Significance
The sequential generation of different types of neurons and glia is a fundamental property of neurogenesis, but little is known about the mechanisms controlling this phenomenon. Conditional deletion of Dicer prevents progenitors from progressing in their competence to generate late cell types, indefinitely generating early cell types. We now elucidate the molecular mechanism for this phenomenon. Three microRNAs, let-7, microRNA-125, and microRNA-9, serve as key regulators of the early to late developmental transition in retinal progenitors. These results show how progenitor temporal identity is controlled, a finding that will impact efforts to generate specific neural types from pluripotent stem cells.
Keywords: heterochronic, progenitor competence
Abstract
Most regions of the vertebrate central nervous system develop by the sequential addition of different classes of neurons and glia. This phenomenon has been best characterized in laminated structures like the retina and the cerebral cortex, in which the progenitor cells in these structures are thought to change in their competence as development proceeds to generate different types of neurons in a stereotypic sequence that is conserved across vertebrates. We previously reported that conditional deletion of Dicer prevents the change in competence of progenitors to generate later-born cell types, suggesting that specific microRNAs (miRNAs) are required for this developmental transition. In this report, we now show that three miRNAs, let-7, miR-125, and miR-9, are key regulators of the early to late developmental transition in retinal progenitors: (i) members of these three miRNA families increase over the relevant developmental period in normal retinal progenitors; (ii) inhibiting the function of these miRNAs produces changes in retinal development similar to Dicer CKO; (iii) overexpression of members of these three miRNA families in Dicer-CKO retinas can rescue the phenotype, allowing their progression to late progenitors; (iv) overexpression of these miRNAs can accelerate normal retinal development; (v) microarray and computational analyses of Dicer-CKO retinal cells identified two potential targets of the late-progenitor miRNAs: Protogenin (Prtg) and Lin28b; and (vi) overexpression of either Lin28 or Prtg can maintain the early progenitor state. Together, these data demonstrate that a conserved miRNA pathway controls a key step in the progression of temporal identity in retinal progenitors.
Complex neural structures, like the retina and the cerebral cortex, develop by the sequential addition of different classes of neurons and glia. In the cerebral cortex, for example, the deeper cortical layers are generated early in development, whereas the cells in the upper cortical layers are generated at progressively later stages. A similar phenomenon was first shown to occur in the developing retina from the birthdating studies of Sidman (1). In all vertebrates, the seven basic types of retinal cells are produced in a conserved sequence: ganglion cells, cones, and horizontal cells are generated early whereas the majority of the amacrine cells, rod photoreceptors, bipolar cells, and Müller glial cells are generated later (Fig. 1A) (2). One of the first steps in this developmental progression is the shift from progenitors that generate retinal ganglion cells to those that do not. In the mouse, this transition occurs with the onset of Ascl1; the earliest retinal progenitors do not express Ascl1, and, with its onset, the progenitors no longer produce ganglion cells (3). The other early-generated cell types, the cones and horizontal cells, are included in the Ascl1+ progenitor lineage, but are overrepresented in clones from Olig2+ progenitors (4). The rods and amacrine cells also are overrepresented in the postnatal Olig2+ progenitors whereas another late progenitor marker, Sox9, is necessary for the generation of Müller glia. Although, as the diagram shows, there is an overall bias in the progression of the different cell types during retinal neurogenesis, there is also considerable overlap in the birthdates of these different cell types and a distinct central-to-peripheral developmental gradient in the retina (1).
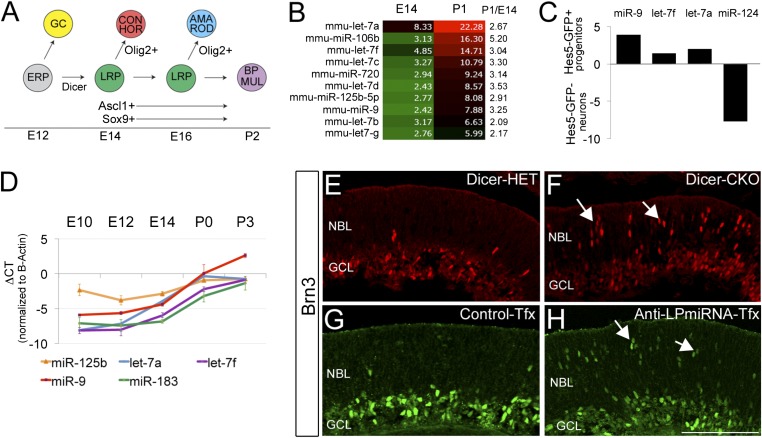
Fig. 1.
miR-125, let-7, and miR-9 are late-progenitor miRNAs. (A) Schematic representation of retinal development. ERP, early retinal progenitor; LRP, late retinal progenitor; RGC, retinal ganglion cell; CON, cone cell; HOR, horizontal cell; AMA, amacrine; ROD, rod photoreceptor; BIP, bipolar cells; MUL, Müller glia. (B) Heat map representation in a green-black-red gradient of miRNA with greatest increase between E14 and P0. (C) RT-qPCR analysis of FACS-sorted Hes5-GFP P0 retinas. Note the enrichment of miR-9, let-7f, and let-7a in progenitor cells. (D) RT-qPCR validation of expression profiles for miR-125, let7a, let7f, miR-9, and miR-183 across five developmental stages. Data indicate mean ± SEM. (E and F) Ganglion cells (Brn3+) increase in the Dicer CKO retina, and (G and H) a similar increase is seen with transfection of antagomirs to late progenitor miRNAs compared to controls. Scale bar, 100 microns.
The change in the cell types generated by retinal progenitors over the period of neurogenesis has led several investigators to propose that the retinal progenitors have an intrinsic “clock” that changes their competence over developmental time (5–7). One of the best candidates to emerge as a regulator of progenitor competence is Ikaros1 (8), a zinc finger transcription factor that is expressed by retinal ganglion cells and early staged progenitors; however, whereas deletion of Ikaros1 in mice leads to a reduction in early generated cell fates, most early cell types are still generated in the Ikaros−/− mouse retina, suggesting that additional factors contribute to the temporal identity of the progenitors.
