Abstract
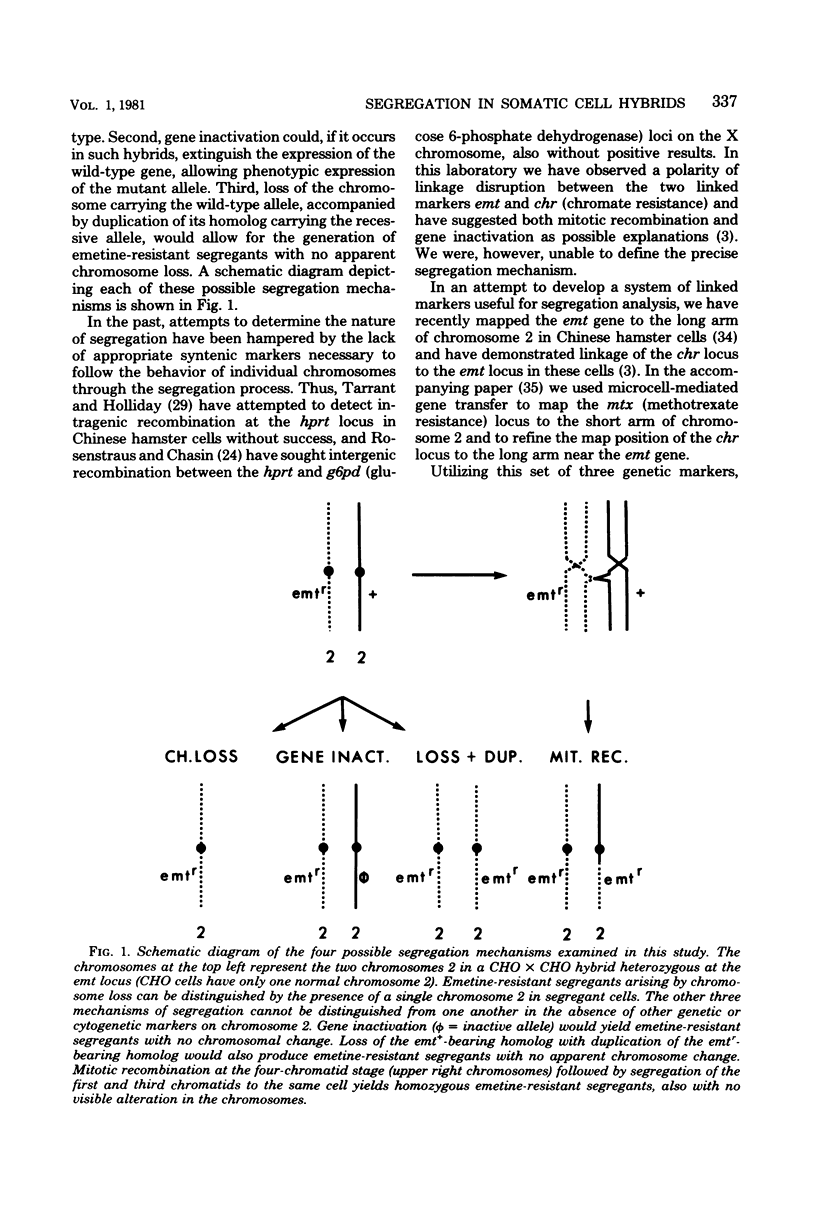
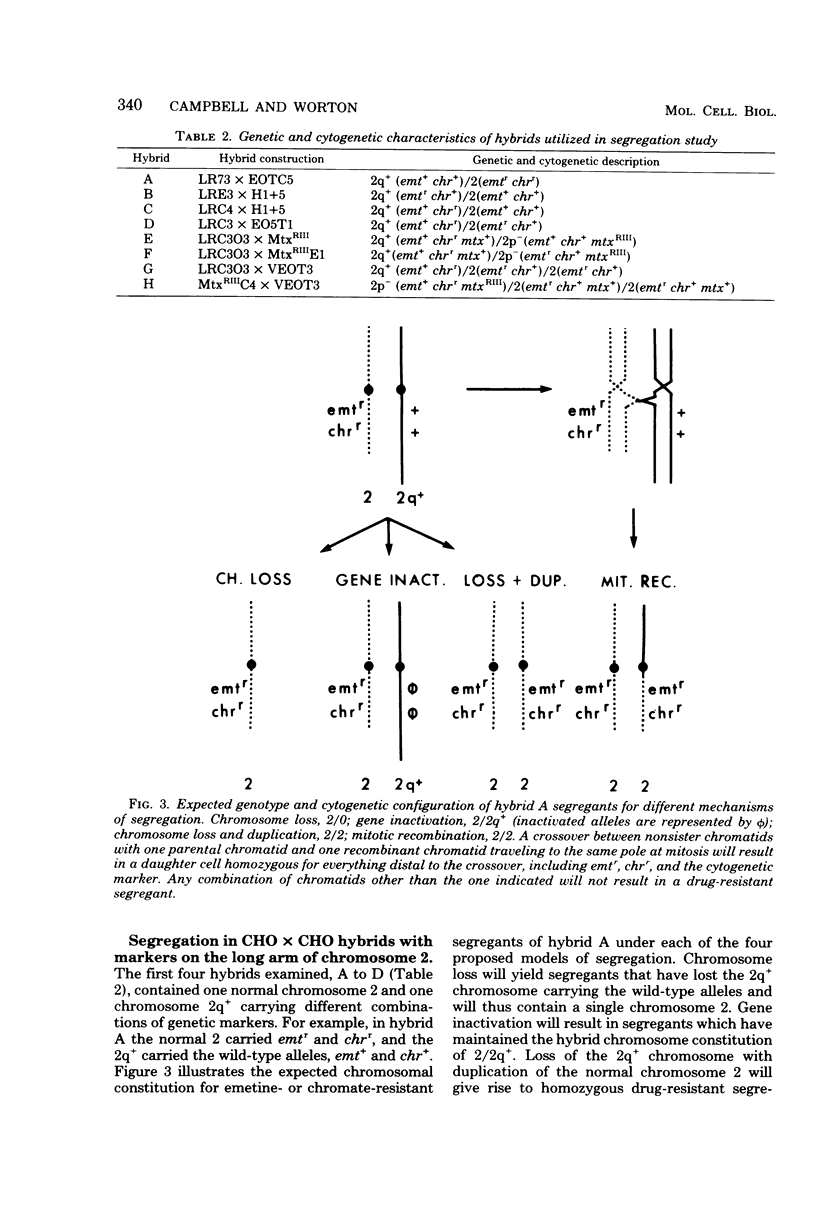
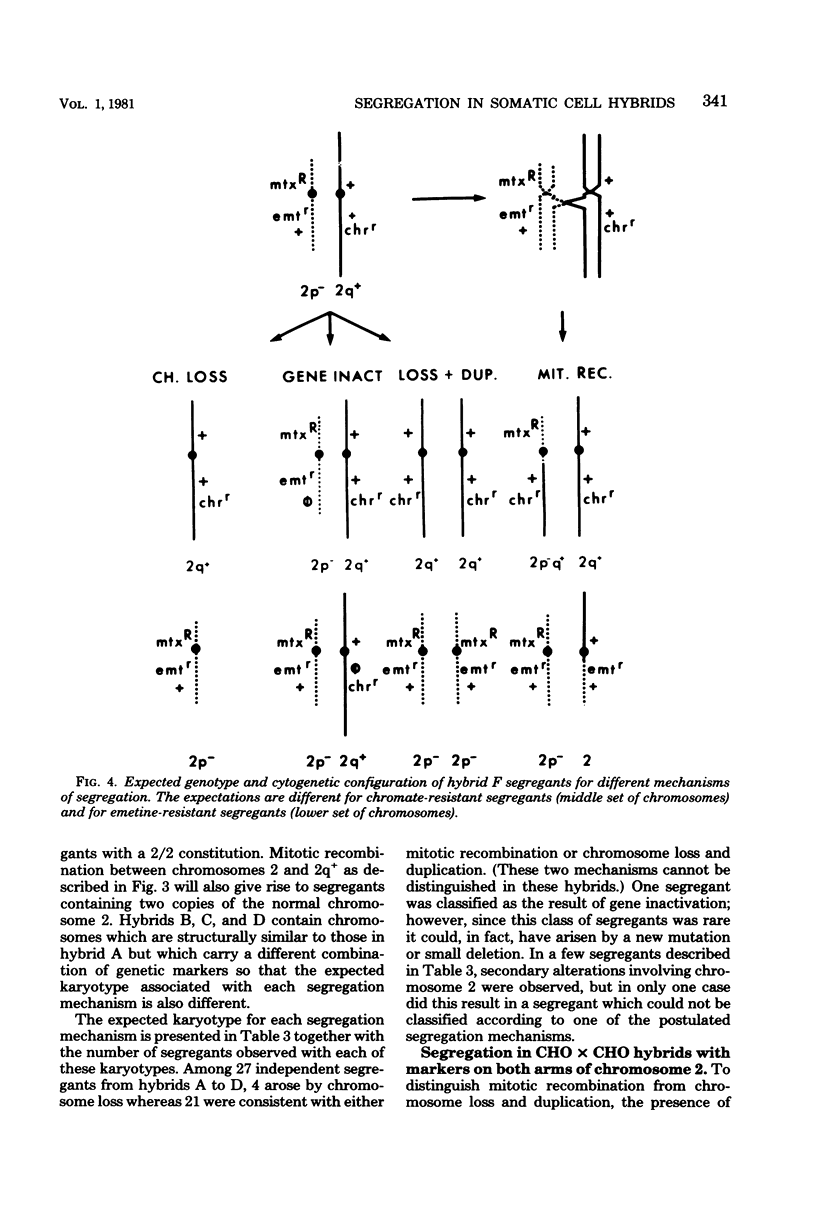
Somatic cell hybrids heterozygous at the emetine resistance locus (emtr/emt+) or the chromate resistance locus (chrr/chr+) are known to segregate the recessive drug resistance phenotype at high frequency. We have examined mechanisms of segregation in Chinese hamster cell hybrids heterozygous at these two loci, both of which map to the long arm of Chinese hamster chromosome 2. To follow the fate of chromosomal arms through the segregation process, our hybrids were also heterozygous at the mtx (methotrexate resistance) locus on the short arm of chromosome 2 and carried cytogenetically marked chromosomes with either a short-arm deletion (2p-) or a long-arm addition (2q+). Karyotype and phenotype analysis of emetine- or chromate-resistant segregants from such hybrids allowed us to distinguish four potential segregation mechanisms: (i) loss of the emt+- or chr+-bearing chromosome; (ii) mitotic recombination between the centromere and the emt or chr loci, giving rise to homozygous resistant segregants; (iii) inactivation of the emt+ or chr+ alleles; and (iv) loss of the emt+- or chr+-bearing chromosome with duplication of the homologous chromosome carrying the emtr or chrr allele. Of 48 independent segregants examined, only 9 (20%) arose by simple chromosome loss. Two segregants (4%) were consistent with a gene inactivation mechanism, but because of their rarity, other mechanisms such as mutation or submicroscopic deletion could not be excluded. Twenty-one segregants (44%) arose by either mitotic recombination or chromosome loss and duplication; the two mechanisms were not distinguishable in that experiment. Finally, in hybrids allowing these two mechanisms to be distinguished, 15 segregants (31%) arose by chromosome loss and duplication, and none arose by mitotic recombination.