Abstract
Mucosal surfaces are a main entry point for pathogens and the principal sites of defense against infection. Both bacteria and phage are associated with this mucus. Here we show that phage-to-bacteria ratios were increased, relative to the adjacent environment, on all mucosal surfaces sampled, ranging from cnidarians to humans. In vitro studies of tissue culture cells with and without surface mucus demonstrated that this increase in phage abundance is mucus dependent and protects the underlying epithelium from bacterial infection. Enrichment of phage in mucus occurs via binding interactions between mucin glycoproteins and Ig-like protein domains exposed on phage capsids. In particular, phage Ig-like domains bind variable glycan residues that coat the mucin glycoprotein component of mucus. Metagenomic analysis found these Ig-like proteins present in the phages sampled from many environments, particularly from locations adjacent to mucosal surfaces. Based on these observations, we present the bacteriophage adherence to mucus model that provides a ubiquitous, but non–host-derived, immunity applicable to mucosal surfaces. The model suggests that metazoan mucosal surfaces and phage coevolve to maintain phage adherence. This benefits the metazoan host by limiting mucosal bacteria, and benefits the phage through more frequent interactions with bacterial hosts. The relationships shown here suggest a symbiotic relationship between phage and metazoan hosts that provides a previously unrecognized antimicrobial defense that actively protects mucosal surfaces.
Keywords: symbiosis, host-pathogen, virus, immunoglobulin, immune system
Mucosal surfaces are the primary zones where animals meet their environment, and thus also the main points of entry for pathogenic microorganisms. The mucus layer is heavily colonized by bacteria, including many symbionts that contribute additional genetic and metabolic potential to the host (1, 2). Bacterial symbionts associated with a variety of other host surfaces also provide goods and services, e.g., nutrients (3–6), bioluminescence (7, 8), and antibiotics (9, 10). These resident symbionts benefit from increased nutrient availability (5, 11–13), as well as the opportunity for both vertical transmission and increased dissemination (14–16).
Within the mucus, the predominant macromolecules are the large (up to 106–109 Da) mucin glycoproteins. The amino acid backbone of these proteins incorporates tandem repeats of exposed hydrophobic regions alternating with blocks bearing extensive O-linked glycosylation (17). Hundreds of variable, branched, negatively charged glycan chains extend 0.5–5 nm from the peptide core outward into the surrounding environment (17, 18). Other proteins, DNA, and cellular debris also are present. Continual secretion and shedding of mucins maintain a protective mucus layer from 10–700 μm thick depending on species and body location (19–22).
By offering both structure and nutrients, mucus layers commonly support higher bacterial concentrations than the surrounding environment (11, 23). Of necessity, hosts use a variety of mechanisms to limit microbial colonization (24–27). Secretions produced by the underlying epithelium influence the composition of this microbiota (12, 27, 28). When invaded by pathogens, the epithelium may respond by increased production of antimicrobial agents, hypersecretion of mucin, or alteration of mucin glycosylation patterns to subvert microbial attachment (29–31).
Also present in the mucus environment are bacteriophage (phage), the most common and diverse biological entities. As specific bacterial predators, they increase microbial diversity through Red Queen/kill-the-winner dynamics (32, 33). Many phages establish conditional symbiotic relationships with their bacterial hosts through lysogeny. As integrated prophages, they often express genes that increase host fitness or virulence (34–36) and protect their host from lysis by related phages. As free phage, they aid their host strain by killing related competing strains (37–39). Phages participate, along with their bacterial hosts, in tripartite symbioses with metazoans that affect metazoan fitness (40–43). However, no direct symbiotic interactions between phage and metazoans are known.
Recently, Minot et al. (44) showed that phages in the human gut encode a population of hypervariable proteins. For 29 hypervariable regions, evidence indicated that hypervariability was conferred by targeted mutagenesis through a reverse transcription mechanism (44, 45). Approximately half of these encoded proteins possessed the C-type lectin fold previously found in the major tropism determinant protein at the tip of the Bordetella phage BPP-1 tail fibers (46); six others contained Ig-like domains. These Ig-like proteins, similar to antibodies and T-cell receptors, can accommodate large sequence variation (>1013 potential alternatives) (47). Ig-like domains also are displayed in the structural proteins of many phage (48, 49). That most of these displayed Ig-like domains are dispensable for phage growth in the laboratory (45, 49) led to the hypothesis that they aid adsorption to their bacterial prey under environmental conditions (49). The possible role and function of these hypervariable proteins remain to be clarified.
Here, we show that phage adhere to mucus and that this association reduces microbial colonization and pathology. In vitro studies demonstrated that this adherence was mediated by the interaction between displayed Ig-like domains of phage capsid proteins and glycan residues, such as those in mucin glycoproteins. Homologs of these Ig-like domains are encoded by phages from many environments, particularly those adjacent to mucosal surfaces. We propose the bacteriophage adherence to mucus (BAM) model whereby phages provide a non–host-derived antimicrobial defense on the mucosal surfaces of diverse metazoan hosts.
Results
Phage Adhere to Mucus.
