Abstract
The scaffolding protein at the neuromuscular junction, rapsyn, enables clustering of nicotinic acetylcholine receptors in high concentration and is critical for muscle function. Patients with insufficient receptor clustering suffer from muscle weakness. However, the detailed organization of the receptor–rapsyn network is poorly understood: it is unclear whether rapsyn first forms a wide meshwork to which receptors can subsequently dock or whether it only forms short bridges linking receptors together to make a large cluster. Furthermore, the number of rapsyn-binding sites per receptor (a heteropentamer) has been controversial. Here, we show by cryoelectron tomography and subtomogram averaging of Torpedo postsynaptic membrane that receptors are connected by up to three rapsyn bridges, the minimum number required to form a 2D network. Half of the receptors belong to rapsyn-connected groups comprising between two and fourteen receptors. Our results provide a structural basis for explaining the stability and low diffusion of receptors within clusters.
Keywords: cryoelectron microscopy, neurotransmitter receptor clustering, synapse, ligand-gated ion channel, tetratricopeptide repeat
The cytoplasmic protein rapsyn, also known as the 43K protein, interacts with nicotinic acetylcholine receptors in the postsynaptic membrane at the neuromuscular junction, forming clusters where receptors are present in high concentration. Such clustering is necessary for efficient neurotransmission and, hence, muscle functioning. Rapsyn-deficient mice fail to form clusters, show severe muscle weakness and breathing difficulties, and die within hours after birth (1). Furthermore, many patients suffering from congenital myasthenic syndrome are found to have mutations in rapsyn (2).
Rapsyn is a 43-kDa protein containing a myristoylation site necessary for submembrane localization, seven tetratricopeptide repeats (TRPs), a putative coiled-coil domain, and a RING-H2 domain (3). Two TPRs are necessary for rapsyn self-association, whereas association with receptor depends on the predicted coiled-coil (3, 4). Rapsyn is also linked through the RING-H2 domain to dystroglycan, a protein that indirectly binds actin cytoskeleton (5). Recombinant rapsyn expressed in heterologous systems spontaneously aggregates underneath the plasma membrane (3), suggesting that rapsyn might first form a wide scaffold to which receptors can subsequently dock (6). On the other hand, rapsyn and the receptor are cotransported in membrane vesicles toward the plasma membrane, and the receptor is necessary for rapsyn clustering in C2 myotubes, raising the possibility that rapsyn only forms short bridges linking receptors together to form a large cluster (7–9). Furthermore, the number of rapsyn-binding sites per receptor (a heteropentamer) has been variously estimated to be between one and five (6, 10, 11). Therefore, the molecular details and organization of the receptor–rapsyn network are poorly understood.
To address these issues, we applied cryoelectron tomography, combined with subtomogram averaging and image classification, to postsynaptic membranes isolated from the electric organ of Torpedo marmorata: a model system for the neuromuscular junction (12, 13). Three-dimensional structures were obtained of acetylcholine receptors complexed with rapsyn molecules in several specific arrangements. We show that the receptors connect to each other by small rapsyn bridges and that each receptor can have up to three bridges, the minimum number required to form a 2D network.
Results
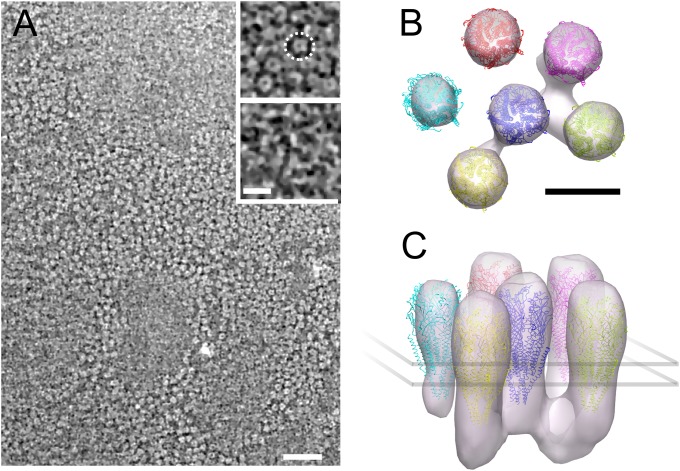
To explore the 3D structure of the receptor–rapsyn network, we performed cryoelectron tomography on the isolated Torpedo postsynaptic membranes (Methods). These membranes formed flat closed vesicles that were thin enough to be plunge-frozen on a support grid and directly imaged. Densely packed receptors are apparent on the postsynaptic membrane surface (Fig. 1A). Their extracellular portions appear as rings (Fig. 1A, Upper Inset), whereas their smaller intracellular portions are associated with filamentous structures (Fig. 1A, Lower Inset). However, even after nonlinear anisotropic diffusion filtering, it was not possible to trace objectively the filamentous structures because of the limited signal-to-noise ratio characteristic of cryotomograms and the small size of the filamentous structures. To gain a clearer view of these structures and their organization, subtomograms (i.e., tomogram subvolumes) were aligned and averaged without using any initial model. For that purpose, we manually picked receptors. Taking advantage of the fact that they were located in relatively flat membranes, we computed the orientation of each receptor long axis defined as the normal vector to the plane that best fitted to the position in three dimensions of the nearby receptors. After correcting for the orientation of the receptor long axis, we were able to compute a first average. We then refined it by aligning each picked particle to the average and computing an updated average in an iterative way. For alignment, we used a spherical mask to include a central receptor and its nearest neighbors.
Fig. 1.