Developmental timing in progenitors also appears to be regulated by microRNAs (miRNAs) (9–11); for example, mice with a retinal-specific deletion of Dicer (αPax6cre/Dicerfl/fl; Dicer-CKO), one of the key enzymes in the miRNA processing pathway, express markers typical of early stage progenitors (e.g., Sox2), but do not go on to express late progenitor markers (e.g., Sox9, Ascl1). In addition, the retinal progenitors in the Dicer-CKO mice generate excess ganglion cells and continue to do so past their normal competence period for this early cell fate. By contrast, those cell types normally generated later in development, like bipolar cells and Müller glia, are not produced in the Dicer-CKO. The Dicer-CKO progenitor cells thus appear to be stuck in an “early” progenitor state. These results led us to hypothesize that there are specific miRNAs that regulate the progression of the progenitors from the early to late competence state. Although a previous study has shown that miRNAs regulate the onset of Xotx2 and Xvsx1 translation in the frog retina (12), the miRNAs analyzed in that report do not affect the competence of the progenitors to generate ganglion cells; and, therefore, to identify specific miRNAs that are developmentally regulated during the relevant developmental period in mice, we carried out miRNA analyses in retinas from both normal and Dicer-CKO animals. We now describe experiments that show that three miRNAs, let-7, microRNA-125 (miR-125), and microRNA-9 (miR-9), are key regulators of the developmental change in competence of retinal progenitors. Members of these three miRNA families increase over the relevant developmental period in normal retinal progenitors whereas overexpression of these three miRNAs in Dicer-CKO retinas can rescue the phenotype, allowing their progression to Ascl1+ late progenitors. Also, overexpression of these miRNAs in normal retina can accelerate their developmental timing whereas knockdown of these miRNAs produces a phenotype similar to the Dicer-CKO, in which excess ganglion cells are generated. Microarray analysis of Dicer-CKO retinal cells, combined with a computational analysis, enabled us to identify two targets of these miRNAs: Protogenin (Prtg) and Lin28b. Both Lin28 and Prtg can maintain the early progenitor state when overexpressed. Together, these data elucidate an miRNA-based molecular mechanism by which retinal progenitor temporal identity is regulated during eye development.
Results
Identification of Late-Progenitor miRNAs.
Conditional deletion of Dicer from early embryonic retinal progenitors causes them to produce ganglion cells in greater than normal numbers and for longer than normal (Fig. 1 E and F; Fig. S1 A–F′). Moreover, the progenitor cells do not progress to express later progenitor markers (Fig. S1 G–J′) and never generate bipolar cells and Müller glia (normally generated late). To determine which miRNAs might be involved in regulating the change in competence of the progenitor cells, we carried out a miRNA array analysis. We found 16 miRNAs that increase by >twofold between embryonic day 14 (E14) and postnatal day 0 (P0); ten of these were either previously shown to be expressed in progenitor cells in other regions of the CNS or in our previous analyses (9). Six of these were members of the let-7 family whereas four were not: miR-125b, miR106b, miR-720, and miR-9 (Fig. 1B). We focused on the let-7 family, miR-125b, and miR-9 because, in our previous analysis of the miRNAs in the Dicer-CKO (9), we found that let-7 family members and miR-9 declined significantly and rapidly with loss of Dicer. By contrast, miR-720 was one of the few miRNAs not reduced in the Dicer-CKO, suggesting either that this miRNA is very stable, or that it is processed independently of Dicer; in either case, it is unlikely to contribute to the Dicer-CKO phenotype. We also did not analyze miR-106b further because a prior analysis of combined deletion of miR-106b and miR106a loci did not report a significant defect in eye development (13).
To validate the microarray data, we used RT-PCR. We found that let-7a, let-7f, and miR-9 all increase over the period of retinal neurogenesis by >5 cycles (>32-fold) whereas miR-125b has a smaller increase (∼fourfold) (Fig. 1D) from E12 to P0. To determine whether these miRNAs are expressed in retinal progenitors, we used FAC sorting to purify progenitors from a Hes5-GFP mouse line that expresses GFP in retinal progenitors (14), and then carried out RT-PCR. We found that let-7 and miR-9 are enriched in the Hes5-GFP(+) progenitors (Fig. 1C) whereas miR-124, an miRNA known to be expressed in postmitotic neurons, is more highly expressed in the Hes5(−) neuronal population. To further confirm progenitor expression of these miRNAs, we used miRNA sensors, specific target sequences in a 3′ UTR added to GFP (Fig. S2A). When these sensors are transfected into HEK cells, they produce a strong GFP signal; however, when cotransfected with mimics or plasmids expressing specific miRNAs, they are degraded as predicted (Fig. S2B), confirming their specificity. We electroporated the sensors for let-7, miR-9, or miR-125 into newborn mouse retinas. In other retinas, we electroporated control sensors: (i) miR-1, which is not expressed in the retina and should not be targeted by an endogenous miRNA, and (ii) miR-183, a miRNA that is highly expressed in photoreceptors, but not in retinal progenitors, and so should only be degraded in the former. We found that let-7, miR-9, and miR-125 sensors are all degraded in the retinal progenitor cells; i.e., there is little GFP signal, despite robust expression of the mCherry transfection control (Fig. S2C). By contrast, the miR-1 sensor is not degraded, due to the absence of miR-1 in the retina. The miR-183 sensor is also not degraded in the cells of the progenitor zone but is degraded in the outer retina, where the developing photoreceptors express miR-183 (Fig. S2D). Although the sensors cannot discriminate between the different miRNA members of the same family, they show that at least some member of the let-7 family is expressed in the retinal progenitors, as are miR-125 and miR-9 family member(s). Interestingly, these miRNAs are also expressed in at least some types of postmitotic retinal cells because we see mCherry expression in amacrine cells and photoreceptors at this age, but no expression of GFP with the let-7, miR-9, or miR-125 sensors.
The above results show that there are members of three miRNA families that are expressed in retinal progenitors and increase in their expression between early embryonic stages and the postnatal period of neurogenesis (we will refer to these as late progenitor miRNAs or LP-miRNAs). To determine whether these miRNAs are required for the change in competence of the progenitors, we knocked-down the activity of these three miRNAs with specific antagomirs (Fig.1 G and H; Figs. S3 and S4). We electroporated antagomirs against each miRNA, and the three in combination, into embryonic retinas, and then analyzed the effects 2 d later. The retinas treated with the combination of miR-125, miR-9 and let-7 show a significant increase in the number of POU class 4 homeobox 3 (Brn3)+ ganglion cells in the electroporated region, similar to what is observed in the Dicer-CKO. Interestingly, let-7 alone causes a smaller, but still significant, increase in ganglion cell numbers. After LP-miRNA antagomir electroporation, we also observed more Prospero homeobox 1 (Prox1)+ horizontal cells, more One cut homeobox 2 (Onecut2)+ cells (ganglion and horizontal cells), and more Recoverin+ photoreceptors (Fig. S4 A–I′). Another aspect of the Dicer-CKO phenotype is an increase in apoptosis (9–11); but we did not observe this in the LP-miRNA antagomir electroporations (Fig. S3K), suggesting that the increase in cell death associated with Dicer deletion is due to the loss of a different miRNA. Interestingly, when we performed the same experiment in P1 retinal explants, we did not observed the same robust effects, and only a handful of Brn3+ ganglion cells were observed in the neuroblastic layer after LP-miRNA inhibition (Fig. S5 A and B′), suggesting that there is a limited temporal window during which RPCs can respond to LP-miRNA regulation.
miRNA Mimics Can Rescue the Dicer CKO Phenotype and Accelerate Developmental Timing.
If the three LP-miRNAs are important in the transition from the early to late progenitor competence states, we reasoned that they should be able to promote this transition in the retinal progenitors from Dicer CKO mice. For these experiments, we used double-stranded RNA “mimics” for these miRNAs. To validate the effectiveness of the mimics for their ability to specifically regulate targets, they were cotransfected with miRNA sensors in HEK cells. When the HEK cells are cotransfected with a particular mimic and the appropriate sensor, the GFP-sensor signal is greatly reduced, but not when cotransfected with a noncognate sensor (Fig. S2B).