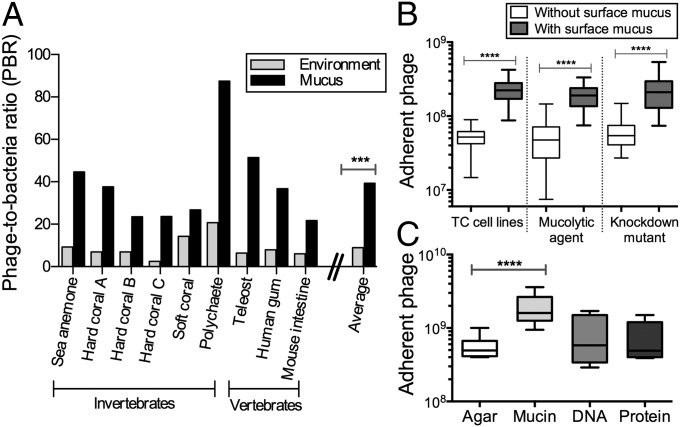
Our preliminary investigations of mucosal surfaces suggested that phage concentrations in the mucus layer were elevated compared with the surrounding environment. Here, we used epifluorescence microscopy to count the phage and bacteria in mucus sampled from a diverse range of mucosal surfaces (e.g., sea anemones, fish, human gum), and in each adjacent environment (SI Materials and Methods and Fig. S1). Comparing the calculated phage-to-bacteria ratios (PBRs) showed that PBRs in metazoan-associated mucus layers were on average 4.4-fold higher than those in the respective adjacent environment (Fig. 1A). The PBRs on these mucus surfaces ranged from 21:1 to 87:1 (average, 39:1), compared with 3:1 to 20:1 for the surrounding milieus (average, 9:1; n = 9, t = 4.719; ***P = 0.0002). Earlier investigations of phage abundance in marine environments reported that phage typically outnumber bacteria by an order of magnitude (50–52), but here we demonstrate that this margin was significantly larger in metazoan-associated mucus surface layers.
Fig. 1.
Phage adhere to cell-associated mucus layers and mucin glycoprotein. (A) PBR for diverse mucosal surfaces and the adjacent environment. On average, PBRs for mucosal surfaces were 4.4-fold greater than for the adjacent environment (n = 9, t = 4.719, ***P = 0.0002, unpaired t test). (B) Phage adherence to TC cell monolayers, with and without surface mucus (unpaired t tests). (Left) Non–mucus-producing Huh-7 liver hepatocyte cells and mucus-producing T84 colon epithelial cells (n > 18, t = 8.366, ****P < 0.0001). (Center) Mucus-producing A549 lung epithelial cells with and without treatment with NAC, a mucolytic agent (n > 40, t = 9.561, ****P < 0.0001). (Right) Mucus-producing shRNA control A549 cells (shControl) and mucus knockdown (MUC–) A549 cells (n > 37, t = 7.673, ****P < 0.0001). (C) Phage adherence to uncoated agar plates and agar coated with mucin, DNA, or protein (n = 12, t = 5.306, ****P < 0.0001, unpaired t test).
To determine whether this enrichment was dependent on the presence of mucus rather than some general properties of the cell surface (e.g., charge), phage adherence was tested with tissue culture (TC) cells with and without surface mucus (SI Materials and Methods). In these assays, T4 phage were washed across confluent cell monolayers for 30 min, after which nonadherent phage were removed by repeated washings and the adherent phage quantified by epifluorescence microscopy. Two mucus-producing cell lines were used: T84 (human colon epithelial cells) and A549 (human lung epithelial cells). For these cells, mucin secretion was stimulated by pretreatment with phorbol 12-myristate 13-acetate (53, 54). Comparison of the T84 cells with the non–mucus-producing Huh-7 human hepatocyte cell line showed that T4 phage adhered significantly more to the mucus-producing T84 cells (Fig. 1B; n > 18, t = 8.366; ****P < 0.0001). To demonstrate the mucus dependence of this adherence, the mucus layer was chemically removed from A549 cells by N-acetyl-l-cysteine (NAC) treatment (55) (Fig. S2). This significantly reduced the number of adherent phage to levels similar to those observed with non–mucus-producing cell lines (Fig. 1B; n > 40, t = 9.561; ****P < 0.0001). We also created an A549 shRNA mucus knockdown cell line (MUC–), reducing mucus production in A549, and a nonsense shRNA control (shControl; Figs. S3 and S4). Again, T4 phage adhered significantly more to the mucus-producing cells (Fig. 1B; n > 37, t = 7.673; ****P < 0.0001).
Although mucin glycoproteins are the predominant component of mucus, other macromolecular components also are present, any of which might be involved in the observed phage adherence. We developed a modified top agar assay to determine whether phage adhered to a specific macromolecular component of mucus. Plain agar plates and agar plates coated with 1% (wt/vol) mucin, DNA, or protein were prepared. That concentration was chosen because it is at the low end of the range of physiological mucin concentrations (56). T4 phage suspensions were incubated on the plates for 30 min, after which the phage suspension was decanted to remove unbound particles. The plates then were overlaid with a top agar containing Escherichia coli hosts and incubated overnight. The number of adherent phage was calculated from the number of plaque-forming units (pfu) observed. Significantly more T4 phage adhered to the 1% mucin-coated agar surface (Fig. 1C; n = 12, t = 5.306; ****P < 0.0001). Combined, these three assays show that phage adhere to mucin glycoproteins.
Phage Adherence and Bacterial Infection.