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradley W. E. Reversible inactivation of autosomal alleles in Chinese hamster cells. J Cell Physiol. 1979 Nov;101(2):325–340. doi: 10.1002/jcp.1041010212. [DOI] [PubMed] [Google Scholar]
- Campbell C. E., Worton R. G. Evidence obtained by induced mutation frequency analysis for functional hemizygosity at the emt locus in CHO cells. Somatic Cell Genet. 1979 Jan;5(1):51–65. doi: 10.1007/BF01538786. [DOI] [PubMed] [Google Scholar]
- Campbell C. E., Worton R. G. Linkage of genetic markers emt and chr in Chinese hamster cells. Somatic Cell Genet. 1980 Mar;6(2):215–224. doi: 10.1007/BF01538797. [DOI] [PubMed] [Google Scholar]
- Chasin L. A. Non-linkage of induced mutations in Chinese hamster cells. Nat New Biol. 1972 Nov 8;240(97):50–52. doi: 10.1038/newbio240050a0. [DOI] [PubMed] [Google Scholar]
- Farrell S. A., Worton R. G. Chromosome loss is responsible for segregation at the HPRT locus in Chinese hamster cell hybrids. Somatic Cell Genet. 1977 Sep;3(5):539–551. doi: 10.1007/BF01539124. [DOI] [PubMed] [Google Scholar]
- Flintoff W. F., Davidson S. V., Siminovitch L. Isolation and partial characterization of three methotrexate-resistant phenotypes from Chinese hamster ovary cells. Somatic Cell Genet. 1976 May;2(3):245–261. doi: 10.1007/BF01538963. [DOI] [PubMed] [Google Scholar]
- Gupta R. S., Siminovitch L. The isolation and preliminary characterization of somatic cell mutants resistant to the protein synthesis inhibitor-emetine. Cell. 1976 Oct;9(2):213–219. doi: 10.1016/0092-8674(76)90112-4. [DOI] [PubMed] [Google Scholar]
