Abstract
The family with sequence similarity 20 (Fam20) kinases phosphorylate extracellular substrates and play important roles in biomineralization. Fam20C is the Golgi casein kinase that phosphorylates secretory pathway proteins within Ser-x-Glu/pSer motifs. Mutations in Fam20C cause Raine syndrome, an osteosclerotic bone dysplasia. Here we report the crystal structure of the Fam20C ortholog from Caenorhabditis elegans. The nucleotide-free and Mn/ADP-bound structures unveil an atypical protein kinase-like fold and highlight residues critical for activity. The position of the regulatory αC helix and the lack of an activation loop indicate an architecture primed for efficient catalysis. Furthermore, several distinct elements, including the presence of disulfide bonds, suggest that the Fam20 family diverged early in the evolution of the protein kinase superfamily. Our results reinforce the structural diversity of protein kinases and have important implications for patients with disorders of biomineralization.
Keywords: amelogenesis imperfecta, Fam20A, hypophosphatemia, Fam20B, enamel renal syndrome
Protein phosphorylation is a fundamental mechanism that regulates numerous physiological processes (1, 2). Protein kinases have evolved to function as dynamic switches relaying signals in response to various stimuli by transferring a phosphate from ATP to target proteins (3–5). Most phosphoproteins are found within the cell, residing in the nuclei and cytosol; however, secreted proteins, including casein and other members of the secretory calcium-binding phosphoprotein family, are phosphorylated as well (6). We recently identified a family of kinases that reside in the secretory pathway and function to phosphorylate extracellular substrates (7, 8). One member of this family, Fam20C (family with sequence similarity 20, member C), is the physiological casein kinase that phosphorylates multiple secreted proteins within a Ser-x-Glu/pSer motif (7, 9, 10). The importance of this discovery is underscored by the fact that some 75% of the phosphoproteins identified in human serum and cerebrospinal fluid contain phosphate within this motif (11–13). Furthermore, mutations in FAM20C cause Raine syndrome, a deadly osteosclerotic bone dysplasia characterized by generalized osteosclerosis, ectopic calcifications, and characteristic facial features (14–16). Most individuals with Raine syndrome die within a few weeks after birth; however, nonlethal cases with dental abnormalities and clinical features of hypophosphatemia have been reported (17, 18). Loss of Fam20C in mice also results in severe bone and tooth anomalies, as well as hypophosphatemia (19–21).
Two other closely related Fam20C paralogs, Fam20A and Fam20B, are present in humans (22). Fam20B is ubiquitously expressed and phosphorylates xylose within the tetrasaccharide linkage region of proteoglycans (23). This phosphorylation event may influence glycosaminoglycan biosynthesis (24). Genetic deletion of Fam20B in mice results in embryonic lethality at E13.5, and mutations in Danio rerio result in reduced cartilage matrix production and skeletal defects (19, 25). The substrates for Fam20A are unknown; however, mutations in FAM20A are known to cause amelogenesis imperfecta (AI) and enamel renal syndrome (ERS) in humans (26–29). AI is a clinically and genetically heterogeneous group of disorders characterized by improper formation of enamel, a highly mineralized tissue comprising the outer surface of the vertebrate tooth (30). Patients with ERS have clinical features of AI along with renal calcifications (31). Fam20A expression in mice is apparently restricted to secretory ameloblasts and the parathyroid gland, the latter of which is important for calcium homeostasis. Loss of Fam20A in mice leads to AI and ectopic calcifications (19).
Here we report the crystal structure of the Fam20C ortholog from Caenorhabditis elegans. C. elegans Fam20 exhibits a protein kinase-like fold contained within a shell-like structure, formed by an N-terminal segment and a novel insertion domain. The Mn/ADP-bound structure reveals critical residues important for nucleotide binding and catalysis. The unique architecture of the kinase suggests that Fam20C is an efficient catalyst as opposed to a dynamically regulated enzyme. Our results highlight the structural diversity of the protein kinase superfamily and have important implications for several human diseases.
Results and Discussion
Overall Structure of C. elegans Fam20.
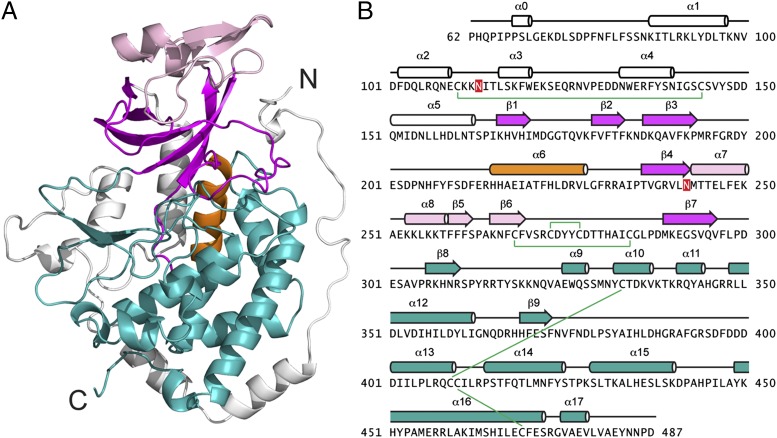
We determined the crystal structure of the single Fam20 ortholog in C. elegans (ceFam20, residues 60–512) at 2.6-Å resolution (Fig. 1A and Table S1). The asymmetric unit contains four ceFam20 molecules that are highly similar (rmsd 0.4–0.5 Å for the Cα atoms). The C-terminal 25 residues are missing from all four molecules and are presumably disordered. Residues 120–129 have poor electron densities and are built in only one molecule.