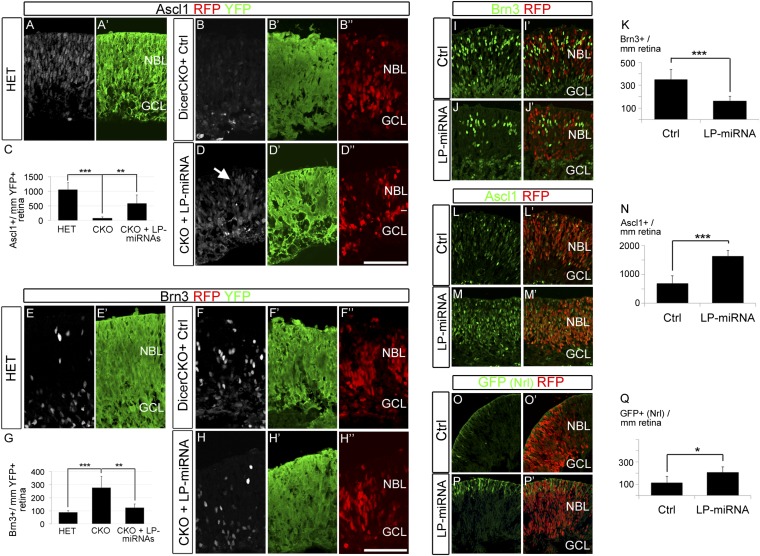
To test whether the mimics are able to substitute for the lack of endogenous miRNAs in the Dicer-deleted regions of the retina (to promote the progression of the progenitors to the late competence state), we electroporated them into the retinas of E16 Dicer-CKO mice. In these mice, the regions where Dicer is deleted are marked with the expression of YFP, due to the presence of a R26EYFP allele, enabling us to confirm that the electroporation was directed to the Dicer-CKO domain (Fig. S6 A–A′′′). At E16, there is an increase in the number of Brn3+ ganglion cells in the Dicer-CKO retinas whereas, at the same time, there is a reduction in expression of the late progenitor marker Ascl1 (9) (Fig. 2 A, B, E, and FfigE). By contrast, when retinas Dicer-CKO mice are transfected with LP-miRNA mimics, the transfected regions of the Dicer CKO retinas now express Ascl1 (Fig. 2 A–D). In addition, the miRNA transfected region of retina shows a reduction in the number of ganglion cells generated in this region (Fig. 2 E–H). Quantification of the number of Brn3+ cells in the Dicer-CKO shows that they are generated at three times their normal level, but transfection of the mimics reduces ganglion cell numbers very close to controls (Fig. 2G). Quantification of the Ascl1+ cells also shows a substantial rescue with the transfection of the mimics (Fig. 2C). These results show that miRNA mimics for the LP-miRNAs are sufficient to promote the transition to the late progenitor state, even when endogenous miRNAs are no longer present.
Fig. 2.
miR-125, let-7, and miR-9 can rescue the Dicer CKO phenotype and accelerate the developmental timing. (A–D′′) LP-miRNA transfection in Dicer-CKO retinas rescued Ascl1 expression. (A and B) Ascl1 (white) was expressed in the developing retina of Dicer-HET controls (A) but was absent in Dicer-CKO transfected with nCherry (red) control construct (B). Coelectroporation of LP-miRNAs together with nCherry (red) rescued Ascl1 expression in Dicer-CKO retina (arrow in D). (E–H′′) LP-miRNA electroporation reduced the number ganglion cells in Dicer-CKO retinal explants. Ganglion cells (Brn3+; white) in Dicer-Het (E) and Dicer-CKO retinas (F–H′′). Coelectroporation of LP-miRNAs with nCherry (labeled with RFP, red) resulted in a reduction in the number of Brn3+ cells (G and H). (Scale bar: A–H′′, 100 μm.) Error bars indicate mean ± SEM. P values: **<0.01; ***<0.001. NBL, neuroblastic layer; GCL, ganglion cell layer. (I–Q) LP-miRNA transfection accelerates development in normal retina. Brn3 (I and J) and Ascl1 (L and M) in E14 retinas transfected with control mimics (I and L) or LP-miRNA (J and M) together with nCherry (red). (O–Q) LP-miRNAs increased the number of Nrl+ photoreceptors in Nrl-GFP E14 retinal explants (P and P′). Error bars indicate mean ± SEM. P values: *<0.05; ***<0.001.NBL, neuroblastic layer; GCL, ganglion cell layer.
The above results show that transfection of mimics for the three LP-miRNAs can substantially rescue the Dicer CKO phenotype; however, if the levels of these miRNAs are critical for the progression of developmental timing in the retinal progenitors, we reasoned that overexpression of one or more of them might accelerate normal developmental timing. We tested this possibility by transfection of the LP-miRNA mimics into early embryonic retina. For these experiments, we used Nrl-GFP mice (15), in which GFP is expressed in rod photoreceptors, a cell type that is primarily generated in the latter half of retinogenesis. Retinas from E14 Nrl-GFP mice were transfected with the three mimics for the LP-miRNAs, and the retinas were cultured for 2 d. The retinas that were transfected with the LP-miRNA mimics had significantly fewer ganglion cells and Prox1+ horizontal cells than the control transfected retinas and, moreover, showed an increase in the number of Ascl1+ cells (Fig. 2 I–N; Figs. S6 B–C′′ and S7 A–B′ and G). We also found that rod photoreceptors, as assessed with Nrl-GFP expression, were increased in the miRNA mimic transfected retinas: at E14 relatively few Nrl-GFP+ rods are present in the retina in the controls (Fig. 2 O and O′); however, we found that transfection of the LP-miRNA mimics caused a significant increase in the number Nrl-GFP+ cells in the retinal explants (Fig. 2 P and Q; Fig. S6 D–E′′). Under our experimental conditions, we did not detect a significant difference in the number of AP2+ cells (Fig. S7 C–D′). We have also analyzed the bipolar cells, a population normally generated at the latest stage of neurogenesis. However, we did not detect Vsx1+ bipolar cells after LP-miRNA overexpression at these early embryonic stages (Fig. S7 E–F′). Together, these results show that the three LP-miRNAs are sufficient to accelerate developmental timing in retinal progenitors to increase the production of the normally late-generated rod photoreceptor fate at the expense of the early generated cell types (ganglion cells and horizontal cells); however, there may be other factors required to produce the last neuronal cell type, i.e., bipolar cells.
Identification of Targets of the LP-miRNAs.