- Harris J. F., Whitmore G. F. Segregation studies in CHO hybrid cells: I. Spontaneous and mutagen-induced segregation events of two recessive drug-resistant loci. Somatic Cell Genet. 1977 Mar;3(2):173–193. doi: 10.1007/BF01551813. [DOI] [PubMed] [Google Scholar]
- Harris M. Genetic and adaptive differences in the expression of drug resistance in hybrid cells. Somatic Cell Genet. 1979 Nov;5(6):793–808. doi: 10.1007/BF01542642. [DOI] [PubMed] [Google Scholar]
- Harris M. Non-mendelian segregation in hybrids between chinese hamster cells. J Cell Physiol. 1975 Oct;86(2 Pt 2 Suppl 1):413–429. doi: 10.1002/jcp.1040860413. [DOI] [PubMed] [Google Scholar]
- Huttner K. M., Ruddle F. H. Study of mitomycin C-induced chromosomal exchange. Chromosoma. 1976 Jun 30;56(1):1–13. doi: 10.1007/BF00293724. [DOI] [PubMed] [Google Scholar]
- Kuhn E. M. Localization by Q-banding of mitotic chiasmata in cases of Bloom's syndrome. Chromosoma. 1976 Aug 4;57(1):1–11. doi: 10.1007/BF00292945. [DOI] [PubMed] [Google Scholar]
- Käfer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- LITTLEFIELD J. W. SELECTION OF HYBRIDS FROM MATINGS OF FIBROBLASTS IN VITRO AND THEIR PRESUMED RECOMBINANTS. Science. 1964 Aug 14;145(3633):709–710. doi: 10.1126/science.145.3633.709. [DOI] [PubMed] [Google Scholar]
- Latt S. A. Localization of sister chromatid exchanges in human chromosomes. Science. 1974 Jul 5;185(4145):74–76. doi: 10.1126/science.185.4145.74. [DOI] [PubMed] [Google Scholar]
- Marin G. Selection of chromosomal segregants in a "hybrid" line of Syrian hamster fibroblasts. Exp Cell Res. 1969 Sep;57(1):29–36. doi: 10.1016/0014-4827(69)90363-2. [DOI] [PubMed] [Google Scholar]
- Merriam J. R., Garcia-Bellido A. A model for somatic pairing derived from somatic crossing over with third chromosome rearrangements in Drosophila melanogaster. Mol Gen Genet. 1972;115(4):302–313. doi: 10.1007/BF00333169. [DOI] [PubMed] [Google Scholar]