Fig. 1.
C. elegans Fam20 exhibits a protein kinase-like fold contained in a shell-like structure. (A) Ribbon representation of ceFam20. The N- and C-lobes are shown in magenta and teal, respectively. The α6 helix (PKA αC equivalent) is highlighted in orange. The N-terminal segment and the insertion domain are in white and pink, respectively. (B) The amino acid sequence of ceFam20 is shown depicting the secondary structural elements, color-coded as in A. The disulfide bonds are indicated with green lines, and the N-linked glycosylation sites are highlighted in red.
ceFam20 displays a relatively large kinase domain, consisting of 18 α-helices and 9 β-strands (Fig. 1 A and B). The kinase core has a two-lobe structure (N-lobe and C-lobe), which is characteristic of all protein kinases. A database search using Dali suggested that ceFam20 is more similar to several atypical kinases, including the bacterial kinases high persistence factor A (HipA) and cell translocating kinase A (CtkA), the slime mold actin-fragmin kinase (AFK), and the phosphatidylinositol 3-kinase–related kinase family (32, 33) (Fig. S1). ceFam20 is contained in a shell-like structure formed by an N-terminal segment (α0-α5) and a novel insertion domain (α7-α8, β5-β6) (Fig. 1 A and B). The N-terminal segment is wrapped around the lower half of the molecule and forms the base for the C-lobe. The Pro-rich N-terminal region, present in all Fam20 family members, has a well-ordered structure containing a helical turn (α0) and a long loop that covers one side of the molecule. The insertion domain has a novel fold and forms a cap-like structure that covers the N-lobe. Primary sequence analyses likely failed to identify the Fam20 family as kinases because this domain lies between β4 and β7 of the N-lobe, disrupting its sequence continuity.
ceFam20 contains five disulfide bonds (Cys110-Cys144, Cys268-Cys284, Cys273-Cys277, Cys333-Cys409, and Cys410-Cys479; Fig. 1B). All but Cys110-Cys144 are evolutionarily conserved among all Fam20 family members. Furthermore, Asn113 and 242 are N-linked glycosylated, a common posttranslational modification in secretory pathway proteins (Fig. 1B and Fig. S2). Notably, two pairs of conserved disulfide bonds (Cys268-Cys284 and Cys273-Cys277) and an N-linked glycosylated Asn (Asn242) are present in the insertion domain (Fig. 1B).
N-lobe.
The N-lobe is composed of a five-stranded β-sheet including β1–β4 and β7. It shows some resemblance to other protein kinases and can be superimposed onto the N-lobe of protein kinase A (PKA) with an rmsd of 3.3 Å over 70 Cα atoms (Fig. S3A). A long loop present between β1 and β2 functions as the lid for the ATP-binding pocket, similar to the Gly-rich loop in other kinases (3). An α-helix, known as αC, is universally present in the N-lobe of eukaryotic protein kinases. It is highly dynamic in nature and plays a critical role in regulating kinase activity. Interestingly, the equivalent helix in ceFam20 (α6) is located predominantly in the C-lobe (Fig. 1A). It occupies a position in the center of the molecule and intimately interacts with several helices in the C-lobe. Many interactions must be broken for this helix to move; thus, this helix is unlikely to play a similar regulatory role as that in canonical protein kinases.
C-lobe.
The C-lobe has a very different organization than that of most kinases (Fig. S3B). Structural superposition of ceFam20 and PKA reveals only two common secondary structural elements, α12 (ceFam20)/αE (PKA) and β9 (ceFam20)/β7 (PKA). The αF helix, which is present in all protein kinases, is displaced in ceFam20. This helix functions as a central scaffold to maintain the kinase structure (3). In ceFam20, α13 corresponds most closely to αF and contains a conserved Pro (Pro405) in the middle, which bends the helix 45 degrees downward compared with the position of αF in PKA. A long loop between β8 and α9 partially occupies the place of αF, and this loop may be involved in substrate recognition (see below). Furthermore, the GHI subdomain (helices αG, αH, and αI in PKA), a hallmark of eukaryotic protein kinases involved in protein–protein interactions and/or allosteric regulation, is absent in ceFam20 (3). Thus, the unique structural features specific to ceFam20 suggest that the Fam20 family diverged from canonical eukaryotic protein kinases at an early stage of evolution.
Fam20C Activity and Specificity Are Evolutionarily Conserved.