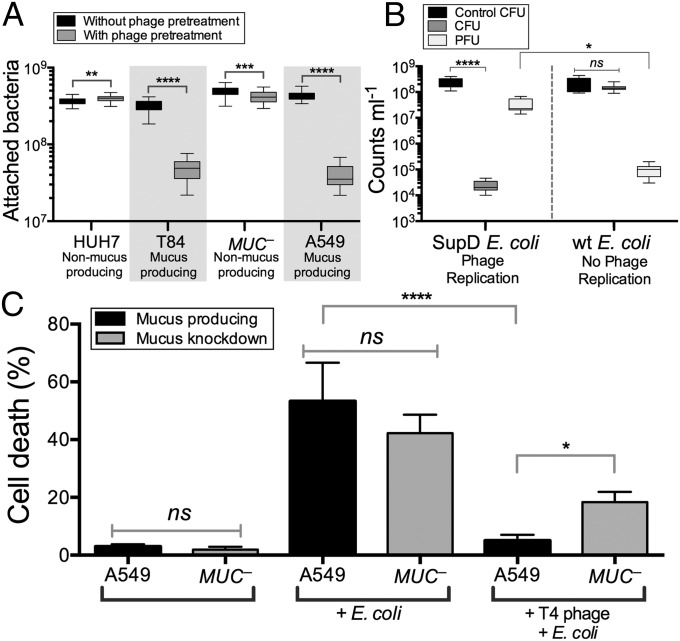
The mucus layer is an optimal environment for microbial growth, providing structure as well as nutrients in the form of diverse, mucin-associated glycans. To limit this growth, the metazoan host retards microbial colonization by diverse antimicrobial mechanisms (24–27). Does the increased number of adherent phage found on mucosal surfaces also reduce microbial colonization? To answer this, bacterial attachment to mucus-producing and non–mucus-producing TC cells was assayed both with and without pretreatment of the cells with the mucus-adherent phage T4. Here confluent monolayers of TC cells were overlaid with T4 phage for 30 min, washed to remove nonadherent phage, and then incubated with E. coli for 4 h. Cells then were scraped from the plates and the attached bacteria were fluorescently stained and counted by epifluorescence microscopy. Phage pretreatment of mucus-producing TC cell lines (T84, A549) significantly decreased subsequent bacterial attachment (Fig. 2A; T84: n > 30, t = 32.05, ****P < 0.0001; A549: n > 30, t = 36.85, ****P < 0.0001). Phage pretreatment of non–mucus-producing cells (Huh-7; MUC–, an A549 mucus knockdown strain) had a much smaller effect on bacterial attachment.
Fig. 2.
Effect of phage adsorption on subsequent bacterial infection of epithelial cells. (A) Bacterial attachment to mucus-producing (T84 and A549) and non–mucus-producing (Huh-7, MUC–) TC cells, with and without phage pretreatment. T4 phage pretreatment significantly decreased subsequent bacterial adherence to mucus-producing TC cell lines (T84: n > 30, t = 32.05, ****P < 0.0001; A549: n > 30, t = 36.85, ****P < 0.0001; unpaired t tests). Less dramatic shifts were seen for non–mucus-producing cells (Huh-7: n > 30, t = 2.72, **P = 0.0098; MUC–: n > 30, t = 3.52, ***P = 0.0007; unpaired t tests). (B) Mucus-producing A549 cells were pretreated with T4 am43–44– phage (Materials and Methods) and then incubated for 4 h with either wild-type (wt) or amber-suppressor (SupD) E. coli. Phage replication in the SupD E. coli strain significantly reduced bacterial colony-forming units (CFU) in the mucus (n = 8, ****P < 0.0001, Tukey’s two-way ANOVA) and increased phage-forming units (PFU) relative to the no-phage replication wt E. coli (n = 8, *P = 0.0227). (C) Mortality of mucus-producing (A549) and mucus knockdown (MUC–) A549 lung epithelial cells following overnight incubation with E. coli. Phage pretreatment completely protected mucus-producing A549 cells from bacterial challenge (n = 12, ****P < 0.0001, Tukey’s one-way ANOVA); protection of MUC– cells was 3.1-fold less (n = 12, *P = 0.0181). ns, not significant.
To determine whether this reduced bacterial attachment depended on bacterial lysis and the production of progeny viruses, we repeated these experiments using an amber mutant T4 phage (T4 am43–44–). When infecting wild-type E. coli, this phage produces no infective progeny virions, but infection of the E. coli amber suppressor strain SupD yields infective virions. For these experiments, mucus-producing A549 cells were pretreated with amber mutant T4 am43–44– phage and then incubated with either wild-type or the amber suppressor strain E. coli (Fig. 2B). Bacterial attachment was reduced by more than four orders of magnitude when phage could replicate and thereby increase the number of infective virions within the mucus (n = 8, ****P < 0.0001). Comparatively, when no phage replication occurred in the mucus, there was no observable change in bacterial colonization and fewer phages were detected (n = 8, *P = 0.0227). These results show that pretreatment of a mucosal surface with phage reduces adherence of a bacterial pathogen and that this protection is mediated by continued phage replication in the mucus.
To test whether the observed reduced bacterial adherence was accompanied by reduced pathology of the underlying TC cells, mucus-producing A549 and non–mucus-producing MUC– TC cells were exposed to E. coli overnight, either with or without a 30-min pretreatment with T4 phage. Infection was quantified as the percentage of cell death. Adherence of phage effectively protected the mucus-producing cells against the subsequent bacterial challenge (Fig. 2C; n = 12, ****P < 0.0001). Phage pretreatment showed a reduced protection to the non–mucus-producing MUC– cells, decreasing cell death only twofold. Evaluating the importance of mucus production for effective protection, we found that phage pretreatment of mucus-producing A549 cells resulted in a 3.6-fold greater reduction in cell death (n = 12, *P = 0.0181) than the same pretreatment of the mucin knockdown MUC– cells.
Role of Capsid Ig-Like Domains in Phage Adherence.
