Abstract
The RAD51 recombinase plays a central role in homologous recombination (HR), which is critical for repair of DNA double-strand breaks, maintenance of genomic stability, and prevention of developmental disorders and cancer. Here, we report the identification of an RAD51-binding protein fidgetin-like 1 (FIGNL1). FIGNL1 specifically interacts with RAD51 through its conserved RAD51 binding domain. Cells depleted of FIGNL1 show defective HR repair. Interestingly, FIGNL1 is recruited to sites of DNA damage in a manner that is independent of breast cancer 2, early onset, RAD51, and probably, RAD51 paralogs. Conversely, FIGNL1 depletion does not affect the loading of RAD51 onto ssDNA. Our additional analysis uncovered KIAA0146, also known as scaffolding protein involved in DNA repair (SPIDR), as a binding partner of FIGNL1 and established that KIAA0146/SPIDR acts with FIGNL1 in HR repair. Collectively, our study uncovers a protein complex, which consists of FIGNL1 and KIAA0146/SPIDR, in DNA repair and provides potential directions for cancer diagnosis and therapy.
Cells are continuously challenged by constant genotoxic pressure from both endogenous and exogenous sources. Severe DNA lesions, including DNA ds breaks (DSBs) and interstrand cross-links, have to be appropriately repaired for cell survival. There are two major pathways involved in the repair of DSBs: the nonhomologous end-joining pathway and homologous recombination (HR) pathway (1). HR is particularly important for the repair of DSBs because of its ability to accurately restore genetic information, whereas repair by the nonhomologous end-joining pathway may potentially lead to deletions and mutations.
The central component in the HR pathway is RAD51, which is the major recombinase in mitotic cells and also plays a critical role in meiotic recombination. RAD51 is the human ortholog of Escherichia coli recombinase protein RecA. It polymerizes onto resected DNA ends to form a nucleoprotein filament and promotes strand exchange between homologous DNA duplexes, therefore ensuring high-fidelity DNA repair (2, 3).
Breast cancer 2, early onset (BRCA2) is encoded by a tumor suppressor gene that, when mutated, greatly elevates risks for breast and ovarian cancer. BRCA2 is another key protein in HR, because it mediates the loading of RAD51 onto ssDNA and stabilizes RAD51 filaments. Another tumor suppressor, partner and localizer of BRCA2 (PALB2), was also found to associate with BRCA2 and participate in the loading of BRCA2–RAD51 repair complex onto DNA (4). In addition to BRCA2/PALB2, other important HR mediators are the five RAD51 paralogs [X-ray repair complementing defective repair in Chinese hamster cells 2 (XRCC2), XRCC3, RAD51B, RAD51C, and RAD51D], which are required for the assembly of DNA damage-induced RAD51 foci and efficient HR repair (5–7). Given the importance of HR in the maintenance of genomic stability, it is not surprising that germ-line mutations in many components involved in HR repair, such as BRCA1, BRCA2, and PALB2, are associated with various human genetic disorders and cancers. Recent studies identified biallelic mutations in RAD51C, which lead to Fanconi anemia-like disorder, and a monoallelic mutation in RAD51C, which is associated with increased risk of breast and ovarian cancer (8, 9).
Although a considerable number of HR factors have been identified, most knowledge of HR pathway comes from studies in prokaryotic and eukaryotic model organisms. It would be particularly interesting to probe the HR pathway in human cells, because genes involved in HR repair are frequently mutated in a variety of human diseases, including cancers. More importantly, investigating how HR is regulated to ensure proper repair and protect against genomic instability is fundamentally important for developing new strategies to prevent and treat breast, ovarian, and other cancers. In this study, we took an unbiased approach and performed tandem affinity purification (TAP) of RAD51-containing protein complexes in human cells. We identified fidgetin-like 1 (FIGNL1) as an RAD51 binding protein that is involved in HR repair. Additional study showed that FIGNL1 specifically interacts with RAD51 through a conserved RAD51 binding domain. However, FIGNL1 is recruited to sites of DNA damage independently of RAD51. Finally, we showed that another previously uncharacterized protein, KIAA0146, is a binding partner of FIGNL1, and both of them are required for efficient HR repair.
Results
FIGNL1 Is an RAD51 Binding Protein Involved in HR Repair.
