010;285(9):6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFadden GI, van Dooren GG. Evolution: Red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14(13):R514–R516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 19.van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci USA. 2008;105(36):13574–13579. doi: 10.1073/pnas.0803862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villarejo A, et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol. 2005;7(12):1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- 21.Burén S, et al. Importance of post-translational modifications for functionality of a chloroplast-localized carbonic anhydrase (CAH1) in Arabidopsis thaliana. PLoS ONE. 2011;6(6):e21021. doi: 10.1371/journal.pone.0021021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faye L, Daniell H. Novel pathways for glycoprotein import into chloroplasts. Plant Biotechnol J. 2006;4(3):275–279. doi: 10.1111/j.1467-7652.2006.00188.x. [DOI] [PubMed] [Google Scholar]
- 23.Kitajima A, et al. The rice alpha-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell. 2009;21(9):2844–2858. doi: 10.1105/tpc.109.068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nanjo Y, et al. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-golgi to the chloroplast through the secretory pathway. Plant Cell. 2006;18(10):2582–2592. doi: 10.1105/tpc.105.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radhamony RN, Theg SM. Evidence for an ER to Golgi to chloroplast protein transport pathway. Trends Cell Biol. 2006;16(8):385–387. doi: 10.1016/j.tcb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Flores-Pérez U, Jarvis P. Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta. 2013;1833(2):332–340. doi: 10.1016/j.bbamcr.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Spork S, et al. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8(8):1134–1145. doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould SB, et al. Protein targeting into the complex plastid of cryptophytes. J Mol Evol. 2006;62(6):674–681. doi: 10.1007/s00239-005-0099-y. [DOI] [PubMed] [Google Scholar]
- 29. Moog D, Stork S, Zauner S, Maier UG (2011) In silico and in vivo investigations of proteins of a minimized eukaryotic cytoplasm. Genome Biol Evol 3:375–382. [DOI] [PMC free article] [PubMed]
- 30.Stork S, et al. Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation. Eukaryot Cell. 2012;11(12):1472–1481. doi: 10.1128/EC.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber T, Gruber A, Kroth PG. The presence and localization of thioredoxins in diatoms, unicellular algae of secondary endosymbiotic origin. Mol Plant. 2009;2(3):468–477. doi: 10.1093/mp/ssp010. [DOI] [PubMed] [Google Scholar]
- 32.Ruprecht M, et al. On the impact of precursor unfolding during protein import into chloroplasts. Mol Plant. 2010;3(3):499–508. doi: 10.1093/mp/ssp116. [DOI] [PubMed] [Google Scholar]
- 33. Gschloessl B, Guermeur Y, Cock JM (2008) HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinformatics 9:393. [DOI] [PMC free article] [PubMed]
- 34.Baïet B, et al. N-glycans of Phaeodactylum tricornutum diatom and functional characterization of its N-acetylglucosaminyltransferase I enzyme. J Biol Chem. 2011;286(8):6152–6164. doi: 10.1074/jbc.M110.175711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hampton RY, Sommer T. Finding the will and the way of ERAD substrate retrotranslocation. Curr Opin Cell Biol. 2012;24(4):460–466. doi: 10.1016/j.ceb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Smith MH, Ploegh HL, Weissman JS. Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wunder T, et al. (2007) The invariant phenylalanine of precursor proteins discloses the importance of Omp85 for protein translocation into cyanelles. BMC Evol Biol 7:236. [DOI] [PMC free article] [PubMed]
- 38.Hinnah SC, Wagner R, Sveshnikova N, Harrer R, Soll J. The chloroplast protein import channel Toc75: Pore properties and interaction with transit peptides. Biophys J. 2002;83(2):899–911. doi: 10.1016/S0006-3495(02)75216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heins L, et al. The preprotein conducting channel at the inner envelope membrane of plastids. EMBO J. 2002;21(11):2616–2625. doi: 10.1093/emboj/21.11.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kovacs-Bogdan E, Benz JP, Soll J, Bolter B (2011) Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol 11:133. [DOI] [PMC free article] [PubMed]
- 41.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3(6):313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 42.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291(5512):2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 43.Hames C, Halbedel S, Schilling O, Stülke J. Multiple-mutation reaction: A method for simultaneous introduction of multiple mutations into the glpK gene of Mycoplasma pneumoniae. Appl Environ Microbiol. 2005;71(7):4097–4100. doi: 10.1128/AEM.71.7.4097-4100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hempel F, Lau J, Klingl A, Maier UG. Algae as protein factories: Expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS ONE. 2011;6(12):e28424. doi: 10.1371/journal.pone.0028424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonzalez NH, et al. A single peroxisomal targeting signal mediates matrix protein import in diatoms. PLoS ONE. 2011;6(9):e25316. doi: 10.1371/journal.pone.0025316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rachel R, et al. (2010) Analysis of the ultrastructure of archaea by electron microscopy. Methods Cell Biol 96:47–69. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.