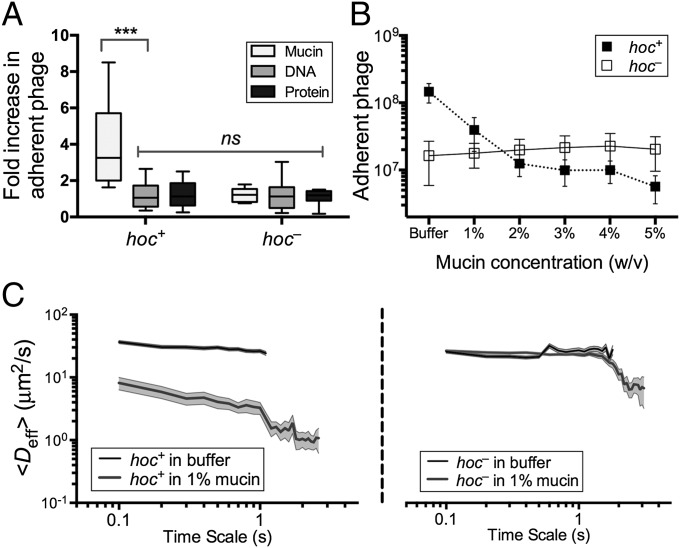
Minot et al. (44) recently reported that phage communities associated with the human gut encode a diverse array of hypervariable proteins, including some with hypervariable Ig-like domains. Four Ig-like domains are found in highly antigenic outer capsid protein (Hoc), a T4 phage structural protein of which 155 copies are displayed on the capsid surface (57, 58). Based on this, and given that most Ig-like domains function in recognition and adhesion processes, we hypothesized that the T4 Hoc protein might mediate the adherence of T4 phage to mucus. To test this, we performed three experiments. First, we compared the adherence of hoc+ T4 phage and a hoc– mutant to mucin-, DNA-, and protein-coated agar plates to an uncoated agar control using the modified top agar assay (see above). Relative to plain agar, the adherence of hoc+ T4 phage increased 4.1-fold for mucin-coated agar (n > 11, t = 3.977, ***P = 0.0007), whereas adherence increased only slightly for agar coated with DNA (1.1-fold) or protein (1.2-fold; Fig. 3A). Unlike the hoc+ T4 phage, the hoc– phage did not adhere preferentially to the mucin-coated agar, but instead showed 1.2-, 1.2-, and 1.1-fold increased adherence for mucin, DNA, and protein coatings, respectively. To ensure that none of the macromolecules directly affected phage infectivity, hoc+ and hoc– T4 phage were incubated in 1% (wt/vol) solutions of mucin, DNA, or protein. Phage suspensions were combined with E. coli top agar as described above and layered over uncoated agar plates. The results confirmed that the macromolecules did not alter phage infectivity (Fig. S5). To provide further evidence that the mucin adherence was dependent on the capsid displayed Ig-like domains rather than some other property of T4 phage, we repeated the modified top agar assay using Ig+ and Ig– T3 phage. As with T4, the Ig-like domains of T3 are displayed on the surface of the major capsid protein (49). Results indicated a similar increase in adherence to mucin for the Ig+, but not the Ig–, T3 phage (Fig. S6). Thus, adherence of these phage to mucus requires the Ig-like protein domains.
Fig. 3.
Effect of Hoc protein on phage–mucin interactions. (A) Adherence of hoc+ and hoc– T4 phage to agar coated with mucin, DNA, or protein reported as an increase relative to plain agar controls (n > 11, t = 3.977, ***P = 0.0007, unpaired t test). (B) Competitive effect of mucin on phage adherence when hoc+ and hoc– T4 phage in 0–5% (wt/vol) mucin solution (1× PBS) were washed over mucus-producing A549 cells (n = 25 per sample). (C) Diffusion of fluorescence-labeled hoc+ (Left) and hoc– (Right) T4 phage in buffer and 1% mucin as determined by MPT. Mucin hindered diffusion of hoc+ T4 phage but not hoc– phage (10 analyses per sample, trajectories of n > 100 particles for each analysis; error bars represent SE).
Second, a competition assay using hoc+ and hoc− T4 phage and mucus-producing TC cells was performed to demonstrate the role of mucin in phage adherence. Phage suspended in mucin solutions ranging from 0% to 5% (wt/vol) were washed over confluent layers of mucus-producing A549 TC cells; phage adherence then was assayed as above. Adherence of hoc+ T4 phage, but not of hoc– T4 phage, was reduced by mucin competition in a concentration-dependent manner (Fig. 3B).
Third, interaction of the Hoc protein domains displayed on the capsid surface with mucin glycoproteins was hypothesized to affect the rate of diffusion of T4 virions in mucus. To evaluate this, we used multiple-particle tracking (MPT) to quantify transport rates of phage particles in buffer and in mucin suspensions. The ensemble average effective diffusivity (Deff) calculated at a time scale of 1 s for both hoc+ and hoc– T4 phage in buffer was compared against that in 1% (wt/vol) mucin suspensions (SI Materials and Methods). Both hoc+ and hoc– phage diffuse rapidly through buffer (Fig. 3C). Whereas hoc– phage diffused in 1% mucin at the same rate as in buffer, the mucin decreased the diffusion rate for hoc+ phage particles eightfold. Thus, all three of these experimental approaches supported our hypothesis that the Hoc proteins displayed on the T4 phage capsid interact with mucin.
Phage Capsid Ig-Like Domains Interact with Glycans.
It is known that ∼25% of sequenced tailed dsDNA phages (Caudovirales) encode structural proteins with predicted Ig-like domains (48). A search of publicly available viral metagenomes for homologs of the Ig-like domains of the T4 Hoc protein yielded numerous viral Ig-like domains from a variety of environments (Fig. 4A). These domains were more likely to be found in samples collected directly from mucus (e.g., sputum samples) or from an environment adjacent to a mucosal surface (e.g., intestinal lumen, oral cavity). All homologs displayed high structural homology (Phyre2 confidence score average, 96 ± 5%) with a plant-sugar binding domain known for its promiscuous carbohydrate binding specificity (SI Materials and Methods and Table S1), suggesting an interaction between these Ig-like domains and glycans.
Fig. 4.