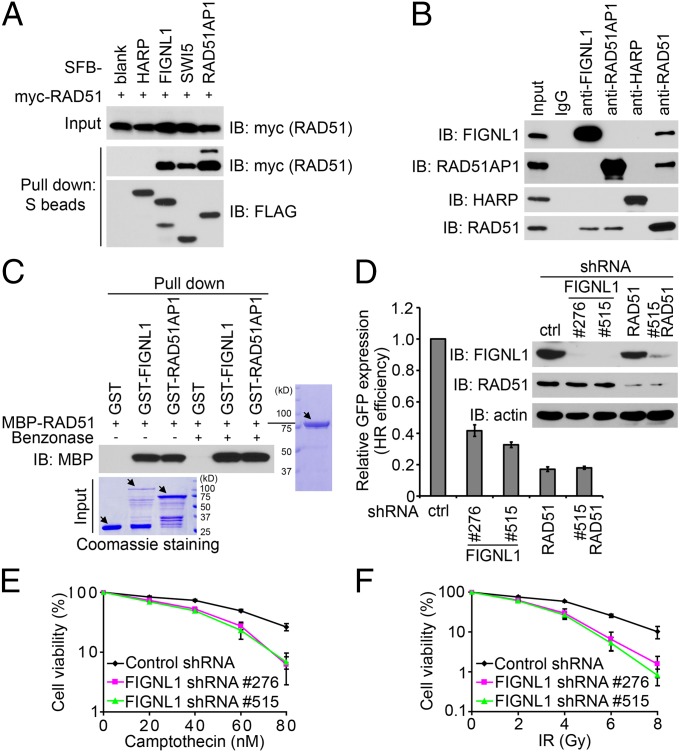
In an attempt to identify RAD51-associated proteins, we performed TAP using whole-cell lysate prepared from 293T cells stably expressing triple-epitope S protein, FLAG, and streptavidin-binding peptide-tagged RAD51 (SFB-RAD51). As shown in Fig. S1A, SFB-RAD51 could clearly form nuclear foci after ionizing radiation (IR) treatment. MS analysis revealed a set of RAD51-associated proteins (Fig. S1B). Because most of the known RAD51-associated proteins localize to DNA damage sites, we asked whether any of the candidate proteins would form DNA damage-induced foci. Excitingly, we found that FIGNL1 was able to localize to IR-induced DNA damage foci (as shown later) and decided to further explore a potential role of FIGNL1 in DNA damage repair. We first confirmed the in vivo interaction of SFB-tagged FIGNL1 with myc-tagged RAD51 (Fig. 1A). The known RAD51 binding proteins SWI5 (10) and RAD51AP1 (11, 12) were included as positive controls, and HepA-related protein (HARP) SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 (SMARCAL1) (13) was used as a negative control (Fig. 1A). Next, we verified the in vivo interaction between endogenous FIGNL1 and RAD51 (Fig. 1B). Furthermore, we used bacterially expressed and purified maltose-binding protein (MBP) -tagged RAD51 and GST-tagged FIGNL1 to show that, like RAD51AP1, FIGNL1 binds directly to RAD51 (Fig. 1C). Benzonase was added to eliminate possible DNA contamination. Together, these data support that FIGNL1 is a direct RAD51 binding protein.
Fig. 1.
FIGNL1 is an RAD51 binding protein involved in homologous recombination repair. (A) The interaction between FIGNL1 and RAD51 was confirmed by coprecipitation with overexpressed proteins. 293T cells were transfected with plasmids encoding myc-tagged RAD51 together with plasmids encoding SFB-tagged HARP, FIGNL1, SWI5, or RAD51AP1. Coprecipitation was carried out using S-protein beads, and immunoblotting was performed using antibodies as indicated. (B) Association of endogenous FIGNL1 with RAD51 in 293T cells was performed by coimmunoprecipitation. Normal rabbit IgG, anti-RAD51AP1, and anti-HARP antibodies were included as controls. (C) FIGNL1 and RAD51 directly interact with each other. MBP- and GST-tagged proteins were expressed and purified from E. coli. Pull-down experiments were performed using glutathione agarose beads and detected by Coommassie staining or immunoblotting as indicated. The main bands are indicated by arrows. The lower bands are caused by protein degradation. Benzonase was added to buffer of 20 mM Tris⋅HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, and 1 μg/mL each pepstatin A and aprotinin before adding glutathione agarose beads to eliminate possible DNA contamination. (D) FIGNL1 depletion impairs HR repair. DR-GFP U2OS cells were electroporated with pCBASce plasmids. The percentage of GFP-positive cells was determined by flow cytometry 48 h after electroporation. The data were normalized to those data obtained from cells infected with control shRNA (set as 1.0). Data are presented as mean ± SD from three different experiments. Knockdown efficiency of FIGNL1 and RAD51 using specific shRNAs was confirmed by immunoblotting. (E and F) Survival curves in response to increasing doses of (E) camptothecin and (F) IR for indicated cell lines are presented. Data are presented as mean ± SD from three different experiments. Ctrl, control; IB, immunoblotting.
Given that RAD51 is the central component in HR, we examined HR efficiency in FIGNL1-depleted cells using the established direct repeat green fluorescent protein (DR-GFP) reporter system (Fig. S1C) (14). Our data clearly revealed that the efficiency of HR repair was impaired in cells with FIGNL1 depletion (Fig. 1D). Although RAD51 depletion significantly reduced HR efficiency, FIGNL1 and RAD51 double depletion did not achieve additional impairment, suggesting that FIGNL1 likely performs in the same HR pathway centered by RAD51. Consistently, down-regulation of FIGNL1 by shRNAs also resulted in increased cellular sensitivity to camptothecin and IR (Fig. 1 E and F). Cells with RAD51 knockdown were unhealthy, dying, and extremely sensitive to IR treatment (Fig. S1D). This finding is consistent with the central role of RAD51 in HR. We did not observe a significant difference in IR sensitivity in cells with RAD51 and FIGNL1 double knockdown (Fig. S1D). Taken together, these data support an important role of FIGNL1 in HR-mediated DNA repair.
FIGNL1 Specifically Interacts with RAD51.
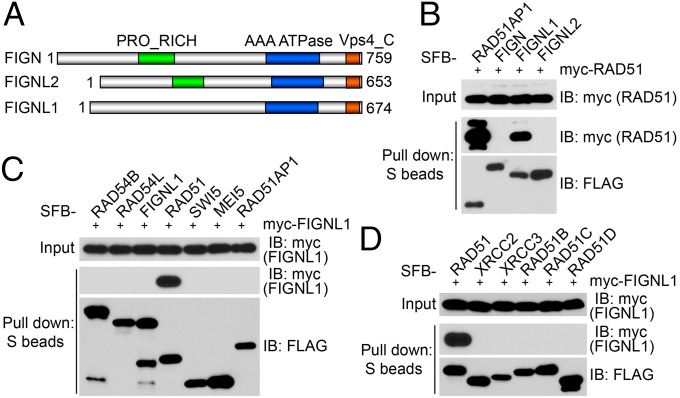
FIGNL1 is one of three fidgetin or fidgetin-like proteins in humans. The mouse mutation fidget arose spontaneously in a heterogeneous albino stock (15). This mutant mouse is characterized by a side-to-side head-shaking and circling behavior caused by reduced or absent semicircular canals (16). By positional cloning, the gene mutated in fidget mice was identified and designated fidgetin (Fign) (17). There are two fidgetin-like proteins in mice, Fignl1 and Fignl2 (17). The domain structures of human FIGN, FIGNL1 and FIGNL2, were shown in Fig. 2A. From our RAD51 TAP results, we did not obtain any peptide derived from FIGN or FIGNL2 (Fig. S1B). Consistently, our coprecipitation experiments showed that RAD51 binds strongly to FIGNL1 and not FIGN and FIGNL2 (Fig. 2B). Interestingly, only RAD51 [but not any of the known RAD51 binding proteins, including RAD54B, RAD54L, SWI5, meiosis protein 5 (MEI5), RAD51AP1, and the five RAD51 paralogs (XRCC2, XRCC3, RAD51B, RAD51C, and RAD51D)] binds to FIGNL1 (Fig. 2 C and D). Together, the above observations show a highly specific interaction between RAD51 and FIGNL1.