To determine what genes might be responsible for the failure of retinal progenitors to progress to the late stage in the Dicer CKO mice, we used cDNA microarrays to compare gene expression between retinal cells FACS-sorted from Dicer CKO and Het littermate retinas from E17 mice (Fig. S1 K–M). Although this approach will measure only the changes in mRNA in the absence of miRNAs, recent studies have shown that, in mammalian cells, the majority of regulation by miRNAs occurs via transcript degradation (16–18). E17 retinas from αPax6-cre+; Dicerfl/fl ; ROSA-YFP (CKO) and αPax6-cre+; Dicerfl/+ ; ROSA-YFP (Het) littermate mouse embryos were dissociated and purified using FACS and subjected to microarray analysis (Fig. S1). We confined our analysis to those genes on the array that were expressed at a higher level than the median expression level (12,075 out of the total 28,301 genes represented on the Affymetrix array). Of these, 200 genes were increased by more than 1.5-fold in the Dicer-CKO retinal cells (Table S1) whereas 382 genes were down-regulated by a similar amount. Many of the genes that were down-regulated in the Dicer CKO cells (Table S2) have been previously shown to be expressed in the late progenitors, e.g., Ascl1, Dll3, Hes6 and Sox9 (3, 14, 19) further confirming that loss of Dicer blocked the developmental progression of the progenitor cells.
Deletion of Dicer is expected to lead to the up-regulation of genes due to the failure in the generation of mature miRNAs that would normally have directly targeted these genes and led to their degradation (20, 21). In addition, other genes may change secondarily from the changes in these direct targets. Within the list of genes that increase most substantially in the Dicer CKO retinal cells, we found several transcription factors (Table S1; Fig. 3A): e.g., members of the T-box transcription factor (Tbx), Iroquois homeobox (Irx) and Onecut families; several genes involved in the circadian cycle, e.g., nuclear receptor subfamily 1, group D, and Period circadian clock 3 (Per3); and two members of the Caenorhabditis elegans heterochronic pathway, Lin28b and Tripartite motif containing 71, E3 ubiquitin protein ligase (Trim71/Lin41). Several of these transcription factors are known to be expressed in ganglion cells, including Tbx5 (22), Irx3 and Irx5 (23), Pou4f2/Brn3b (24), and Onecut1 and Onecut2 (25), consistent with the increase of this cell type in the Dicer CKO. Other genes that increased in the Dicer CKO cells have been previously identified as “early progenitor” genes [e.g., Secreted frizzled-related protein 2 (Sfrp2)]. However, one of the most highly up-regulated genes, Protogenin (Prtg), has not been studied in the retina although it is important in spinal cord development. In addition, the heterochronic genes Lin28b and Trim71/Lin41 have not been studied in the developing retina.
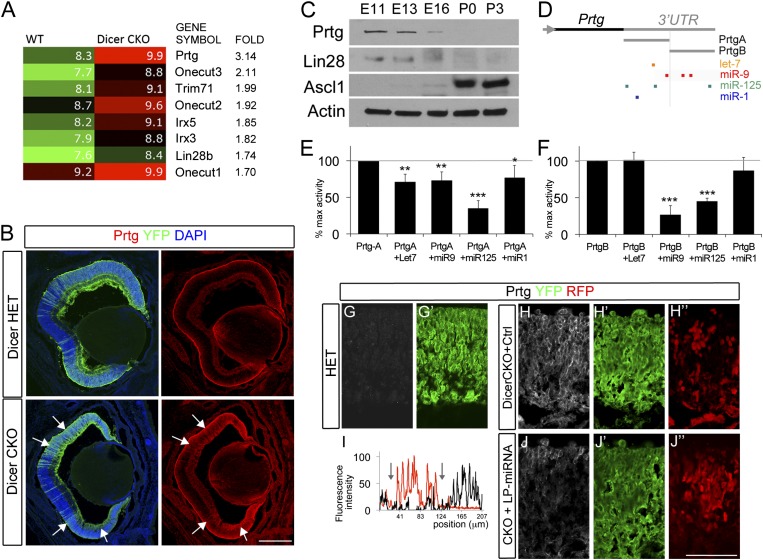
Fig. 3.
Prtg and Lin28 are targets of LP-miRNAs. (A) Gene changes in Dicer-CKO retinas. Log2 expression levels are shown as heat maps. Numbers indicate fold change relative to controls. (B) Prtg labeling in E18 Dicer-Het (B, Upper) or Dicer-CKO (B, Lower). YFP staining (green) indicates areas of cre-mediated recombination. Prtg is up-regulated in Dicer-CKO cells (arrows). (Scale bar: 200 μm.) (C) Western blot analyses of wild-type retinal homogenates at five developmental ages. Equal loading was confirmed by beta-Actin immunoblot (Bottom). (D) Schematic representation of Prtg 3′ UTR region and predicted miRNA-target sites. PrtgA and PrtgB fragments were cloned in dual luciferase reporter vectors. (E and F) Prtg is directly regulated by miRNAs. The luciferase reporter constructs were cotransfected together with control vectors or with miRNA expression constructs in HEK293T cells. Firefly luciferase activity was measured and normalized with Renilla luciferase activity in the same well. Data are expressed as percentage of induction relative to control. Values are mean ± SD of four independent experiments. (G–J) Prtg is a direct target of LP-miRNA in retinal cells. Endogenous Prtg protein (white) in Dicer-HET (G) or Dicer-CKO retinas transfected with control mimics (H–H′′) or LP-miRNA (J–J′′) together with nCherry (red). YPF staining (green) indicates areas of cre-mediated recombination. (I) Line scan measurements for nCherry (red) and Prtg (black) represented as fluorescence intensity (arbitrary units). Note the reduction in Prtg intensity in LP-miRNA electroporated areas (area between arrows, I).
The nature of the changes in gene expression in the retinal progenitors with the loss of Dicer could be direct or indirect. We further narrowed the list to those genes that increase in the Dicer CKO and that also have conserved sites in their 3′ UTR for the three LP-miRNAs, in all three of the following computational databases for miRNA prediction: miRanda, PicTar, and MiRtarget2. Of the top 100 genes that increase in the Dicer CKO, there were only two genes that fit these criteria, Lin28b and Protogenin. Therefore, we focused our further analyses on these two genes.
Prtg and Lin28b Are Targets of LP-miRNAs in the Developing Retina.
We verified that Prtg and Lin28b are expressed in the developing retina and are down-regulated from the early to late stages of development. Fig. 3C shows a Western blot for Prtg at several stages of retinal development; Prtg is expressed at E11 and E13, but only a faint band can be seen at E16 and it was not detectable at P0 or P3. Lin28b has a similar pattern of developmental expression. Lin28b is a known target of miR-125 (26) and let-7 (27), and the increase in expression of LP-miRNAs is consistent with the observed decline in Lin28b. By contrast, Ascl1 is expressed in progenitor cells only after ganglion cell production is complete (3, 28) and is strongly up-regulated over these same ages. We confirmed that Prtg and Lin28b are expressed in early progenitors by immunostaining (Fig. S8 A–F′). At E12, both Prtg and Lin28 were highly expressed in retinal progenitors (Fig. S8 A and D, arrows). The retina develops in a central-to-peripheral pattern, and E14 defines the initial onset of Ascl1 in a subset of late progenitors in the central retina (3). By E14, Prtg and Lin28b were down-regulated in the central retina but still highly expressed in peripheral regions (Fig. S8 B and E). These results show that the two putative targets of the LP-miRNAs are expressed in the retina at the appropriate ages and decline in a manner consistent with the hypothesis that they are targeted by these miRNAs.