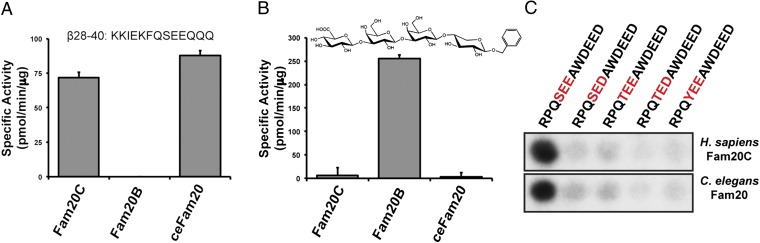
Fam20C and Fam20B have a high degree of sequence similarity, but drastically different substrate specificities. Because ceFam20 shares comparable sequence similarity with both Fam20C (63%) and Fam20B (57%) within their kinase domains, we asked whether ceFam20 was a protein kinase or a xylosyl kinase (Fig. S4). ceFam20 efficiently phosphorylated the Fam20C-specific peptide substrate, KKIEKFQSEEQQQ (β28–40), but not a model Fam20B substrate containing the tetrasaccharide proteoglycan linkage glucuronic acid-β1–3-galactose-β1–3-galactose-β1–4-xylose-benzene (GlcAβ1–3Galβ1–3Galβ1–4Xylβ1-O-benzyl) (Fig. 2 A and B). Similar to human Fam20C, ceFam20 phosphorylated Ser more efficiently than either Thr or Tyr and required Glu at the n+2 position (Fig. 2C). Steady-state kinetic analyses found Km values for Mn/ATP of 0.8 μM for Fam20C and 1.2 μM, for ceFam20 (Fig. S5 A and C). The Km values of both kinases for Mg/ATP were ∼100-fold higher, indicating a strong preference for Mn/ATP (Fig. S5 B and D). It is not uncommon for Golgi-localized enzymes to prefer Mn2+ as a cofactor (34). The remarkably low Km values are consistent with the idea that these kinases have evolved to function in the lumen of the secretory pathway and/or in the extracellular space, where ATP concentrations are likely to be much lower compared with the cytosol.
Fig. 2.
Fam20C activity and specificity are evolutionarily conserved. (A) Human Fam20C and ceFam20 phosphorylated the Fam20C model peptide substrate β28–40, whereas Fam20B did not. (B) Fam20B phosphorylated a model substrate representing the tetrasaccharide core linkage region in proteoglycans (GlcAβ1–3Galβ1–3Galβ1–4Xylβ1-O-benzyl), whereas Fam20C and ceFam20 did not. (C) Peptides containing variants of the Ser-x-Glu Fam20C consensus motif were synthesized to assess the specificity of ceFam20. The Ser was changed to Thr or Tyr, and the n+2 residue was changed to Asp. Incorporation of phosphate was monitored by autoradiography.
Nucleotide-Binding Site.
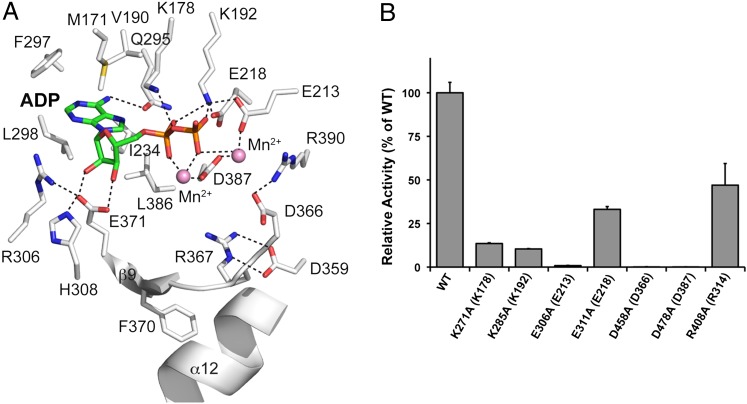
We determined the crystal structure of ceFam20 in complex with Mn/ADP at 3.05 Å resolution (Fig. S6 A and B and Table S1). The Mn/ADP-bound structure is highly similar to the nucleotide-free (apo) structure, with an rmsd of 0.5 Å for the Cα atoms. The adenine nucleotide is located in the cleft between the N-lobe and C-lobe. In-depth analysis revealed a common, yet intricately distinct mechanism of nucleotide binding that highlights the structural diversity of the protein kinase family (Fig. 3A).
Fig. 3.
The Mn/ADP-bound structure of ceFam20 reveals critical residues required for catalysis. (A) Enlarged image of the nucleotide-binding pocket showing the detailed molecular interactions important for catalysis. The ADP molecule and the two Mn2+ ions are shown as sticks and spheres, respectively. Salt bridge and hydrogen bond interactions are shown as dashed lines. (B) Human Fam20C mutants display reduced kinase activity. Mutant Fam20C proteins were assayed against β28–40 peptide, and their activity relative to that of the WT enzyme is depicted graphically. The amino acids in brackets indicate the corresponding residues in ceFam20.
β1-β2 loop.
On nucleotide binding, the β1-β2 loop moves slightly toward the ADP molecule to shelter the nucleotide-binding pocket (Fig. S6A). The β1-β2 loop is not as rich in Gly residues as canonical protein kinases and features a highly conserved Gxx(L/V)K motif (Fig. S4). The L/V residue (Val177 in ceFam20) anchors the C-terminal end of the loop to the insertion domain through hydrophobic interactions. We hypothesize that the insertion domain might impact the conformation of the β1-β2 loop and function to regulate nucleotide binding. The highly conserved Lys (Lys178 in ceFam20) forms hydrogen bonds with both ADP and the main chain atoms of the β1-β2 loop in the ADP-bound structure (Fig. 3A). Mutation of this Lys impairs kinase activity, consistent with its critical role in the structural organization of this region (Fig. 3B). This Lys is not commonly present in canonical kinases but is found in HipA and CtkA, two bacterial kinases that are remotely related to the Fam20 family (7, 35, 36).