Fig. 2.
FIGNL1 specifically interacts with RAD51. (A) Schematic representation of the FIGN family proteins, including FIGN, FIGNL1, and FIGNL2. (B) RAD51 only binds to FIGNL1 and not FIGN or FIGNL2. (C) FIGNL1 only binds to RAD51 and not other HR factors, including RAD54B, RAD54L, SWI5, MEI5, and RAD51AP1. (D) FIGNL1 does not bind to any of the RAD51 paralogs. Coprecipitation assays shown in B–D were carried out as described in Fig. 1A. IB, immunoblotting.
Defining a Conserved RAD51 Binding Domain in FIGNL1.
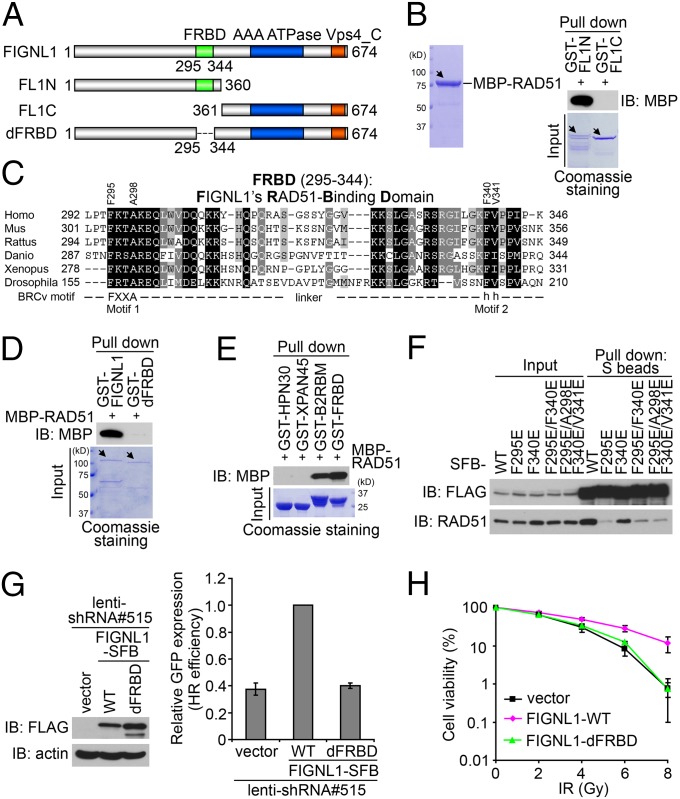
The specific and direct binding of FIGNL1 with RAD51 encouraged us to explore the nature of FIGNL1–RAD51 interaction. We took advantage of the FIGNL1–RAD51 direct binding assays and showed that RAD51 binds to the N-terminal one-half of FIGNL1 (Fig. 3 A and B). We aligned the human FIGNL1 protein with its orthologs from other species and found that, other than the C-terminal AAA (ATPases Associated with diverse cellular Activities) ATPase domain, there is another conserved region at residues 295–344 in human FIGNL1 (Fig. 3C and Fig. S2). Excitingly, both our direct binding and coprecipitation assays showed that this conserved region (residues 295–344) is required for the binding of FIGNL1 to RAD51 (Fig. 3D and Fig. S3 A and B). Therefore, we designated this region as FIGNL1’s RAD51 binding domain (FRBD). Additional direct binding and coprecipitation experiments confirmed that the FRBD domain alone binds to RAD51 (Fig. 3E and Fig. S3C). Although the positive control, the BRCA2 C-terminal RAD51 binding motif, also binds to RAD51 (Fig. 3E), the two negative controls, replication protein A (RPA) binding domains from HARP (HPN30) and XPA (XPAN45), showed no appreciable binding to RAD51 (Fig. 3E and Fig. S3C). We also checked one of the FIGNL1-interacting proteins, KIAA0146 (will be introduced later), and showed that the binding of KIAA0146 to FIGNL1 does not require the FRBD domain (Fig. S3D). In addition, the FRBD domain strongly binds to RAD51 but not KIAA0146 (Fig. S3E). These experiments suggest that FRBD in FIGNL1 is specific for its interaction with RAD51.
Fig. 3.