To determine whether Prtg and Lin28b are directly regulated by miRNAs in the retina, we used several different approaches. First, we analyzed their expression in the Dicer-CKO mouse retina. Using immunofluorescence labeling, we found that Prtg labeling was increased in the regions of recombination in the Dicer-CKO retina, as indicated by the YFP-fluorescence (Fig. 3B). At low magnification, the domains of Dicer deletion can be seen to correspond to regions of increased fluorescence. At high magnification (Fig. 3 G and H), the predominantly membrane localization of the Prtg labeling can be observed in the Dicer deleted regions of the Dicer-CKO retinas (Fig. 3H) whereas the labeling is very faint in a corresponding region of the Dicer-Het retina (Fig. 3G). A similar pattern was observed when sections were labeled with an antibody against Lin28b (Fig. S9 A–B′′; other genes up-regulated in the array also show increases in the Dicer-CKO). These data are consistent with the hypothesis that both Prtg and Lin28b are targets of the LP-miRNAs in the developing retina because their expression is increased when those miRNAs are absent.
The above results show a relationship between miRNA levels during development and in the Dicer-CKO and the expression of Prtg and Lin28b. Several previous studies have shown that Lin28b is a target of the three LP-miRNAs (29, 30); however, no study to date has analyzed regulation of Prtg by miRNAs. Therefore, to directly test whether Prtg is a direct target of miR-125, miR-9, and let-7, we used a plasmid containing the 3′ UTR of Prtg cloned downstream of a luciferase reporter. Because the Prtg 3′ UTR is 5 kb, we used two separate clones, which we named Prtg-UTR_A and Prtg-UTR_B. Prtg-UTR_A contains sites for let-7, miR-9, and miR-125, and Prtg-UTR_B contains sites for miR-9 and miR-125, but not let-7 (Fig. 3D). When Prtg-UTR_A reporter plasmid was transfected into HEK cells along with plasmids expressing let-7, miR-9, or miR-125, there was a significant decline in the luciferase levels (Fig. 3E). A similar decline was observed when Prtg-UTR_B was cotransfected with plasmids expressing miR-9 or miR-125, but not let-7, consistent with the lack of let-7 sites in the Prtg-UTR_B 3′ UTR (Fig. 3F). Similar results were obtained when the reporter plasmids were cotransfected with miRNA mimics, instead of miRNA containing plasmids (Fig. S10 A and B). When the sites for these miRNAs were mutated to remove the miRNA consensus sites from the 3′ UTR, the reporter no longer reduced by the LP-miRNAs (Fig. S10 C and D), confirming that the 3′ UTR for Prtg is targeted by these miRNAs. Finally, to determine whether Prtg is directly regulated by the LP-miRNAs in the retina, we assayed Prtg expression in the Dicer CKO after transfection with mimics for let-7, miR-9, and miR-125. This approach is similar to our rescue experiment described in the previous section. We transfected E16 retinas with mimics for let-7, miR-9, and miR-125 and analyzed the expression of Prtg with immunofluorescence. The three LP-miRNAs substantially reduced Prtg in the transfected region (indicated by cotransfection with nCherry; Fig. 3 H–J). Together, our data demonstrate that Prtg is a target of let-7, miR-9, and miR-125.
Prtg and Lin28b Maintain Progenitors in an Early Competence State.
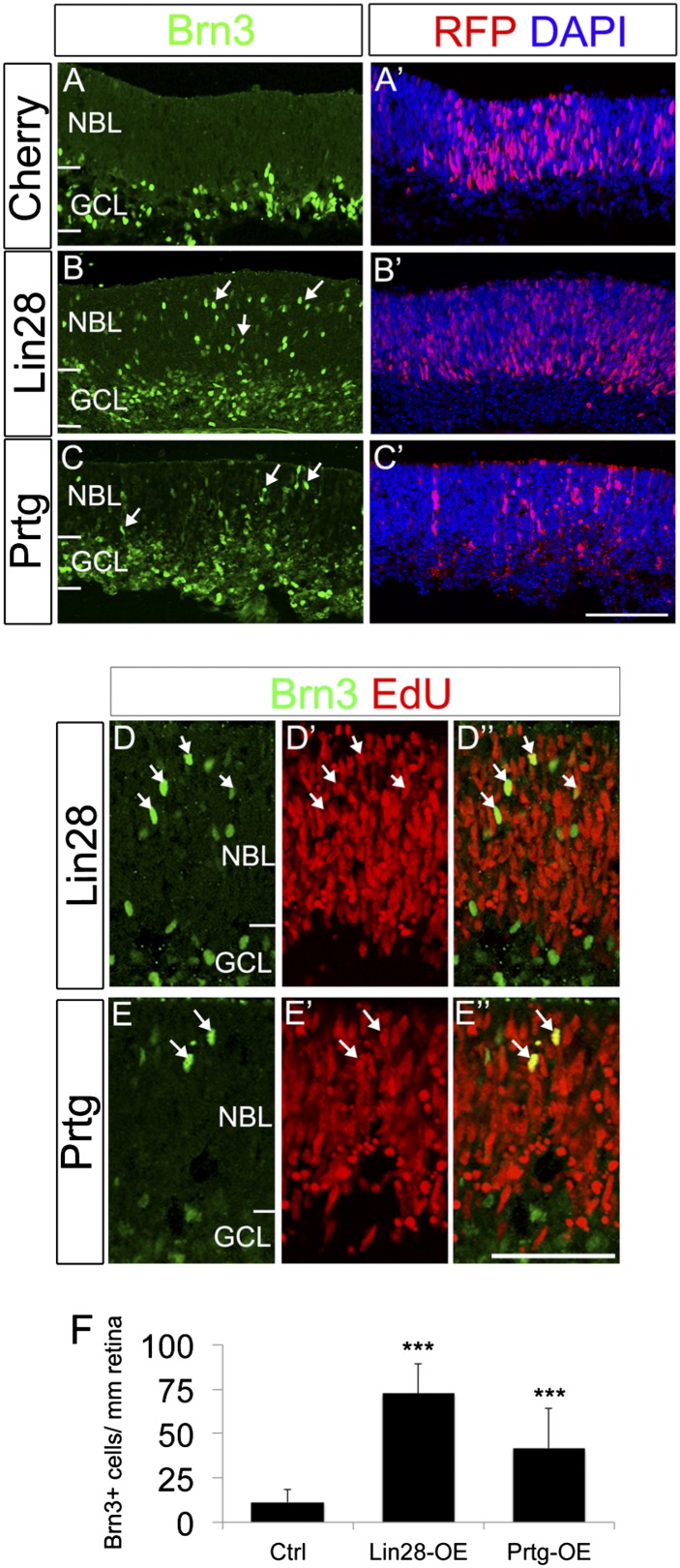
The above results show that both Prtg and Lin28b are targets of the LP-miRNAs in the developing retina. This data suggests that these proteins may be important regulators of the transition from the early to late competence state in retinal progenitors. To directly determine whether Prtg and/or Lin28 are sufficient to maintain retinal progenitors in an early state, we used an overexpression strategy. If miRNA-mediated degradation of Prtg and Lin28b is necessary for maintaining the early progenitor competence state, overexpression of either or both might overcome this inhibition. We overexpressed Prtg or Lin28b in embryonic retinal progenitors starting at E16 and analyzed the effects 2 d later. When either Lin28 (Fig. 4 B and F) or Prtg (Fig. 4 C and F) were ectopically expressed in the E16 retinal progenitors (a stage when they would no longer be producing ganglion cells in the central retina), there was a significant increase in the number of Brn3+ ganglion cells produced in the region of transfection, compared with untransfected regions of the same retina or retinas transfected with control constructs. The additional Brn3+ cells were frequently found in the neuroblastic layer in the regions of transfection, suggesting that they were newly produced and had yet to migrate to the ganglion cell layer. To directly demonstrate that the Brn3+ cells were newly generated, we cultured the retinas with EdU to label proliferating progenitors and their progeny (Fig. 4 D and E). Nearly all of the Brn3+ cells in the neuroblastic layer of the retinas transfected with Prtg and Lin28b were also labeled with EdU, indicating that the effect of overexpression of either Lin28b or Prtg was to cause the progenitors to maintain an early competence state. To address whether late progenitors could reacquire an early competence state at later time points, we overexpressed Lin28b mRNA in P1 retinal explants (Fig. S11). In these experiments, we did not observe a significant number of newly born Brn3+ ganglion cells in the neuroblastic layer, after Lin28 overexpression. These data suggest that there is a temporal window of competence over which the progenitors can be shifted to an earlier stage, but, once they passed this temporal window, Lin28 is no longer capable to reset them into early competence states.