- PONTECORVO G., KAFER E. Genetic analysis based on mitotic recombination. Adv Genet. 1958;9:71–104. [PubMed] [Google Scholar]
- Parry J. M., Zimmerman F. K. The detection of monosomic colonies produced by mitotic chromosome non-disjunction in the yeast Saccharomyces cerevisiae. Mutat Res. 1976 Jul;36(1):49–66. doi: 10.1016/0027-5107(76)90020-8. [DOI] [PubMed] [Google Scholar]
- Pollard J. W., Stanners C. P. Characterization of cell lines showing growth control isolated from both the wild type and a leucyl-tRNA synthetase mutant of Chinese hamster ovary cells. J Cell Physiol. 1979 Mar;98(3):571–585. doi: 10.1002/jcp.1040980315. [DOI] [PubMed] [Google Scholar]
- RONEN A. INTERCHROMOSOMAL EFFECTS ON SOMATIC RECOMBINATION IN DROSOPHILA MELANOGASTER. Genetics. 1964 Oct;50:649–658. doi: 10.1093/genetics/50.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M., Mohandas T. Proposed banding nomenclature for the Chinese hamster chromosomes (Cricetulus griseus). Cytogenet Cell Genet. 1976;16(1-5):83–91. doi: 10.1159/000130559. [DOI] [PubMed] [Google Scholar]
- Rosenstraus M. J., Chasin L. A. Separation of linked markers in Chinese hamster cell hybrids: mitotic recombination is not involved. Genetics. 1978 Dec;90(4):735–760. doi: 10.1093/genetics/90.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddle F. H., Creagan R. P. Parasexual approaches to the genetics of man. Annu Rev Genet. 1975;9:407–486. doi: 10.1146/annurev.ge.09.120175.002203. [DOI] [PubMed] [Google Scholar]
- SHAW M. W., COHEN M. M. CHROMOSOME EXCHANGES IN HUMAN LEUKOCYTES INDUCED BY MITOMYCIN C. Genetics. 1965 Feb;51:181–190. doi: 10.1093/genetics/51.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch L. On the nature of hereditable variation in cultured somatic cells. Cell. 1976 Jan;7(1):1–11. doi: 10.1016/0092-8674(76)90249-x. [DOI] [PubMed] [Google Scholar]