The β1-β2 loop is different in the Fam20A subfamily, containing an extra segment comprising 17 residues that include a pair of invariant cysteines (Fig. S4). This extension appears to be exclusive to Fam20A, because BLAST searches using this sequence as a query only detected Fam20A homologs. How this segment may affect the conformation of the β1-β2 loop and nucleotide binding in Fam20A is unclear.
Ion pair.
Almost all protein kinases contain a conserved Lys in β3 and a Glu in αC (Lys72 and Glu91 in PKA). Formation of the ion pair between these two residues is considered a hallmark of an active kinase. The Lys coordinates the α- and β-phosphates of ATP, and the Glu stabilizes the conformation of the Lys. In ceFam20, Lys192 and Glu218 form the corresponding ion pair, and mutations in these residues impair kinase activity (Fig. 3 A and B). Interestingly, Lys192 also forms a salt bridge with Glu213, which coordinates a Mn2+ ion (Fig. 3A). Consistent with Glu213’s dual function, its mutation has a more drastic effect on kinase activity compared with Glu218 mutation (Fig. 3B). The coordination between Lys192 (nucleotide binding) and Glu213 (metal binding) reflects another unique property of the Fam20 family of kinases. Importantly, the configuration of the active site in both the Apo- and Mn/ADP-bound structures suggest that the kinase adopts an active conformation.
DFG motif.
In canonical protein kinases, the DFG motif plays an important role in metal binding and catalysis. A variant of this motif, D(N/H)(A/G), is present in the Fam20 family members and is located between β9 and α13 in ceFam20. In the Mn/ADP-bound structure, the Asp in this motif (Asp387) has an altered side chain conformation to engage both Mn2+ ions (Fig. 3A). Mutation of the corresponding Asp abolishes the kinase activity of human Fam20C in vitro and in vivo (7). His388 and Gly389 play important structural roles in establishing the position of Asp387. The conserved Leu (Leu386) preceding Asp387 is part of the hydrophobic pocket that accommodates the adenosine moiety of the adenine nucleotide.
Adenine-binding pocket and gatekeeper.
A network of residues, including Met171, Val190, Ile234, Gln295, Phe297, Leu298, Glu371, and Leu386, participate in binding to the adenosine group, mainly through hydrophobic interactions (Fig. 3A). Met171 lies at the beginning of the β1-β2 loop. A Lys often occupies this position in other Fam20 homologs and likely interacts with the adenine ring through a cation–π interaction. Val190, Ile234, Phe297, Leu298, and Leu386 are conserved or substituted by similar hydrophobic residues in the Fam20 family members (Fig. S4).
Most protein kinases contain a bulky hydrophobic residue in the adenine-binding pocket, known as the “gatekeeper” (37). This residue dictates the selectivity for nucleotides and small-molecule inhibitors. Chemical genetic studies have shown that substitution of this residue to a smaller amino acid, such as Gly or Ala, permits the use of bulky ATP analogs as substrates (38). In ceFam20, a Gln (Gln295) occupies the gatekeeper position and forms a hydrogen bond with the adenine ring (Fig. 3A); however, this residue is not a typical hydrophobic gatekeeper and is not highly conserved (Fig. S4). An Ala is often present in Fam20C and may provide a unique opportunity to design compounds that can modulate kinase activity.
Fam20C is remarkably insensitive to the broad-spectrum ATP-competitive kinase inhibitor staurosporine (7, 39). Close examination of the nucleotide-binding pocket of ceFam20 suggests that staurosporine likely will not be accommodated (Fig. S7). The long β1-β2 loop (Gly-rich loop) will impede entry of the compound. Furthermore, several residues, including Lys178 and Lys192, would be expected to clash with the aromatic regions of staurosporine. Thus, the structure of ceFam20 provides molecular insight into this unusual insensitivity.
Catalytic segment.
The catalytic segment includes the putative catalytic Asp (Asp366) and is located between α12 and β9 in ceFam20. Mutation of this Asp completely abolishes kinase activity (Fig. 3B). Interestingly, in both the Apo- and Mn/ADP-bound structures, a salt bridge is formed between Asp366 and a conserved Arg (Arg390) that immediately follows the DFG motif (Fig. 3A). This interaction likely prevents Asp366 from acting as a general base to activate the serine hydroxyl in the protein substrate. We anticipate that this ion pair will be broken once the protein substrate and/or ATP bind, and that Arg390 may engage the γ-phosphate of ATP. In any event, the ion pair between this conserved Arg and the catalytic Asp may represent a unique mechanism for regulating Fam20 kinases.
Arg367 following the catalytic Asp forms bidentate interactions with another invariant Asp (Asp359) (Fig. 3A). The importance of this interaction is underscored by the fact that mutation of this Asp to Asn causes Raine syndrome (Table S2) (17). This substitution is predicted to destabilize the conformation of the catalytic segment and alter both the stability and activity of the kinase. Indeed, this mutation was found to prevent Fam20C secretion and diminish its ability to phosphorylate osteopontin (7).