Defining a conserved RAD51 binding domain in FIGNL1. (A) Schematic representation of WT FIGNL1 and the mutants used in the study. (B) N-terminal one-half of FIGNL1 binds to RAD51. Direct binding assay was carried out as described in Fig. 1C. The main bands are indicated by arrows. The lower bands are caused by protein degradation. (C) Alignment of FRBD from different species. The conserved BRCv motif (including Motif 1 and 2) is indicated. (D) FRBD is required for FIGNL1 binding to RAD51. (E) FRBD alone binds to RAD51. HPN30 (HARP RPA binding domain) and XPAN45 (XPA RPA binding domain) were used as negative controls. BRCA2 C-terminal RAD51-binding motif (B2RBM) was used as a positive control. (F) The conserved BRCv motif is critical for RAD51 binding. BRCv motif mutants (F295E, F340E, F295E/F340E, and F295E/A298E/F340E/V341E) were generated. (G) FRBD deletion mutant could not rescue HR defect in FIGNL1-depleted DR-GFP U2OS cells. (H) FRBD deletion mutant could not rescue IR hypersensitivity of FIGNL1-depleted cells. Data are presented as mean ± SD from three different experiments. IB, immunoblotting.
The BRC repeats in BRCA2 are essential for BRCA2 to bind to RAD51 and promote HR (18). More recently, a BRC repeat variant, termed as BRCv, was identified in RecQ protein-like 5 (RECQL5) helicase, which is required for its binding to RAD51 (19). Interestingly, the FRBD domain seems to contain a motif similar to BRCv (Fig. 3C). We generated single, double, and combination mutations of key residues in BRCv motif (F295E, F340E, F295E/F340E, and F295E/A298E/F340E/V341E) (Fig. 3 C and F). Interestingly, coprecipitation experiments showed that all of these mutants, especially a point mutation (F295E) within the putative BRCv motif, greatly reduced the interaction between FIGNL1 and RAD51 (Fig. 3F). However, none of these mutants abolished the FIGNL1–RAD51 interaction, suggesting that residues beyond the putative BRCv motif within FRBD may also contribute to RAD51 binding.
We next asked whether RAD51 binding is essential for the in vivo function of FIGNL1. We generated constructs encoding shRNA-resistant SFB-tagged WT or FRBD deletion mutant of FIGNL1, and therefore, we were able to express exogenous FIGNL1 when the endogenous FIGNL1 was depleted by shRNA. As shown in Fig. 3G, although the expression of WT FIGNL1 rescued HR repair defect in FIGNL1-depleted cells, the FRBD deletion mutant and the F295E mutant failed to do so (Fig. S4A). The FRBD deletion mutant or F295E mutant also failed to rescue cellular sensitivity to IR in FIGNL1-depleted cells (Fig. 3H and Fig. S4A). These data indicate that FIGNL1 participates in the HR repair pathway through its association with RAD51.
FIGNL1 Is Recruited to Sites of DNA Damage Independently of RAD51.
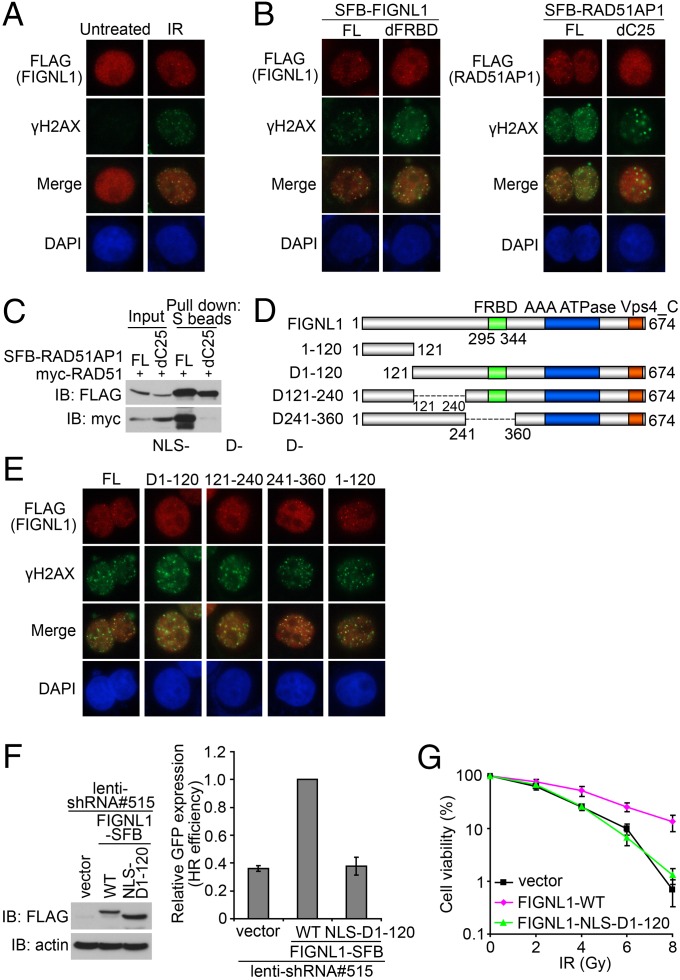
As shown in Fig. 4A and Fig. S5 A and B, SFB-tagged FIGNL1 localized to discrete foci, which colocalized with a subset of H2A histone family, member X (γH2AX) after IR. Unexpectedly, the FRBD deletion mutant, which cannot bind to RAD51, was still recruited to DNA damage sites after IR (Fig. 4B and Fig. S5C). This result is distinctly different from the result of RAD51AP1. As for RAD51AP1, deletion of its C-terminal RAD51 binding domain abolished not only its interaction with RAD51 (Fig. 4C) but also, its focus localization after DNA damage (Fig. 4B). These results suggest that FIGNL1 and RAD51AP1 accumulate at sites of DNA damage by different mechanisms. Thus, it would be interesting to know which region is important for the recruitment of FIGNL1 to DNA damage sites. We generated three internal deletion mutants that cover the N-terminal one-half of FIGNL1 (Fig. 4D). Deletion of the N-terminal 120 residues resulted in cytoplasmic localization of this mutant; therefore, a nuclear localization signal (NLS) was added to the N terminus of this deletion mutant (NLS-D1-120) to ensure its proper nuclear localization. We found that, although the full-length D121-240 and D241-360 mutants of FIGNL1 could localize to DNA damage sites after IR, the D1-120 mutant of FIGNL1 failed to do so, but this mutant could still interact with RAD51 (Fig. 4E and Fig. S5 D–F). In addition, the N-terminal 120 residues alone are sufficient to localize to damage-induced foci (Fig. 4E). Thus, the N-terminal region of FIGNL1 (residues 1–120) is necessary and sufficient for FIGNL1 foci formation.