Fig. 4.
Overexpression of Lin28 or Prtg maintains an early progenitor competence. (A–F) Prtg or Lin28 overexpression increased the number of ganglion cells generated in the neuroblastic layer. E16 wild-type retinas were electroporated with nCherry alone (A and A′) or together with Lin28 (B and B′; D and D′′) or Prtg (C and C′; E and E’) and labeled with Brn3 (green) and RFP (red) antibodies. Brn3+ ganglion cells increase in the neuroblastic layer after Prtg or Lin28 overexpression (arrows) compared with controls. (D–E′′) Labeling for EdU (red) colabeled Brn3+ ganglion cells (green) in the neuroblastic layer (arrows). (F) Graph of effects of Prtg or Lin28 overexpression; mean ± SEM. P values: ***<0.001. NBL, neuroblastic layer; GCL, ganglion cell layer; L, lenses. (Scale bars: A–C′, 200 μm; D–E′′, 100 μm.)
Discussion
We have previously reported that a retinal-specific conditional deletion of Dicer leads to a stall in the developmental progression of the progenitors (9, 11). In this report, we demonstrate that overexpression of mimics for three “late-progenitor” miRNAs is sufficient to rescue this phenotype and allow the progenitors to progress from the early to the late competence state. We have also found that overexpression of these mimics into normal retina can accelerate developmental timing; late progenitor markers are acquired prematurely and the transfected progenitors increase their production of late-born neurons, like rod photoreceptors. Lastly, we have identified two of the key targets that are regulated by let-7, miR-125, and miR-9: protogenin (Prtg) and Lin28b. These genes (i) are up-regulated in the Dicer CKO retinal progenitors, (ii) have conserved sites for binding these miRNAs in their 3′ UTRs, and (iii) can induce an early progenitor competence state in late progenitors, such that they renew their production of ganglion cells. Together, these data show that miRNAs regulate developmental timing in the vertebrate retina, using a conserved miRNA heterochronic pathway.
The developmental increase in expression of miR-9, miR-125b, and let-7 family members that we observe in the retinal progenitors confirms and extends earlier analyses of retinal miRNAs (31–36). The most recent comprehensive microarray and in situ analyses (31, 35) show that miR-9, miR-125, and several members of the let-7 family (including let-7a, -b, -c, -d, -e, -f, -g, and -i) are expressed in the retina and increase over the developmental period that we have analyzed. Moreover, previous in situ localization studies have demonstrated that there is widespread expression of let-7 family members in all retinal layers from E16.5 to adulthood (ref. 33; http://mirneye.tigem.it), and miR-9 is expressed from optic cup stages to all retinal layers at P0 (33, 35). Our results now show that several members of the let-7 family, miR-9, and miR-125 increase quantitatively over the period of retinal development when the progenitors change their competence, consistent with the hypothesis that miRNAs regulate this process. Previous analysis of the phenotype in the Dicer CKO revealed that, in highly mosaic regions of cre-recombinase expression, there was an apparent rescue of the effect on developmental timing in the retinal progenitors. It is possible that some of the effects we report in these study with mimic and antagomir expression might also be non-cell autonomous; however, at the present time we do not have a direct way to test this possibility.
Developmental timing in other regions of the CNS may also be regulated by miRNAs. In the cerebral cortex, deletion of Dicer leads to an increase in the early-born neurons by E15.5 (37), and a reduction in the number of late-born neurons and glia (38, 39), a phenotype analogous to that which occurs in the Dicer-CKO retina. Moreover, some of the same genes (e.g., Ascl1, Sox9) decline in the Dicer CKO progenitors in cerebral cortex and retina (38). Deletion of Dicer in spinal cord causes defects in the production of glia (40), similar to the loss of Müller glia in the retina. In embryonic stem cells and P19 cells, miR-9 and miR-125 increase during neural differentiation (41–43), and overexpression of miR-125 potentiates neural specification of human ESCs. Lin28 is also expressed in progenitor cells in the neural tube (44) and is down-regulated as they differentiate. Although none of these earlier studies tested whether the effects of miRNA misexpression or inhibition altered the developmental “time” of the progenitor cells, overexpression of Lin28 in P19 cells inhibits generation of astrocytes (41), a cell type normally generated by late progenitors in the CNS. It is also likely that other developmental pathways may also be affected by the LP-miRNAs. In developing mouse and chick CNS, miR-9 regulates cell cycle exit inhibiting components of the Notch pathway in the zebrafish hindbrain (45), targeting the transcription factor T-cell leukemia homeobox 1 (Tlx) (45) in the cerebral cortical progenitors, and Forkhead box P1 (FoxP1) in the neural tube to regulate motorneuron specification (46).
It is not clear how Prtg and Lin28, the key targets of the LP-miRNAs, maintain progenitors in an early state. In the chick spinal cord, Prtg expression is inversely correlated with Ascl1 expression, similar to what we have found in the retina (48). Prtg is a member of the DCC/Neogenin family (49, 50) and contains four Ig and five fibronectin III domains in the extracellular domain. Ligands for Prtg have not been identified, and the intracellular domain does not share significant homology with the other four members of this family (DCC/Neogenin/Punc/Nope). Interestingly, the intracellular domain of Prtg is cleaved by gamma-secretase and can localize to the nucleus, and therefore may directly regulate transcription (51). As noted above, Ikaros1 has been shown to be a regulator of progenitor competence. Although the LP-miRNAs do not appear to directly regulate the expression of Ikaros1 (Ikzf1)—there are no conserved sites for any of the LP-miRNAs in the mouse Ikfz1 3′ UTR, and the expression of Ikfz1 does not change in the Dicer-CKO microarray analysis—Ikzf3 and Ikzf5 show small increases in the Dicer-CKO array analysis, and there is a conserved site for miR-125 in the 3′ UTRs of both Ikzf3 and Ikzf5, although at present it is not known whether either of these Ikaros family members plays a role in retinal development. A recent study of Ikaros in cerebral cortex development (52) has provided further evidence for a role of this transcription factor in the regulation of developmental timing in the nervous system. Although Ikaros knockout mice do not show defects in cortical development, overexpression of Ikaros in the early stages of cortical development prolongs the production of the early born cell fates at the expense of the later born fates. Although it is possible that the effects we observe in our manipulations of Dicer, LP-miRNAs, and their targets Prtg and Lin28 are mediated in part through Ikaros, we do not observe the changes in cell cycle genes in the Dicer CKO (e.g., cyclin D1 and D2 and cdkn1c) that were reported after Ikaros overexpression. Therefore, our data are more consistent with the possibility that the miRNA-mediated changes in Lin28 and Prtg constitute a parallel pathway for the control of developmental timing.