- Stern C. Somatic Crossing over and Segregation in Drosophila Melanogaster. Genetics. 1936 Nov;21(6):625–730. doi: 10.1093/genetics/21.6.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant G. M., Holliday R. A search for allelic recombination in Chinese hamster cell hybrids. Mol Gen Genet. 1977 Nov 18;156(3):273–279. doi: 10.1007/BF00267182. [DOI] [PubMed] [Google Scholar]
- Therman E., Kuhn E. M. Cytological demonstration of mitotic crossing-over in man. Cytogenet Cell Genet. 1976;17(5):254–267. doi: 10.1159/000130721. [DOI] [PubMed] [Google Scholar]
- Thompson L. H., Baker R. M. Isolation of mutants of cultured mammalian cells. Methods Cell Biol. 1973;6:209–281. doi: 10.1016/s0091-679x(08)60052-7. [DOI] [PubMed] [Google Scholar]
- Wasmuth J. J., Chu L. Y. Linkage in cultured Chinese hamster cells of two genes, emtB and leuS, involved in protein synthesis and isolation of cell lines with mutations in three linked genes. J Cell Biol. 1980 Dec;87(3 Pt 1):697–702. doi: 10.1083/jcb.87.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worton R. G., Duff C. Karyotyping. Methods Enzymol. 1979;58:322–344. doi: 10.1016/s0076-6879(79)58148-8. [DOI] [PubMed] [Google Scholar]
- Worton R. G., Ho C. C., Duff C. Chromosome stability in CHO cells. Somatic Cell Genet. 1977 Jan;3(1):27–45. doi: 10.1007/BF01550985. [DOI] [PubMed] [Google Scholar]
- Worton R., Duff C., Flintoff W. Microcell-mediated cotransfer of genes specifying methotrexate resistance, emetine sensitivity, and chromate sensitivity with Chinese hamster chromosome 2. Mol Cell Biol. 1981 Apr;1(4):330–335. doi: 10.1128/mcb.1.4.330. [DOI] [PMC free article] [PubMed] [Google Scholar]