A conserved Phe (Phe370) is also part of the catalytic segment and interacts with α12 although hydrophobic interactions, thereby stabilizing the conformation of the segment (Fig. 3A). Furthermore, Glu371 forms two hydrogen bonds with the ribose group of ADP, and its conformation is stabilized by salt bridge interactions from Arg306 and His308 in β8. Thus, extensive molecular interactions are present to stabilize the catalytic segment and coordinate nucleotide binding.
Insights into Substrate Selectivity.
The Fam20C subfamily phosphorylates Ser-x-Glu/pSer motifs in secreted proteins. In addition, Fam20C can phosphorylate peptides from proline-rich phosphoprotein-1 (PRP-1) within a related motif, Ser-x-Gln-x-x-Asp-Glu-Glu (40). Lacking a structure with peptide bound, we cannot describe the exact molecular basis for these specificities; however, it is interesting that three basic residues (Arg314, Lys319, and Lys320) were present near the active site within the β8-α9 loop (Fig. 4A). Importantly, these residues were not present in Fam20A and Fam20B yet, are highly conserved in Fam20C (Fig. 4B). Mutation of Arg314 (Arg408 in human Fam20C) diminished the activity of Fam20C, consistent with an important role in substrate binding (Fig. 3B). Based on the location of Arg314 relative to the catalytic Asp (Asp366), we anticipate that Arg314 may interact with the Glu or pSer residues at the n+2 position in the peptide substrate. In the PRP1 peptide, a Gln at the n+2 position would still be able to form a hydrogen bond with Arg314; however, additional interactions in the distal region of the peptide would be required for efficient binding (n+5, 6, and 7 positions). Lys319 and Lys320 are optimally positioned to facilitate such interactions (Fig. 4A). This idea is consistent with the fact that Ser-x-Gln-x-x-Asp-Glu-Glu is phosphorylated and requires the Gln at the n+2 position and the three acidic residues at the n+5, 6, and 7 positions (40). Substitution of these residues with alanines prevented phosphorylation by Fam20C.
Fig. 4.
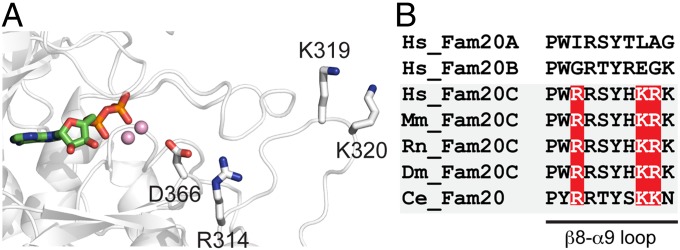
Insights into substrate specificity. (A) Structural representation of the active site of ceFam20, highlighting residues potentially involved in substrate recognition. The Mn/ADP, the catalytic Asp (D366), and three Fam20C-specific basic residues (R314, K319, and K320) are shown. (B) Sequence alignment of the β8-α9 loop within the Fam20 family members. The three Fam20C-specific basic residues are highlighted.
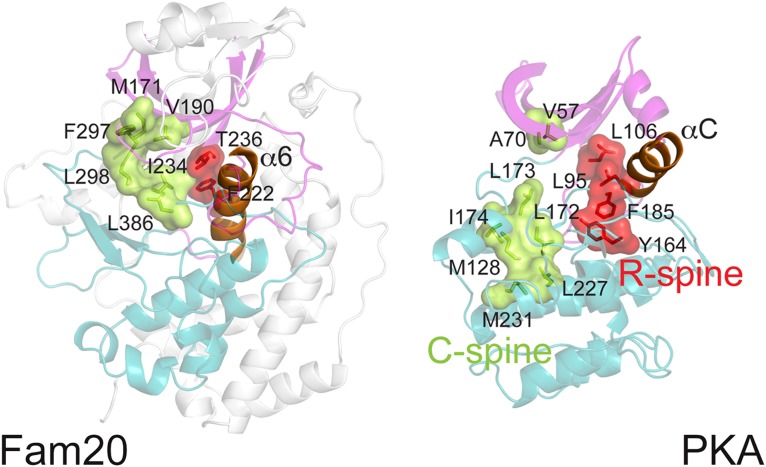
Regulatory and Catalytic Spines.
Two hydrophobic “spine” structures exist in the active conformation of protein kinases: a regulatory spine (R-spine) and a catalytic spine (C-spine) (41) (Fig. 5). The dynamic nature of the R-spine plays a critical role in the regulation of protein kinases. In PKA, the R-spine consists of four hydrophobic residues, two from each lobe, including the Phe in the DFG motif. Two N-lobe residues are present in ceFam20, Thr236 from β4 and Phe222 from α6 (αC equivalent); however, Fam20 family members contain a His or Asn instead of a Phe in the DFG motif. These hydrophilic residues do not contribute to formation of the R-spine. The α6 helix appears to function as the C-terminal portion of the spine. This unique architecture likely restrains ceFam20 from undergoing highly dynamic conformational changes. To the best of our knowledge, the sole active protein kinase with a hydrophilic residue (Ser) at the Phe position in the DFG motif is AFK from slime mold (42). Of note, the αC in AFK is situated at a similar position as the α6 in ceFam20 (Fig. S1).