The progressive production of different types of neurons and glia is characteristic of the vertebrate CNS. Our data reveal that the temporal identity of the progenitors in the mammalian retina is controlled by an miRNA-dependent pathway. It is interesting that a similar mechanism, using miRNAs let-7, miR-125/lin-4, and Lin28, regulates developmental timing in C. elegans seam cells (53). miRNAs are well conserved across animals, and, remarkably, the core miRNAs for the regulation of developmental transitions appear to be conserved as well. Although some of the targets of these miRNAs differ between the retinal progenitors and seam cells, Lin28 and the LP-miRNAs regulate developmental timing in both systems. Possibly, the combinatorial nature of miRNA targeting provides a flexible system for gene regulation necessary for the sequential fate-determination events that characterize a cell’s temporal identity during development.
Materials and Methods
All animals were used in accordance with University of Washington Institutional Animal Care and Use Committee approved protocols (UW-IACUC). Generation and genotyping of Dicer-CKO and Nrl-GFP mice have been described previously (9, 15, 54). Immunofluorescence labeling and antibodies used in this study have been described previously (3, 9) and are described more fully in SI Materials and Methods. Western blotting was carried out with standard protocols. Primary antibodies were as follows: sheep anti-Prtg (1:1,000) (R&D Systems), rabbit anti-Lin28 (1:2,000) (sc-67266; Santa Cruz), mouse anti-Ascl1 (1:1,000) (BD Pharmingen), and mouse anti–beta-Actin (1:10,000) (Abcam). Retinas from E14 or E16 mouse embryos or P0 pups were dissected free of surrounding ocular tissue and lens. Whole retinas were placed into 24-well plates and incubated for 1 or 2 d in DMEM/F12 (1:1; Invitrogen/Gibco) containing 10% (vol/vol) heat-inactivated FBS and 5 ng/mL BDNF (R&D Systems).
For transfection, we used RNA electroporation. As described previously, RNA transfection results in an extremely high rate of transfection compared with DNA methods. Cherry mRNA was used as an electroporation control to reveal the region of transfection of miRNA mimics and miRNA antagomirs, which are also RNA molecules. To generate mRNA for transfection, we used the pSLU plasmid as described previously (53). We used the Ambion mMessage mMachine (AM1345; Ambion) according to the manufacturer's instructions. Before culture, retinas were placed in an electroporation chamber filled with HBSS; the positive electrode was placed below the explant and the negative electrode above. The RNA was carefully added over the explant. Voltage was then applied: three 50-ms pulses of 40 V using an ECM 830 BTX electroporator. The explants were then returned to the tissue culture medium for up to 48 h.
For the luciferase assays, HEK 293T cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 10% heat-inactivated FBS. Transfection was performed with the Lipofectamine 2000 transfection reagent, using 0.8 μg of the PrtgUTR constructs (Prtg-A, Prtg-B, Prtg-Amutant, or Prtg-Bmutant) or sensor vectors and 0.8 μg of the miRNA expression constructs or 10 nM of the miRNA mimics (Thermo). For luciferase assays, the cells were transferred to a 96-well plate 18–20 h after transfection and cultured for another 4 h. Both Firefly luciferase and Renilla luciferase activities were measured following the manufacturer’s instructions (GeneCopoeia) and data were recorded. Firefly luciferase activity was then normalized with Renilla luciferase activity in the same well. For sensor experiments, cells were cultured for 24 h after transfection and fixed with 4% paraformaldehyde for 20 min. Fluorescent images were captured in an inverted fluorescent microscope. For the miRNA microarray studies, RNA was labeled and hybridized onto miRCURY v.11.0 LNA microRNA Arrays (Exiqon) according to the manufacturer’s instructions by the Genomics Resource DNA Array Laboratory at the Fred Hutchinson Cancer Research Center (Seattle, WA). Background subtraction and normalization were carried out as described in ref. 9. The Affymetrix analysis was carried out on Dicer-CKO and Dicer-Het FACS-sorted cells from several animals across two litters. E16 or E17 retinas were dissected in HBSS (Sigma), rinsed in calcium and magnesium free HBSS, and dissociated by trituration after incubation in 0.25% Trypsin-EDTA (Invitrogen). Cells were collected by centrifugation at 180 × g and resuspended in HBSS. Cells were sorted for YFP by using a BD Influx flow cytometer (Becton Dickinson) with 60 mW of 488-nm argon excitation and a bandpass filter of 525/30 nm. Cells were collected by centrifugation and placed in TRIzol (Invitrogen) or Qiazol (Qiagen). RNA was extracted and processed for hybridization to an Affymetrix Mouse Gene ST 1.0 Array according to the manufacturer’s protocols. Array hybridization and postprocess normalization were performed by the Institute for Systems Biology (Seattle, WA). Data analysis was performed in Microsoft Excel and MultiExperiment Viewer 4.6 Version 10.2 (Dana-Farber Cancer Institute). Real-time reverse-transcription PCR was performed as described in ref. 56. For miRNA RT, whole RNA was reverse transcribed in multiplex using custom designed stemloop reverse transcription primers as described previously (57). Primer sequences are listed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge the help and advice of T.A.R. laboratory and Olivia Bermingham-McDonogh laboratory members. We also acknowledge helpful comments on the manuscript from Drs. Olivia Bermingham-McDonogh, Rachel Wong, and W. A. Harris. This work was supported by National Institutes of Health Grants 1 PO1 GM081619-01 and 1R01EY021482 and by the imaging core of Vision Core Grant P30EY01730 (to the University of Washington).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301837110/-/DCSupplemental.