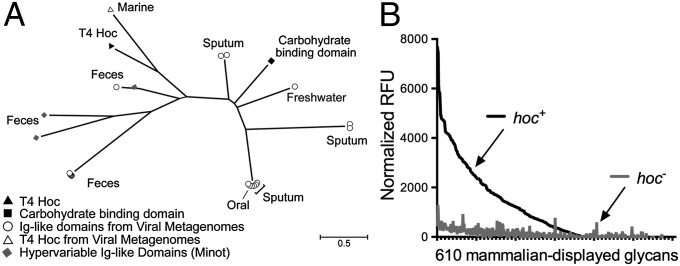
Hoc-mediated glycan binding and Hoc-related phylogeny. (A) Phylogenetic tree of sequences from viral metagenomes with high-sequence homology to Ig-like domains. Many of the identified homologs are from mucus-associated environments (e.g., human feces, sputum). Also included are the Hoc protein of T4 phage and the hypervariable Ig-like domains previously obtained by deep sequencing of phage DNA from the human gut (44). The scale bar represents an estimated 0.5 amino acid substitutions per site. See SI Materials and Methods for methods. (B) Binding of fluorescence-stained hoc+ and hoc– T4 phage to a microarray of 610 mammalian glycans. Normalized relative fluorescence units (RFU) were calculated from mean fluorescence minus background binding.
Mucins are complex glycoproteins with highly variable glycan groups exposed to the environment. To investigate whether Hoc interacts with glycans and, if so, to determine whether it interacts with a specific glycan or with a diverse array of glycans, we assayed phage adherence to microarrays printed with 610 mammalian glycans. The hoc+ T4 phage adhered to many diverse glycans and showed a preference for the O-linked glycan residues typically found in mucin glycoproteins (Fig. 4B, SI Materials and Methods, and Table S2). The hoc– T4 phage exhibited significantly lower affinity for all tested glycans. This indicates that Hoc mediates interactions between T4 phage and varied glycan residues.
Discussion
In diverse metazoans, body surfaces that interact with the environment are covered by a protective layer of mucus. Because these mucus layers provide favorable habitats for bacteria, they serve as the point of entry for many pathogens and support large populations of microbial symbionts. Also present are diverse phages that prey on specific bacterial hosts. Moreover, phage concentrations in mucus are elevated relative to the surrounding environment (an average 4.4-fold increase for a diverse sample of invertebrate and vertebrate metazoans; Fig. 1A). The increased concentration of lytic phage on mucosal surfaces provides a previously unrecognized metazoan immune defense affected by phage lysis of incoming bacteria.
Working with a model system using T4 phage and various TC cell lines, we demonstrated that the increased concentration of phage on mucosal surfaces is mediated by weak binding interactions between the variable Ig-like domains on the T4 phage capsid and mucin-displayed glycans. The Ig protein fold is well known for its varied but essential roles in the vertebrate immune response and cell adhesion. Ig-like domains also are present in approximately one quarter of the sequenced genomes of tailed DNA phages, the Caudovirales (48). Notably, these domains were found only in virion structural proteins and typically are displayed on the virion surface. Thus, they were postulated to bind to bacterial surface carbohydrates during infection (48, 49). However, mucin glycoproteins, the predominant macromolecular constituent of mucus, display hundreds of variable glycan chains to the environment that offer potential sites for binding by phage Ig-like proteins. Furthermore, we speculate that phage use the variability of the Ig-like protein scaffold (supporting >1013 potential alternatives) to adapt to the host’s ever-changing patterns of mucin glycosylation.
The presence of an Ig-like protein (Hoc) displayed on the capsid of T4 phage significantly slowed the diffusion of the phage in mucin solutions. In vivo, similar phage binding to mucin glycans would increase phage residence time in mucus layers. Because bacterial concentrations typically are enriched in mucus (Fig. S1), we predict that mucus-adherent phage are more likely to encounter bacteria, potentially increasing their replicative success. If so, phage Ig-like domains that bind effectively to the mucus layer would be under positive selection. Likely, Hoc and other phage proteins with Ig-like domains interact with other glycans with different ramifications, as well (49, 58).
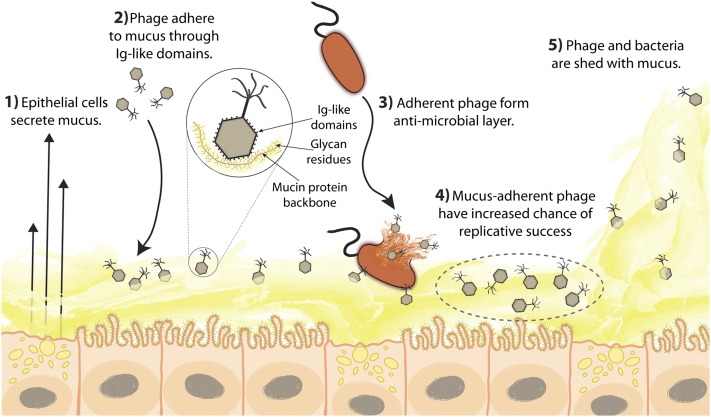
Previous metagenomic studies documented the ubiquity and diversity of bacteria and phage within mucus-associated environments (e.g., human gut, human respiratory tract, corals) (52, 59–64). Known also were some of the essential but adaptable services provided by symbiotic bacteria in these environments (65). However, only recently have efforts been made to investigate the dynamic influences of phage within host-associated ecosystems (37, 44, 66). In this work, we used an in vitro model system to demonstrate a mechanism of phage adherence to the mucus layers that shield metazoan cells from the environment. Furthermore, adherent phage protected the underlying epithelial cells from bacterial infection. Based on these observations and previous research, we proposed the BAM model of immunity, in which the adherence of phage to mucosal surfaces yields a non–host-derived, antimicrobial defense. According to this model (summarized in Fig. 5), the mucus layer, already considered part of the innate immune system and known to provide physical and biochemical antimicrobial defenses (18, 27, 67), also accumulates phage.