Fig. 4.
FIGNL1 is recruited to sites of DNA damage independent of RAD51. (A) HeLa cells were infected with lentiviral SFB-tagged FIGNL1. Immunostaining experiments were performed 6 h after IR treatment (10 Gy) using indicated antibodies. (B) FRBD is not required for FIGNL1 foci formation. dC25, deleted with the C-terminal 25 aa of RAD51AP1. (C) Coprecipitation was performed to confirm that the dC25 mutant of RAD51AP1 does not bind to RAD51. (D) Schematic representation of WT and mutants of FIGNL1 used in the following study. (E) The N-terminal 120 aa of FIGNL1 are required for the recruitment of FIGNL1 to sites of DNA damage. An NLS was added to the N terminus of mutant D1-120 (NLS-D1-120) to ensure its nuclear localization. (F) FIGNL1 N-terminal domain (amino acids 1–120) is required to rescue HR defect in FIGNL1-depleted DR-GFP U2OS cells. (G) FIGNL1 N-terminal domain deletion mutant could not rescue IR hypersensitivity of FIGNL1-depleted cells. Data are presented as mean ± SD from three different experiments. FL, full length; IB, immunoblotting.
As shown in Fig. 4F, the D1-120 mutant of FIGNL1 failed to rescue the HR repair defect in FIGNL1-depleted cells. Consistently, this mutant also could not rescue cellular sensitivity to IR in FIGNL1-depleted cells (Fig. 4G). These observations indicate that the ability to localize at DSB sites is important for FIGNL1 function in HR repair.
KIAA0146 Is a Binding Partner of FIGNL1 and Participates in HR Repair.
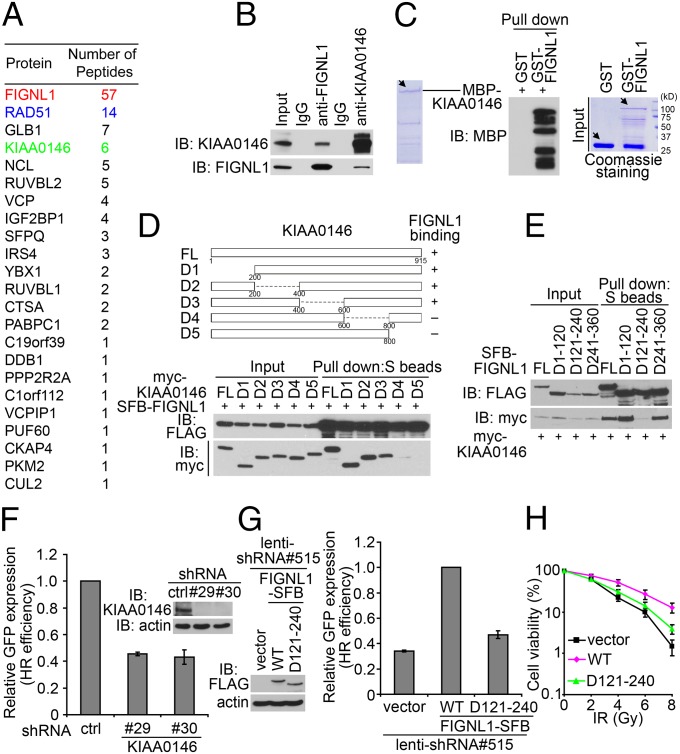
To understand exactly how FIGNL1 participates in HR repair, we performed TAP of FIGNL1-containing protein complexes. We identified RAD51 as the major FIGNL1-associated protein (Fig. 5A), which confirmed that FIGNL1 is a bone fide RAD51 binding protein. In addition, we uncovered a candidate FIGNL1 binding protein, KIAA0146, which is a nuclear protein with high peptide hits as revealed by the MS analysis.
Fig. 5.
KIAA0146 is a binding partner of FIGNL1 and participates in homologous recombination repair. (A) TAP was performed using 293T cells stably expressing SFB-tagged FIGNL1. The data from MS analysis are shown. (B) Association of endogenous KIAA0146 with FIGNL1 in 293T cells was performed by coimmunoprecipitation. (C) KIAA0146 and FIGNL1 interact directly with each other. The main bands are indicated by arrows. The lower bands are caused by protein degradation. (D) FIGNL1 binds to the C terminus of KIAA0146. Schematic representation of the full-length and deletion mutants of KIAA0146 used in this study is shown. FL, full length. (E) Amino acids 121–240 are required for FIGNL1 binding to KIAA0146. (F) KIAA0146 depletion impairs HR repair. Data are presented as mean ± SD from three different experiments. (G) Amino acids 121–240 deletion mutant could not rescue HR defect in FIGNL1-depleted DR-GFP U2OS cells. (H) Amino acids 121–240 deletion mutant could not rescue IR hypersensitivity of FIGNL1-depleted cells. Data are presented as mean ± SD from three different experiments. Ctrl, control; IB, immunoblotting.
KIAA0146 is an uncharacterized protein with no known functional domains. We first verified the interaction between endogenous KIAA0146 and FIGNL1 using coimmunoprecipitation experiments (Fig. 5B). Furthermore, these two proteins bind directly to each other (Fig. 5C). We next generated a series of truncation or internal deletion mutants of KIAA0146 (Fig. 5D), and the C terminus of KIAA0146 is involved in its binding to FIGNL1 (Fig. 5D). However, KIAA0146 binds to the N-terminal one-half of FIGNL1 (Fig. S6A). We also narrowed down the region that is necessary and sufficient for binding to KIAA0146 to residues 121–240 of FIGNL1 (Fig. 5E and Fig. S6B).