References
- 1.Sidman RL. Histogenesis of mouse retina studied with thymidine H3. In: Smelser GK, editor. Structure of the Eye. New York: Academic; 1961. [Google Scholar]
- 2.Wallace VA. Making a retina: From the building blocks to clinical applications. Stem Cells. 2011;29(3):412–417. doi: 10.1002/stem.602. [DOI] [PubMed] [Google Scholar]
- 3.Brzezinski JA, 4th, Kim EJ, Johnson JE, Reh TA. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011;138(16):3519–3531. doi: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hafler BP, et al. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci USA. 2012;109(20):7882–7887. doi: 10.1073/pnas.1203138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reh TA, Cagan RL. Intrinsic and extrinsic signals in the developing vertebrate and fly eyes: Viewing vertebrate and invertebrate eyes in the same light. Perspect Dev Neurobiol. 1994;2(2):183–190. [PubMed] [Google Scholar]
- 6.Cepko CL. The patterning and onset of opsin expression in vertebrate retinae. Curr Opin Neurobiol. 1996;6(4):542–546. doi: 10.1016/s0959-4388(96)80062-6. [DOI] [PubMed] [Google Scholar]
- 7.Reh TA, Kljavin IJ. Age of differentiation determines rat retinal germinal cell phenotype: Induction of differentiation by dissociation. J Neurosci. 1989;9(12):4179–4189. doi: 10.1523/JNEUROSCI.09-12-04179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60(1):26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci. 2010;30(11):4048–4061. doi: 10.1523/JNEUROSCI.4982-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iida A, Shinoe T, Baba Y, Mano H, Watanabe S. Dicer plays essential roles for retinal development by regulation of survival and differentiation. Invest Ophthalmol Vis Sci. 2011;52(6):3008–3017. doi: 10.1167/iovs.10-6428. [DOI] [PubMed] [Google Scholar]
- 11.Davis N, Mor E, Ashery-Padan R. Roles for Dicer1 in the patterning and differentiation of the optic cup neuroepithelium. Development. 2011;138(1):127–138. doi: 10.1242/dev.053637. [DOI] [PubMed] [Google Scholar]
- 12.Decembrini S, et al. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci USA. 2009;106(50):21179–21184. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He J, et al. How variable clones build an invariant retina. Neuron. 2012;75(5):786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson BR, et al. Genome-wide analysis of Müller glial differentiation reveals a requirement for Notch signaling in postmitotic cells to maintain the glial fate. PLoS ONE. 2011;6(8):e22817. doi: 10.1371/journal.pone.0022817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akimoto M, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci USA. 2006;103(10):3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331(6017):550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson BR, et al. Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development. Dev Dyn. 2009;238(9):2163–2178. doi: 10.1002/dvdy.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol. 2008;18(17):1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu L, Belasco JG. Examining the influence of microRNAs on translation efficiency and on mRNA deadenylation and decay. Methods Enzymol. 2008;449:373–393. doi: 10.1016/S0076-6879(08)02418-X. [DOI] [PubMed] [Google Scholar]
- 22.Sowden JC, Holt JK, Meins M, Smith HK, Bhattacharya SS. Expression of Drosophila omb-related T-box genes in the developing human and mouse neural retina. Invest Ophthalmol Vis Sci. 2001;42(13):3095–3102. [PubMed] [Google Scholar]
- 23.Cohen DR, Cheng CW, Cheng SH, Hui CC. Expression of two novel mouse Iroquois homeobox genes during neurogenesis. Mech Dev. 2000;91(1-2):317–321. doi: 10.1016/s0925-4773(99)00263-4. [DOI] [PubMed] [Google Scholar]
- 24.Wang SW, et al. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002;129(2):467–477. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- 25.Wu F, Sapkota D, Li R, Mu X. Onecut 1 and Onecut 2 are potential regulators of mouse retinal development. J Comp Neurol. 2012;520(5):952–969. doi: 10.1002/cne.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 27.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 28.Jasoni CL, Reh TA. Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J Comp Neurol. 1996;369(2):319–327. doi: 10.1002/(SICI)1096-9861(19960527)369:2<319::AID-CNE11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 29.Zhong X, Li N, Liang S, Huang Q, Coukos G, Zhang L. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem. 2010;285(53):41961–41967. doi: 10.1074/jbc.M110.169607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 31.Hackler L, Jr, Wan J, Swaroop A, Qian J, Zack DJ. MicroRNA profile of the developing mouse retina. Invest Ophthalmol Vis Sci. 2010;51(4):1823–1831. doi: 10.1167/iovs.09-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora A, et al. Prediction of microRNAs affecting mRNA expression during retinal development. BMC Dev Biol. 2010;10:1. doi: 10.1186/1471-213X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48(2):509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 34.Sanuki R, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14(9):1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 35.Karali M, et al. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282(34):25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 37.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersson T, et al. Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS ONE. 2010;5(10):e13453. doi: 10.1371/journal.pone.0013453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowakowski TJ, Mysiak KS, Pratt T, Price DJ. Functional dicer is necessary for appropriate specification of radial glia during early development of mouse telencephalon. PLoS ONE. 2011;6(8):e23013. doi: 10.1371/journal.pone.0023013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J Neurosci. 2010;30(24):8245–8250. doi: 10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139(7):1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- 42.Delaloy C, et al. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6(4):323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eda A, Tamura Y, Yoshida M, Hohjoh H. Systematic gene regulation involving miRNAs during neuronal differentiation of mouse P19 embryonic carcinoma cell. Biochem Biophys Res Commun. 2009;388(4):648–653. doi: 10.1016/j.bbrc.2009.08.040. [DOI] [PubMed] [Google Scholar]
- 44.Yang DH, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003;3(6):719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 45.Coolen M, Thieffry D, Drivenes O, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Dev Cell. 2012;22(5):1052–1064. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16(4):365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J Neurosci. 2011;31(3):809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong YH, et al. Protogenin defines a transition stage during embryonic neurogenesis and prevents precocious neuronal differentiation. J Neurosci. 2010;30(12):4428–4439. doi: 10.1523/JNEUROSCI.0473-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toyoda R, Nakamura H, Watanabe Y. Identification of protogenin, a novel immunoglobulin superfamily gene expressed during early chick embryogenesis. Gene Expr Patterns. 2005;5(6):778–785. doi: 10.1016/j.modgep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Vesque C, Anselme I, Couvé E, Charnay P, Schneider-Maunoury S. Cloning of vertebrate Protogenin (Prtg) and comparative expression analysis during axis elongation. Dev Dyn. 2006;235(10):2836–2844. doi: 10.1002/dvdy.20898. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe Y, Nakamura H. Nuclear translocation of intracellular domain of Protogenin by proteolytic cleavage. Dev Growth Differ. 2012;54(2):167–176. doi: 10.1111/j.1440-169X.2011.01315.x. [DOI] [PubMed] [Google Scholar]
- 52.Alsiö JM, Tarchini B, Cayouette M, Livesey FJ. Ikaros promotes early-born neuronal fates in the cerebral cortex. Proc Natl Acad Sci USA. 2013;110(8):E716–E725. doi: 10.1073/pnas.1215707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21(4):511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Georgi SA, Reh TA. Dicer is required for the maintenance of notch signaling and gliogenic competence during mouse retinal development. Dev Neurobiol. 2011;71(12):1153–1169. doi: 10.1002/dneu.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayashi T, Lamba DA, Slowik A, Reh TA, Bermingham-McDonogh O. A method for stabilizing RNA for transfection that allows control of expression duration. Dev Dyn. 2010;239(7):2034–2040. doi: 10.1002/dvdy.22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol. 2007;304(2):479–498. doi: 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.