Fig. 5.
The BAM model. (1) Mucus is produced and secreted by the underlying epithelium. (2) Phage bind variable glycan residues displayed on mucin glycoproteins via variable capsid proteins (e.g., Ig-like domains). (3) Phage adherence creates an antimicrobial layer that reduces bacterial attachment to and colonization of the mucus, which in turn lessens epithelial cell death. (4) Mucus-adherent phage are more likely to encounter bacterial hosts, thus are under positive selection for capsid proteins that enable them to remain in the mucus layer. (5) Continual sloughing of the outer mucus provides a dynamic mucosal environment.
The model system we used involved a single lytic phage and host bacterium; the situation in vivo undoubtedly is more complex. Within the mucosal layer reside diverse bacterial lineages and predictably an even greater diversity of phage strains, both enmeshed within complex phage–bacterial infection networks and engaged in a dynamic arms race (68, 69). These and other factors lower the probability that any given phage–bacterium encounter will result in a successful infection. The time dimension adds further complexity. The mucus layer is dynamic. Mucins are secreted continually by the underlying epithelium while mucus is sloughed continually from the outer surface. As a result, there is an ongoing turnover of both the bacterial and phage populations in the mucus layer. Driven by kill-the-winner dynamics, the population of phage types that can infect the dominant bacterial types present will cycle along with the populations of their hosts. Through such mechanisms, we envision that adherent lytic phages provide a dynamic and adaptable defense for their metazoan hosts—a unique example of a metazoan–phage symbiosis.
We posit that BAM immunity reduces bacterial pathogenesis and provides a previously unrecognized, mucosal immunity. This has far-reaching implications for numerous fields, such as human immunity, gastroenterology, coral disease, and phage therapy. Meanwhile, key questions remain. For instance, what role do temperate phages play in the dynamics of BAM immunity? When integrated into the bacterial chromosome as prophages, they protect their bacterial hosts from infection by related phages; as free phages, they infect and kill sensitive related bacterial strains that compete with their bacterial hosts (37–39). Both mechanisms may benefit their metazoan host by contributing to the maintenance of a selected commensal mucosal microbiota. These possibilities remain to be investigated. Likewise, in vivo investigations are needed to characterize the bacterial and phage diversity present and the consequent effects on BAM immunity. As of now, the relationships shown here open an arena for immunological study, introduce a phage–metazoan symbiosis, and recognize the key role of the world’s most abundant biological entities in the metazoan immune system.
Materials and Methods
Bacterial Strains, Phage Stocks, TC Cell Lines, and Growth Conditions.
E. coli 1024 strain was used for all E. coli experiments and was grown in LB (10 g tryptone, 5 g yeast extract, 10 g NaCl, in 1 L dH2O) at 37 °C overnight. E. coli amber-suppressor strain SupD strain CR63 was used as a host for amber mutant phage and grown as above. Bacteriophage T4 was used at ∼109 pfu⋅mL–1. Hoc– T4 phage were kindly supplied by Prof. Venigalla Rao (58), The Catholic University of America, Washington, D.C. T3 am10 Ig– amber mutant phage were kindly supplied by Prof. Ian J. Molineux (70), University of Texas, Austin, TX. T4 replication-negative 43– (amE4332: DNA polymerase) 44– (amN82: subunit of polymerase clamp holder) amber mutant phage were kindly supplied by Prof. Kenneth Kreuzer (71), Duke University School of Medicine, Durham, NC. The human tumorigenic colon epithelial cell line, T84, was obtained from the American Type Culture Collection (ATCC) and cultured in DMEM/F12-K media with 5% FBS and 100 μg⋅mL–1 penicillin–streptomycin (PS). The human tumorigenic lung epithelial cell line A549 was kindly supplied by Prof. Kelly Doran, San Diego State University, San Diego, CA and cultured in F12-K media with 10% FBS, 100 μg⋅mL–1 PS. The human tumorigenic liver epithelial cell line Huh7 was kindly supplied by Prof. Roland Wolkowicz, San Diego State University, San Diego, CA, and cultured in F12-K media with 10% FBS, 100 μg⋅mL–1 PS. All TC cell lines initially were grown in 50 mL Primaria Tissue Culture Flasks (Becton Dickinson) at 37 °C and 5% CO2.
Phage Adherence to Mucus-Associated Macromolecules.
LB agar plates were coated with 1 mL of 1% (wt/vol) of one of the following in 1× PBS: type III porcine stomach mucin, DNA from salmon testes, or BSA (all three from Sigma–Aldrich) and then allowed to dry. Stocks of hoc+ and hoc– T4 phage (109 pfu⋅mL−1) were serially diluted to 1 × 10−7 and 1 × 10−8 per milliliter in LB, and a 5-mL aliquot of each dilution was washed across the plates for 30 min at 37 °C on an orbital shaker. After the phage suspensions were decanted from the plates, the plates were shaken twice to remove excess liquid and dried. Each plate then was layered with 1 mL of overnight E. coli culture (109 mL–1) in 3 mL of molten top agar and incubated overnight at 37 °C. The number of adherent phage was calculated from the number of plaque-forming units observed multiplied by the initial phage dilution. To determine whether mucus macromolecules directly affected phage infectivity, hoc+ and hoc– T4 phage (109 pfu⋅mL−1) were serially diluted as described above into 1 mL LB solutions containing 1% (wt/vol) mucin, DNA, or BSA. After incubation for 30 min at 37 °C, the phage suspensions were combined with E. coli top agar as described above and layered over uncoated agar plates (Fig. S5).
Phage Treatment of TC Cells.