Fig. 5.
ceFam20 has unique spine structures. The N-lobe, C-lobe, and αC helix of PKA are colored in magenta, teal, and orange, respectively. Corresponding regions in ceFam20 are highlighted in the same color scheme. The regulatory and catalytic spines are depicted in red and green, respectively. Residues involved in the formation of the spines are indicated.
Most protein kinases are activated by a phosphorylation event within an activation loop following the DFG motif (43). The analogous loop in ceFam20 is not phosphorylated, however (Fig. S8). Furthermore, this loop interacts with several regions of the protein and does not have the flexibility to transiently regulate kinase activity. Collectively, these features suggest that Fam20C is constitutively active and does not require phosphorylation or significant conformational changes for catalysis.
The C-spine in ceFam20 also displays some unique features (Fig. 5). In canonical protein kinases, the C-spine contains a gap that is filled by the adenine ring of ATP on nucleotide binding. In ceFam20 this gap is absent, and the C-spine is continuous in the absence of nucleotide. A cradle-like structure accommodates the adenosine group of the adenine nucleotide, as described above (Fig. 3A).
Implications for Human Diseases.
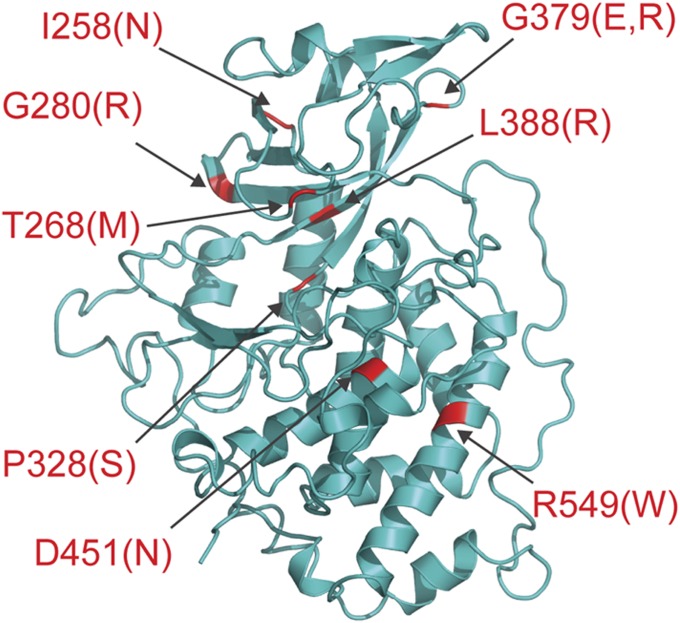
Mutations in Fam20C cause Raine syndrome, an autosomal recessive osteosclerotic bone dysplasia. Most individuals with Raine syndrome succumb within the first few weeks of life; however, mutations associated with nonlethal forms of the disease have been documented, and some of these patients present with dental anomalies and hypophosphatemia. To understand how these mutations lead to disease, we modeled the structure of human Fam20C using the ceFam20 structure (Fig. 6 and Table S2). We mapped the eight known missense mutations to the structure model and examined their potential impacts.
Fig. 6.
Structure modeling provides insights into Fam20C-related human disorders. The homology structure model of human Fam20C is shown as a ribbon diagram. The model was generated using Swiss-Model (44). Disease mutations are highlighted in red.
Ile258, Gly280, Pro328, Gly379, and Leu388 all reside in hydrophobic environments. Our ceFam20 structures suggest that introduction of a hydrophilic residue at any of these positions would disrupt the local region and have a detrimental effect on protein folding/stability. Indeed, the mutant proteins were not secreted from the osteosarcoma cell line U2OS (7). As described above, mutation of Asp451 to an Asn likely would destabilize the catalytic segment by disrupting the Asp-Arg salt bridge present in the WT protein. Arg549 is located in α16 and has multiple interactions within the molecule. Mutation to a Trp would severely interfere with packing of the C-lobe and disrupt protein integrity. Recently, Rafaelsen et al. (18) identified compound heterozygous mutations in FAM20C in siblings with clinical presentation of dental abnormalities, ectopic calcifications, and fibroblast growth factor 23-related hypophosphatemia. A novel missense mutation, Thr268Met, was reported in this study. This Thr is highly conserved and located in the β1-β2 loop (Gly-rich loop). Mutation of this residue likely would inhibit ATP binding and lead to impaired kinase activity.
Mutations in Fam20A are linked to the human disorders AI and ERS. Most mutations are inactivating and result in truncated versions of the protein; however, two missense mutations, Gly331Asp and Leu173Arg, have been reported in patients with ERS (Fig. S9). Interestingly, Gly331 corresponds to Gly379 in Fam20C, a residue that is also mutated in Raine syndrome (Fig. 6 and Table S2). Similar to the situation in Fam20C, mutation of this residue in Fam20A to an Asp would affect the local hydrophobic environment and result in formation of an unstable/unfolded protein. Structural modeling suggests that Leu173 is part of the N-terminal segment and has extensive interactions with the kinase core. An Arg substitution at this position would disrupt this interaction and perturb the kinase. Collectively, these observations provide a molecular explanation of how Fam20C and Fam20A mutations can negatively impact kinase structure and/or activity, leading to disorders of biomineralization.