Similar to FIGNL1, KIAA0146 also participates in HR repair, because HR repair efficiency was impaired in KIAA0146-depleted cells (Fig. 5F). Moreover, the D121-240 mutant of FIGNL1, which disrupts its binding to KIAA0146, was defective in HR repair (Fig. 5G) and fails to rescue cellular sensitivity to IR in FIGNL1-depleted cells (Fig. 5H). Consistently, although the expression of WT KIAA0146 rescued HR efficiency and cellular sensitivity to IR in KIAA0146-depleted cells, the C-terminal deletion mutant (D5), which cannot bind FIGNL1, failed to do so (Fig. S6C). We did not observe any additive effects on HR repair or cell survival after IR in cells with both FIGNL1 and KIAA0146 depletion (Fig. S6D), suggesting that FIGNL1 and KIAA0146 may affect HR repair through the same pathway. Taken together, these data established that KIAA0146 is an FIGNL1 binding protein and that the interaction between KIAA0146 and FIGNL1 is important for HR repair.
FIGNL1 and KIAA0146 Are Components of the H2AX-Dependent DNA Damage Response and HR Repair Pathway.
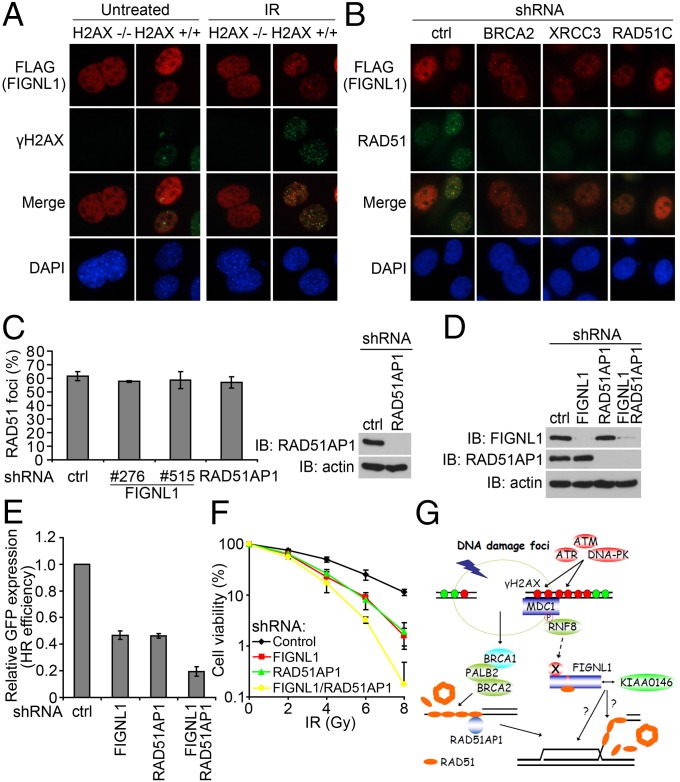
It remains elusive exactly how FIGNL1 accumulates at DNA damage sites and acts with RAD51 and KIAA0146 in HR repair. Phosphorylation of histone variant H2AX at serine 139, which produces γH2AX, has been widely used as a sensitive marker for DSBs. Moreover, γH2AX is required for the accumulation of many DNA damage response proteins at DSBs and thus, plays a role in DNA repair (reviewed in ref. 20). To determine whether FIGNL1 depends on H2AX for its foci formation, we introduced SFB-tagged FIGNL1 into H2AX+/+ and H2AX−/− mouse embryonic fibroblasts (MEFs). As shown in Fig. 6A and Fig. S7A, SFB-tagged FIGNL1 only formed IR-induced foci in H2AX+/+ MEFs and did not form them in H2AX−/− MEFs, suggesting that FIGNL1 requires γH2AX for its localization to DNA damage sites. Using laser-induced microirradiation in WT, MDC1−/−, or RNF8−/− MEFs, we also showed that FIGNL1 recruitment also requires mediator of DNA-damage checkpoint 1 (MDC1) and ring finger protein 8 (RNF8), two downstream effectors of γH2AX, in the DNA damage signaling pathway (Fig. S7B).
Fig. 6.
FIGNL1 and KIAA0146 are components of the H2AX-dependent DNA damage response and HR repair pathway. (A) FIGNL1 is retained at sites of DNA damage in an H2AX-dependent manner. H2AX−/− and H2AX+/+ MEFs were infected with lentiviral plasmids encoding SFB-tagged FIGNL1. (B) BRCA2, XRCC3, or RAD51C depletion does not affect FIGNL1 foci formation after IR treatment. (C) FIGNL1 depletion does not impair RAD51 foci formation after DNA damage. The quantification of foci-positive cells was performed by counting a total of 200 cells per sample. Data are presented as mean ± SD from three different experiments. (D) The knockdown of FIGNL1 and RAD51AP1 in DR-GFP U2OS cells was confirmed with immunoblotting. (E) FIGNL1 and RAD51AP1 depletion has an additive effect on HR repair. Data are presented as mean ± SD from three different experiments. (F) FIGNL1 and RAD51AP1 depletion have an additive effect on cell sensitivity after IR treatment. Data are presented as mean ± SD from three different experiments. (G) A proposed model for FIGNL1-containing protein complex in DNA damage response and homologous recombination repair. Details in the text. Ctrl, control; IB, immunoblotting.