TC cells were washed twice with 5 mL of serum-free media to remove residual antibiotics, layered with 2 mL of serum-free media containing T4 phage (107 or 109 mL–1), and incubated at 37 °C and 5% CO2 for 30 min. Cells then were washed five times with 5 mL of serum-free media to remove nonadherent phage.
Phage Adherence to TC Cells.
TC cells were treated with phage (109 mL–1; see above), then scraped from plates using Corning Cell Scrapers (Sigma–Aldrich). Adherent phage were counted by epifluorescence microscopy as described above.
Bacterial Adherence to TC Cells With/Without Phage Pretreatment.
TC cells with or without pretreatment with T4 phage (107 mL–1) were layered with 2 mL serum-free media containing E. coli (107 mL–1), incubated at 37 °C and 5% CO2 for 4 h, and then washed five times with 5 mL serum-free medium to remove nonadherent phage and bacteria. Cells were scraped from plates, and adherent phage and bacteria were counted by either epifluorescence microscopy, as described above, or colony-forming and plaque-forming units. Then, 100 μL of a relevant dilution was spread onto an agar plate and incubated overnight at 37 °C, and the number of adherent bacteria was calculated from the colony-forming units observed multiplied by the initial dilution. Plaque-forming units were counted by a top agar assay as described above.
TC Cell Death from Bacterial Infection.
Mucus-producing A549 and MUC– A549 TC cells were grown to confluence. T4 phage were cleaned using Amicon 50-kDa centrifugal filters (Millipore) and saline magnesium buffer (SM) to remove bacterial lysis products. Cells, with or without T4 phage pretreatment (107 mL–1), were incubated with E. coli (107 mL–1) overnight. Afterward, TC cells were recovered from the plates by trypsin/EDTA solution (Invitrogen). Cells were pelleted by centrifugation and resuspended in 1× PBS. Dead cells were identified by staining with 1 mg/mL of propidium iodide (Invitrogen). Samples then were analyzed on a FACSCanto II flow cytometer (BD Biosciences) with excitation at 488 nm and emission detected through a 670 long pass filter. The forward scatter threshold was set at 5,000, and a total of 10,000 events were collected for each sample.
Mucin Competition Assay.
Mucus-producing A549 TC cells were grown to confluence. Hoc+ and hoc– T4 phage (109 mL–1) were diluted into mucin solutions ranging between 0% and 5% (wt/vol) in 1× PBS then washed over TC cells for 30 min at 37 °C and 5% CO2. Cells were washed five times with 5 mL serum-free media to remove nonadherent phage, scraped from plates, and adherent phage were quantified as described above.
Graphing and Statistics.
Graphing and statistical analyses were performed using GraphPad Prism 6 (GraphPad Software). All error bars represent 5–95% confidence intervals. The midline represents the median and the mean for box plots and bar plots, respectively.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants R01: GM095384, GM073898, and R21: AI094534 from the National Institute of General Medical Sciences. The authors thank the Protein-Glycan Interaction Resource at Emory University School of Medicine, Atlanta, GA (funded by NIH Grant GM98791), for support of the glycan microarray analyses. The authors acknowledge the San Diego State University (SDSU) Flow Cytometry Core Facility and the SDSU Electron Microscopy Facility for assistance with sample analysis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 10475.
Data deposition: Raw glycan array data are available from the Consortium for Functional Glycomics (accession no. 2621).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305923110/-/DCSupplemental.
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clay K, Holah J. Fungal endophyte symbiosis and plant diversity in successional fields. Science. 1999;285(5434):1742–1745. doi: 10.1126/science.285.5434.1742. [DOI] [PubMed] [Google Scholar]
- 4.Douglas AE. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 1989;64(4):409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 5.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22(1):283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 6.Hosokawa T, Koga R, Kikuchi Y, Meng X-Y, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci USA. 2010;107(2):769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nyholm SV, McFall-Ngai MJ. The winnowing: Establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2(8):632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 8.Ruby EG. Lessons from a cooperative, bacterial-animal association: The Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50(1):591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 9.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398(6729):701–704. [Google Scholar]
- 10.Kaltenpoth M, Göttler W, Herzner G, Strohm E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 2005;15(5):475–479. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 11.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 13.Berry D, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci USA. 2013;110(12):4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: Microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53(1):71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 16.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cone RA. Barrier properties of mucus. Adv Drug Deliv Rev. 2009;61(2):75–85. doi: 10.1016/j.addr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1(3):183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clunes MT, Boucher RC. Cystic fibrosis: The mechanisms of pathogenesis of an inherited lung disorder. Drug Discov Today Dis Mech. 2007;4(2):63–72. doi: 10.1016/j.ddmec.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strugala V, Allen A, Dettmar PW, Pearson JP. Colonic mucin: Methods of measuring mucus thickness. Proc Nutr Soc. 2003;62(1):237–243. doi: 10.1079/pns2002205. [DOI] [PubMed] [Google Scholar]
- 21.Garren M, Azam F. Corals shed bacteria as a potential mechanism of resilience to organic matter enrichment. ISME J. 2012;6(6):1159–1165. doi: 10.1038/ismej.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Button B, et al. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 2012;337(6097):937–941. doi: 10.1126/science.1223012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poulsen LK, et al. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun. 1994;62(11):5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phalipon A, et al. Secretory component: A new role in secretory IgA-mediated immune exclusion in vivo. Immunity. 2002;17(1):107–115. doi: 10.1016/s1074-7613(02)00341-2. [DOI] [PubMed] [Google Scholar]
- 25.Raj PA, Dentino AR. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol Lett. 2002;206(1):9–18. doi: 10.1111/j.1574-6968.2002.tb10979.x. [DOI] [PubMed] [Google Scholar]