Conclusion
We have demonstrated that Fam20C activity and specificity are evolutionarily conserved. The crystal structure of the Fam20C ortholog from C. elegans reveals the unique architecture of a secretory pathway-specific kinase. The distinctive structural features of the Fam20 family suggest that this family diverged early from canonical protein kinases and is constitutively active. Our results expand the repertoire of the structural kinome and have important implications for human disorders of biomineralization.
Methods
Protein Expression and Purification.
C. elegans Fam20 (residues 60–512) was cloned into a modified pI-secSUMOstar vector (LifeSensors) containing a tobacco etch virus (TEV) protease cleavage site. The protein was expressed and purified using recombinant baculovirus (20). More details on the protein purification procedure are presented in SI Methods.
Enzyme Assays.
Fam20C, Fam20B, and ceFam20 activity was monitored against the peptide KKIEKFQSEEQQQ (β28–40) as described previously (7) and GlcAβ1–3Galβ1–3Galβ1–4Xylβ1-O-benzyl (a generous gift from Dr. Jeffrey Esko, University of California at San Diego). The enzymatic assays are described in SI Methods.
Crystallization and Structure Determination.
The ceFam20 crystals were grown at 20 °C using the sitting-drop vapor diffusion method. Details of the crystallization and structure determination procedures are provided in SI Methods.
Supplementary Material
Acknowledgments
We are grateful to the Advanced Light Source (beamlines 8.2.1 and 8.2.2) staff at Lawrence Berkeley National Laboratory for beam access and help with data collection. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under Contract DE-AC02-05CH11231. We thank David King of the Howard Hughes Medical Institute’s Mass Spectrometry Laboratory (University of California Berkeley) for peptide synthesis and James Engel for technical assistance. We also thank Dr. Gregory Taylor, Dr. Alexandr Kornev, Dr. Lisa Kinch, and Dr. Carolyn Worby for their critical reading of the manuscript, and members of the Dixon laboratory for helpful discussions. Special thanks to Dr. Susan Taylor and Dr. Philip Bourne for their insightful comments. This work was supported by National Institutes of Health Grants DK018849 and DK018024 (to J.E.D.) and National Institutes of Health/National Cancer Institute Training Grant T32 CA009523 (to V.S.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4KQA and 4KQB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309211110/-/DCSupplemental.
References
- 1.Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4(5):E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 2.Fischer EH. Cellular regulation by protein phosphorylation. Biochem Biophys Res Commun. 2013;430(2):865–867. doi: 10.1016/j.bbrc.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 3.Taylor SS, Kornev AP. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36(2):65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109(3):275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 5.Endicott JA, Noble ME, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- 6.Hammarsten O. Zur frage ob caseín ein einheitlicher stoff sei. Zeitschrift Physiol Chemie. 1883;7:227–273. [Google Scholar]
- 7.Tagliabracci VS, et al. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 2012;336(6085):1150–1153. doi: 10.1126/science.1217817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tagliabracci VS, Pinna LA, Dixon JE. Secreted protein kinases. Trends Biochem Sci. 2013;38(3):121–130. doi: 10.1016/j.tibs.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lolli G, et al. Inhibition of protein kinase CK2 by flavonoids and tyrphostins: A structural insight. Biochemistry. 2012;51(31):6097–6107. doi: 10.1021/bi300531c. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa HO, Xu A, Ogura E, Manning G, Irvine KD. The Raine syndrome protein FAM20C is a Golgi kinase that phosphorylates bio-mineralization proteins. PLoS ONE. 2012;7(8):e42988. doi: 10.1371/journal.pone.0042988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahl JM, Jensen SS, Larsen MR, Heegaard NH. Characterization of the human cerebrospinal fluid phosphoproteome by titanium dioxide affinity chromatography and mass spectrometry. Anal Chem. 2008;80(16):6308–6316. doi: 10.1021/ac800835y. [DOI] [PubMed] [Google Scholar]
- 12.Zhou W, et al. An initial characterization of the serum phosphoproteome. J Proteome Res. 2009;8(12):5523–5531. doi: 10.1021/pr900603n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvi M, Cesaro L, Tibaldi E, Pinna LA. Motif analysis of phosphosites discloses a potential prominent role of the Golgi casein kinase (GCK) in the generation of human plasma phospho-proteome. J Proteome Res. 2010;9(6):3335–3338. doi: 10.1021/pr100058r. [DOI] [PubMed] [Google Scholar]
- 14.Raine J, Winter RM, Davey A, Tucker SM. Unknown syndrome: Microcephaly, hypoplastic nose, exophthalmos, gum hyperplasia, cleft palate, low-set ears, and osteosclerosis. J Med Genet. 1989;26(12):786–788. doi: 10.1136/jmg.26.12.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson MA, et al. Mutations in FAM20C are associated with lethal osteosclerotic bone dysplasia (Raine syndrome), highlighting a crucial molecule in bone development. Am J Hum Genet. 2007;81(5):906–912. doi: 10.1086/522240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fradin M, et al. Osteosclerotic bone dysplasia in siblings with a Fam20C mutation. Clin Genet. 2011;80(2):177–183. doi: 10.1111/j.1399-0004.2010.01516.x. [DOI] [PubMed] [Google Scholar]