Because FIGNL1 is involved in HR repair, we also asked whether FIGNL1 would act downstream of the main HR factors, like BRCA2 or RAD51 paralogs. Although depletion of BRCA2, XRCC3, or RAD51C dramatically reduced RAD51 foci formation as previously reported (5, 21), FIGNL1 foci formation was not impaired in these knockdown cells (Fig. 6B and Fig. S7C). Thus, we concluded that FIGNL1 likely localizes to DNA damage sites independently of BRCA2 or RAD51 paralogs.
The formation of nuclear foci involving RAD51 most likely represents loading of RAD51 onto ssDNA, one of the critical events in the faithful repair of DNA damage by HR. Because FIGNL1 binds directly to RAD51 and participates in HR, we also examined whether FIGNL1 would affect RAD51 foci formation. As shown in Fig. 6C, we did not observe any quantitative reduction of RAD51 foci formation in FIGNL1-depleted cells, which is similar to the result of RAD51AP1 as previously reported (22, 23). These results suggested that FIGNL1 may act similarly to RAD51AP1 and assist RAD51-mediated HR through a mechanism different from BRCA2 or RAD51 paralogs.
RAD51AP1 helps to maintain genomic integrity by enhancing RAD51 recombinase activity (23). Specifically, RAD51AP1 stimulates joint molecule formation through its structure-specific DNA binding and its physical association with RAD51 (22). Given that both FIGNL1 and RAD51AP1 act in HR repair without influencing RAD51 foci formation, we also tested whether FIGNL1 and RAD51AP1 would be involved in the same pathway. We depleted FIGNL1, RAD51AP1, or both in DR-GFP U2OS cells using lentiviral shRNAs (Fig. 6D). FIGNL1 and RAD51AP1 depletion had an additive effect on HR repair (Fig. 6E). Consistently, they also had an additive effect on cell survival after IR (Fig. 6F), suggesting that FIGNL1 and RAD51AP1 may affect HR repair through different pathways. As a matter of fact, we did not detect an interaction between FIGNL1 and RAD51AP1 (Fig. 2C), although both of them exist in the list of RAD51 binding proteins (Fig. S1B). Together, our data indicate that the FIGNL1-containing protein complex participates in HR repair, which requires the binding of FIGNL1 to RAD51 but not any of the known HR proteins that we tested here (Discussion and Fig. 6G).
Discussion
Our study has revealed a function for FIGNL1 and KIAA0146 in cellular responses to DNA damage and HR repair. We showed that (i) FIGNL1 is a specific RAD51 binding protein involved in HR repair, (ii) FIGNL1 contains a conserved RAD51 binding domain, (iii) FIGNL1 is likely recruited to sites of DNA damage independently of RAD51, BRCA2, and some and probably all of the RAD51 paralogs, and (iv) KIAA0146 interacts with FIGNL1 and participates in HR repair.
Although it is clear that RAD51 plays critical roles in HR repair, so far, we still do not have a cell-free system that can faithfully reproduce HR repair in vitro. This situation may be because of a number of reasons, and one of them is that we may not have all of the components that act with RAD51 in various steps of HR repair. Thus, it is imperative to identify all of the protein partners of RAD51 and understand how they may function with RAD51 in HR repair. TAP of RAD51-associated protein complexes allowed us to recover most of the known HR proteins that bind to and act with RAD51. These proteins include BRCA2, PALB2, RAD54L, RAD54B, XRCC3, RAD51C, RAD51AP1, and MEI5. In this study, we focused on a previously uncharacterized protein complex, which consists of FIGNL1, KIAA0146, and a yet-to-be-identified protein. We expect that our study on the identification of this protein complex involved in HR repair will lead to further understanding of the complexity of HR repair in mammalian cells, which may provide directions for the development of tools for cancer diagnosis and therapy.
In response to DSBs, the MRE11–RAD50–NBS1 (MRN) complex recruits CTBP-interacting protein (CtIP) and BRCA1/PALB2/BRCA2 to initiate its preferred classic homologous recombination. At the same time, the MRN complex also recruits and activates ataxia telangiectasia mutated (ATM), which is involved in the initiation of a series of phosphorylation events including γH2AX. γH2AX and H2AX-dependent signal transduction events are required for the sustained accumulation of many DNA damage response and repair proteins at DSB sites to facilitate DNA repair processes (reviewed in ref. 20). However, it remains confusing exactly how the H2AX-dependent pathway participates in HR repair. Here, we propose that FIGNL1 may participate in the regulation of HR repair, which acts downstream of the H2AX-dependent signaling pathway (Fig. 6G). We showed that FIGNL1 is recruited to DSBs in an H2AX-dependent manner. At DSB sites, FIGNL1 interacts with RAD51 through its RAD51 binding domain (FRBD), which contains a BRC repeat variant, and plays a role after the presynapsis phase of HR repair. Although biochemical data are needed to further understand the roles of FIGNL1 in HR repair, which may include promoting RAD51-dependent D-loop formation, removal of RAD51 after homology search, or affecting nucleosome remodeling, our preliminary experiments interestingly showed that the ATPase activity of FIGNL1 is critical for its role in HR repair. As shown in Fig. S4B, the FIGNL1 ATPase mutant (with both Walker A motif mutation K447A and Walker B motif mutation D500A) failed to rescue HR repair defect and cellular sensitivity to IR in FIGNL1-depleted cells. FIGNL1 also interacts with KIAA0146, which is also required for HR repair. This protein complex seems to act independently of the known HR proteins, including BRCA2 and RAD51 paralogs, because these proteins do not interact with FIGNL1 and are not involved in the recruitment of FIGNL1 to DSB sites. Thus, we hypothesize that, although the BRCA2 and RAD51 paralogs are the major regulators and act upstream of RAD51 in DNA repair, the FIGNL1-containing protein complex may function at later steps of the HR repair process (Fig. 6G).