- 26.Vaishnava S, et al. The antibacterial lectin RegIII{gamma} promotes the spatial segregation of microbiota and host in the intestine. Sci STKE. 2011;334(6053):255. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10(11):e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96(17):9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerken TA. Kinetic modeling confirms the biosynthesis of mucin core 1 (β-Gal(1-3) α-GalNAc-O-Ser/Thr) O-glycan structures are modulated by neighboring glycosylation effects. Biochemistry. 2004;43(14):4137–4142. doi: 10.1021/bi036306a. [DOI] [PubMed] [Google Scholar]
- 30.Jentoft N. Why are proteins O-glycosylated? Trends Biochem Sci. 1990;15(8):291–294. doi: 10.1016/0968-0004(90)90014-3. [DOI] [PubMed] [Google Scholar]
- 31.Schulz BL, et al. Glycosylation of sputum mucins is altered in cystic fibrosis patients. Glycobiology. 2007;17(7):698–712. doi: 10.1093/glycob/cwm036. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Brito B, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4(6):739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- 33.Thingstad T, Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol. 1997;13:19–27. [Google Scholar]
- 34.Groisman EA, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12(10):3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansen BK, Wasteson Y, Granum PE, Brynestad S. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology. 2001;147(Pt 7):1929–1936. doi: 10.1099/00221287-147-7-1929. [DOI] [PubMed] [Google Scholar]
- 36.Willner D, et al. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci USA. 2012;109(43):17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuse K, et al. Bacteriophage distribution in human faeces: Continuous survey of healthy subjects and patients with internal and leukaemic diseases. J Gen Virol. 1983;64(Pt 9):2039–2043. doi: 10.1099/0022-1317-64-9-2039. [DOI] [PubMed] [Google Scholar]
- 39.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28(2):127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. The players in a mutualistic symbiosis: Insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci USA. 2005;102(47):16919–16926. doi: 10.1073/pnas.0507029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325(5943):992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roossinck MJ. The good viruses: Viral mutualistic symbioses. Nat Rev Microbiol. 2011;9(2):99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 44.Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proc Natl Acad Sci USA. 2012;109(10):3962–3966. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMahon SA, et al. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 2005;12(10):886–892. doi: 10.1038/nsmb992. [DOI] [PubMed] [Google Scholar]
- 46.Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10(4):388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halaby DM, Mornon JPE. The immunoglobulin superfamily: an insight on its tissular, species, and functional diversity. J Mol Evol. 1998;46(4):389–400. doi: 10.1007/pl00006318. [DOI] [PubMed] [Google Scholar]
- 48.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J Mol Biol. 2006;359(2):496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 49.Fraser JS, Maxwell KL, Davidson AR. Immunoglobulin-like domains on bacteriophage: Weapons of modest damage? Curr Opin Microbiol. 2007;10(4):382–387. doi: 10.1016/j.mib.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 50.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399(6736):541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 51.Danovaro R, Serresi M. Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl Environ Microbiol. 2000;66(5):1857–1861. doi: 10.1128/aem.66.5.1857-1861.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breitbart M, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99(22):14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong DH, Petrovics G, Anderson WB, Forstner J, Forstner G. Induction of mucin gene expression in human colonic cell lines by PMA is dependent on PKC-ε. Am J Physiol. 1999;277(5 Pt 1):G1041–G1047. doi: 10.1152/ajpgi.1999.277.5.G1041. [DOI] [PubMed] [Google Scholar]
- 54.Forstner G, Zhang Y, McCool D, Forstner J. Mucin secretion by T84 cells: Stimulation by PKC, Ca2+, and a protein kinase activated by Ca2+ ionophore. Am J Physiol. 1993;264(6 Pt 1):G1096–G1102. doi: 10.1152/ajpgi.1993.264.6.G1096. [DOI] [PubMed] [Google Scholar]
- 55.Alemka A, et al. Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect Immun. 2010;78(6):2812–2822. doi: 10.1128/IAI.01249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lieleg O, Vladescu I, Ribbeck K. Characterization of particle translocation through mucin hydrogels. Biophys J. 2010;98(9):1782–1789. doi: 10.1016/j.bpj.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sathaliyawala T, et al. Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol Microbiol. 2010;77(2):444–455. doi: 10.1111/j.1365-2958.2010.07219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fokine A, et al. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J Virol. 2011;85(16):8141–8148. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marhaver KL, Edwards RA, Rohwer F. Viral communities associated with healthy and bleaching corals. Environ Microbiol. 2008;10(9):2277–2286. doi: 10.1111/j.1462-2920.2008.01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willner D, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. 2009;4(10):e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9(11):2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 63.Willner D, et al. Case studies of the spatial heterogeneity of DNA viruses in the cystic fibrosis lung. Am J Respir Cell Mol Biol. 2012;46(2):127–131. doi: 10.1165/rcmb.2011-0253OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minot S, et al. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res. 2011;21(10):1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lieleg O, Lieleg C, Bloom J, Buck CB, Ribbeck K. Mucin biopolymers as broad-spectrum antiviral agents. Biomacromolecules. 2012;13(6):1724–1732. doi: 10.1021/bm3001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 69.Weitz JS, et al. Phage-bacteria infection networks. Trends Microbiol. 2013;21(2):82–91. doi: 10.1016/j.tim.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 70.Condreay JP, Wright SE, Molineux IJ. Nucleotide sequence and complementation studies of the gene 10 region of bacteriophage T3. J Mol Biol. 1989;207(3):555–561. doi: 10.1016/0022-2836(89)90464-6. [DOI] [PubMed] [Google Scholar]
- 71.Benson KH, Kreuzer KN. Plasmid models for bacteriophage T4 DNA replication: Requirements for fork proteins. J Virol. 1992;66(12):6960–6968. doi: 10.1128/jvi.66.12.6960-6968.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.