- 17.Simpson MA, et al. Mutations in FAM20C also identified in non-lethal osteosclerotic bone dysplasia. Clin Genet. 2009;75(3):271–276. doi: 10.1111/j.1399-0004.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- 18.Rafaelsen SH, et al. Exome sequencing reveals FAM20c mutations associated with FGF23-related hypophosphatemia, dental anomalies and ectopic calcification. J Bone Miner Res. 2013;28(6):1378–1385. doi: 10.1002/jbmr.1850. [DOI] [PubMed] [Google Scholar]
- 19.Vogel P, et al. Amelogenesis imperfecta and other biomineralization defects in Fam20a and Fam20c null mice. Vet Pathol. 2012;49(6):998–1017. doi: 10.1177/0300985812453177. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, et al. Inactivation of a novel FGF23 regulator, FAM20C, leads to hypophosphatemic rickets in mice. PLoS Genet. 2012;8(5):e1002708. doi: 10.1371/journal.pgen.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, et al. FAM20C plays an essential role in the formation of murine teeth. J Biol Chem. 2012;287(43):35934–35942. doi: 10.1074/jbc.M112.386862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nalbant D, et al. FAM20: An evolutionarily conserved family of secreted proteins expressed in hematopoietic cells. BMC Genomics. 2005;6:11. doi: 10.1186/1471-2164-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koike T, Izumikawa T, Tamura J, Kitagawa H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem J. 2009;421(2):157–162. doi: 10.1042/BJ20090474. [DOI] [PubMed] [Google Scholar]
- 24.Nadanaka S, et al. EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J Biol Chem. 2013;288(13):9321–9333. doi: 10.1074/jbc.M112.416909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eames BF, et al. Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 2011;7(8):e1002246. doi: 10.1371/journal.pgen.1002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Sullivan J, et al. Whole-exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am J Hum Genet. 2011;88(5):616–620. doi: 10.1016/j.ajhg.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho SH, et al. Novel FAM20A mutations in hypoplastic amelogenesis imperfecta. Hum Mutat. 2012;33(1):91–94. doi: 10.1002/humu.21621. [DOI] [PubMed] [Google Scholar]
- 28.Jaureguiberry G, et al. Nephrocalcinosis (enamel renal syndrome) caused by autosomal recessive FAM20A mutations. Nephron, Physiol. 2012;122(1-2):1–6. doi: 10.1159/000349989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang SK, et al. FAM20A mutations can cause enamel-renal syndrome (ERS) PLoS Genet. 2013;9(2):e1003302. doi: 10.1371/journal.pgen.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephanopoulos G, Garefalaki ME, Lyroudia K. Genes and related proteins involved in amelogenesis imperfecta. J Dent Res. 2005;84(12):1117–1126. doi: 10.1177/154405910508401206. [DOI] [PubMed] [Google Scholar]
- 31.Lubinsky M, Angle C, Marsh PW, Witkop CJ., Jr Syndrome of amelogenesis imperfecta, nephrocalcinosis, impaired renal concentration, and possible abnormality of calcium metabolism. Am J Med Genet. 1985;20(2):233–243. doi: 10.1002/ajmg.1320200205. [DOI] [PubMed] [Google Scholar]
- 32.Scheeff ED, Bourne PE. Structural evolution of the protein kinase-like superfamily. PLOS Comput Biol. 2005;1(5):e49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulberti S, et al. The functional glycosyltransferase signature sequence of the human beta 1,3-glucuronosyltransferase is a XDD motif. J Biol Chem. 2003;278(34):32219–32226. doi: 10.1074/jbc.M207899200. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher MA, et al. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323(5912):396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, et al. Helicobacter pylori proinflammatory protein up-regulates NF-kappaB as a cell-translocating Ser/Thr kinase. Proc Natl Acad Sci USA. 2010;107(50):21418–21423. doi: 10.1073/pnas.1010153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–795. doi: 10.1146/annurev-biochem-090308-173656. [DOI] [PubMed] [Google Scholar]
- 38.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407(6802):395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 39.Lasa M, Marin O, Pinna LA. Rat liver Golgi apparatus contains a protein kinase similar to the casein kinase of lactating mammary gland. Eur J Biochem. 1997;243(3):719–725. doi: 10.1111/j.1432-1033.1997.00719.x. [DOI] [PubMed] [Google Scholar]
- 40.Brunati AM, Marin O, Bisinella A, Salviati A, Pinna LA. Novel consensus sequence for the Golgi apparatus casein kinase, revealed using proline-rich protein-1 (PRP1)-derived peptide substrates. Biochem J. 2000;351(Pt 3):765–768. [PMC free article] [PubMed] [Google Scholar]
- 41.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103(47):17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinbacher S, et al. The crystal structure of the Physarum polycephalum actin-fragmin kinase: An atypical protein kinase with a specialized substrate-binding domain. EMBO J. 1999;18(11):2923–2929. doi: 10.1093/emboj/18.11.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kornev AP, Taylor SS. Defining the conserved internal architecture of a protein kinase. Biochim Biophys Acta. 2010;1804(3):440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37(Database issue):D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.