The determinant region for FIGNL1 foci formation is located at its conserved N terminus (residues 1–120). There may be other uncharacterized components that bind to the N terminus of FIGNL1 and are responsible for recruiting FIGNL1 to sites of DNA damage (indicated as protein X in Fig. 6G). Understanding how FIGNL1 is recruited to DNA damage sites through the H2AX-dependent pathway will reveal mechanistically how the H2AX-dependent DNA damage signaling pathway contributes to HR repair. Nevertheless, our data showed that, although the expression of WT FIGNL1 rescued HR repair defect or cellular sensitivity to IR in FIGNL1-depleted cells, cells expressing the FRBD deletion mutant or the D1-120 mutant behaved similarly to FIGNL1-depleted cells and failed to rescue any of these phenotypes (Figs. 3 G and H and 4 F and G), suggesting that deleting one of these domains (FRBD or residues 1–120) is sufficient to inhibit FIGNL1 HR function in vivo. We propose that these two domains act together and are required for carrying out FIGNL1 HR function at DNA damage sites.
Given the important roles of RAD51 and its regulators in DNA repair and prevention of inappropriate recombination, it is not surprising that the mutations of genes encoding these proteins lead to predisposition to a variety of cancers. Moreover, mutations of genes involved in HR repair also provide potential targets for anticancer therapy, because tumors with HR deficiency are more sensitive to DNA-damaging agents or chemicals that inhibit other repair or checkpoint pathways. A promising strategy for treating tumors with HR deficiency is the use of poly (ADP-ribose) polymerase (PARP) inhibitors, which have great efficacy in treating cancers associated with BRCA1 or -2 mutations (24, 25). However, PARP inhibitors are not without flaws. Studies of platinum refractory or PARP inhibitor-resistant patients with BRCA mutations have suggested the existence of intragenic reversion mutations, which may allow tumor cells to restore their HR repair capability and therefore, become resistant to these agents (26, 27). Alternatively, 53BP1 loss can also restore HR repair and lead to drug resistance in BRCA1-deficient cells (28, 29). However, these mechanisms are unlikely the only mechanisms for the development of drug resistance. The identification of a unique protein complex, which consists of FIGNL1, KIAA0146, and most likely, other uncharacterized components, provides additional understanding of the complex regulation of HR repair. Future studies will reveal whether this FIGNL1-containing protein complex is involved in tumor initiation and sensitivity or resistance of tumors to chemotherapies and PARP inhibitors.
Materials and Methods
TAP was performed as previously described (13, 30). Anti-FIGNL1, anti-KIAA0146, anti-RAD51, and anti-RAD51AP1 antibodies were raised in rabbits. Detailed descriptions of the reagents and protocols used in this study can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Maria Jasin for providing U2OS cells with integrated DR-GFP substrate and DR-GFP and pCBASce constructs. We thank the Cytometry and Cellular Imaging Core Facility of the MD Anderson Cancer Center for technical assistance. We thank all members of the J.C. laboratory, especially Wenqi Wang, for their advice and technical assistance. J.C. is a recipient of an Era of Hope Scholar Award from the Department of Defense (W81XWH-05-1-0470) and member of the MD Anderson Cancer Center (CA016672). This work was supported, in part, by National Institutes of Health Grants CA089239, CA092312, and CA100109 (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220662110/-/DCSupplemental.
References
- 1.Pierce AJ, et al. Double-strand breaks and tumorigenesis. Trends Cell Biol. 2001;11(11):S52–S59. doi: 10.1016/s0962-8924(01)02149-3. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259(5103):1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 3.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82(3):453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 4.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Takata M, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21(8):2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deans B, Griffin CS, O’Regan P, Jasin M, Thacker J. Homologous recombination deficiency leads to profound genetic instability in cells derived from Xrcc2-knockout mice. Cancer Res. 2003;63(23):8181–8187. [PubMed] [Google Scholar]
- 7.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 8.Meindl A, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 9.Vaz F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 10.Yuan J, Chen J. The role of the human SWI5-MEI5 complex in homologous recombination repair. J Biol Chem. 2011;286(11):9888–9893. doi: 10.1074/jbc.M110.207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovalenko OV, Golub EI, Bray-Ward P, Ward DC, Radding CM. A novel nucleic acid-binding protein that interacts with human rad51 recombinase. Nucleic Acids Res. 1997;25(24):4946–4953. doi: 10.1093/nar/25.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuta R, et al. RAB22 and RAB163/mouse BRCA2: Proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci USA. 1997;94(13):6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23(20):2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruneberg H. Two new mutant genes in the house mouse. J Genet. 1943;45(1):22–28. [Google Scholar]
- 16.Truslove GM. The anatomy and development of the fidget mouse. J Genet. 1956;54(1):64–86. [Google Scholar]
- 17.Cox GA, Mahaffey CL, Nystuen A, Letts VA, Frankel WN. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat Genet. 2000;26(2):198–202. doi: 10.1038/79923. [DOI] [PubMed] [Google Scholar]
- 18.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272(51):31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 19.Islam MN, et al. A variant of the breast cancer type 2 susceptibility protein (BRC) repeat is essential for the RECQL5 helicase to interact with RAD51 recombinase for genome stabilization. J Biol Chem. 2012;287(28):23808–23818. doi: 10.1074/jbc.M112.375014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: To be or not to be. FEBS Lett. 2010;584(17):3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59(15):3547–3551. [PubMed] [Google Scholar]
- 22.Modesti M, et al. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell. 2007;28(3):468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 23.Wiese C, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell. 2007;28(3):482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 25.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 26.Edwards SL, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451(7182):1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 27.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451(7182):1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17(6):688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan J, Ghosal G, Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol Cell. 2012;47(3):410–421. doi: 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.