Highly concentrated receptors and associated cytoplasmic proteins in an isolated fragment of postsynaptic membrane. (A) Tomographic slice cross-sectioning the extracellular domain of many receptors. (Upper Inset) Magnified view of a detail in the same slice: receptors appear as dense rings with a central cavity. One receptor is circled by a dashed ring. (Lower Inset) Detail with the same xy coordinates as in the Upper Inset but taken 8.2 nm under it, at the level of the narrow cytoplasmic ends of receptors. Filamentous structures interconnecting receptors are visible. Electron-dense structures appear brighter than the low-density background. (B and C) Organization of receptors and associated cytoplasmic proteins obtained by subtomogram averaging as viewed from the synaptic cleft (B) and near parallel to the membrane (C). Receptors are connected to up to three other receptors through proteins at their base, ∼3 nm underneath the membrane surface. [Scale bars: 50 nm (A, main frame); 10 nm (A, Inset; B; and C)]
The refined density map and docked-in atomic model (14) (Fig. 1 B and C) resembles the close packed arrangement of dimers of receptors observed in electric organ synapses and in tubular vesicles of isolated postsynaptic membranes (15, 16). Based on the strength of the densities, which reflects the level of reproducibility, most receptors are consistently surrounded by three to five neighbors in defined positions. Densities are apparent connecting the central receptor with three other receptors on the cytoplasmic side of the membrane. These can be identified with the filaments shown in Fig. 1A (Lower Inset). As Fig. 1 B and C shows, the connections lie well below the membrane surface, creating a submembrane protein layer. This layer can be identified with the line structure consisting of rapsyn in contact with receptors observed in 2D electron microscopic images of cross-sectioned postsynaptic membranes (17, 18). However, the visibility of the filamentous features is sensitive to the threshold used for isosurface representation, suggesting that both their presence and their orientation with respect to the receptor lattice could vary. No density interconnects receptors on the extracellular side of the membrane, as occurs in clustering of acetylcholine receptors in Caenorhabditis elegans membranes (19).
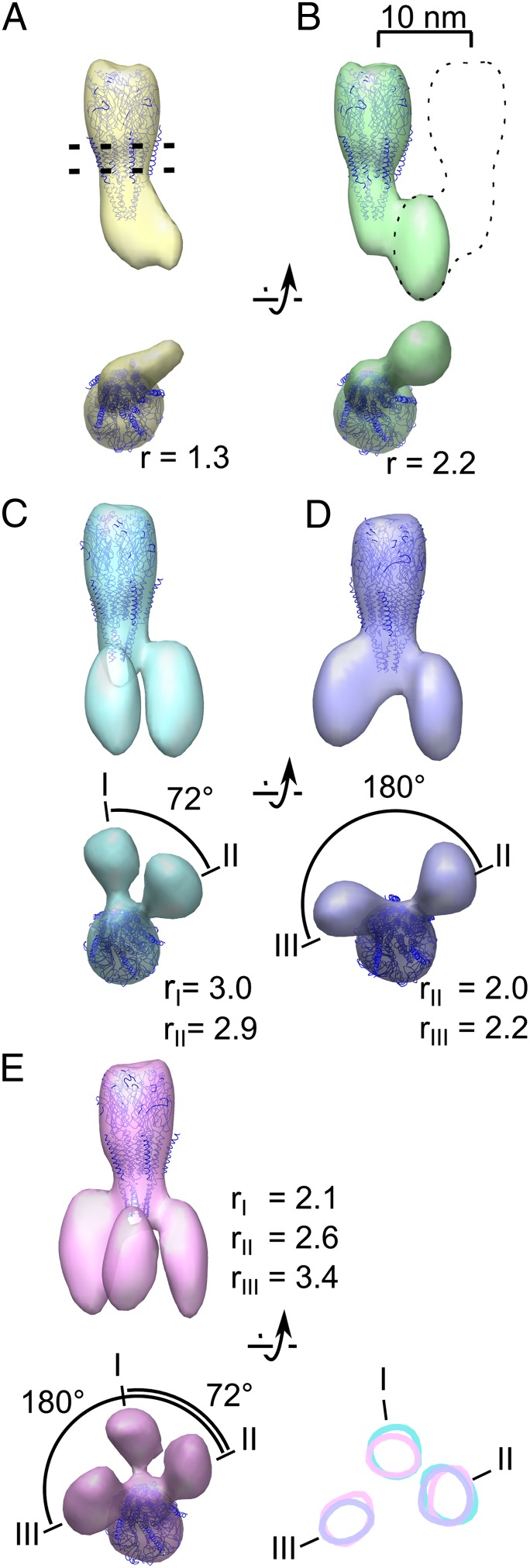
We next analyzed the cytoplasmic densities associated with individual receptors by designing a tighter mask that includes only one receptor and all of the potentially associated cytoplasmic densities to prevent neighboring receptors from driving the alignment (Fig. S1). Subtomograms were aligned and classified simultaneously without initial models to obtain seven class averages. Out of these, five exhibit well-defined lobes extending from the base of the receptor and are presented in Fig. 2. The atomic model of the receptor (14) can be fitted within the central density of every resulting class average (Fig. 2). In addition, the class averages possess a variable number of lobes associated with the receptor in the cytoplasmic region. The first class (class A; Fig. 2, yellow) shows a single lobe occupying 70 nm3. Class B (Fig. 2, green) has a lobe occupying 124 nm3, which is approximately twice the volume of the first. The remaining three classes show two or three lobes, which are roughly the volume of the lobe in class B and the azimuthal position of which varies (see below).
Fig. 2.
Receptors are bound to up to three rapsyn lobes. (A–E) Averages obtained after classification as viewed parallel to the membrane (top of each pair) or from the cytoplasm (bottom of each pair). The position of the plasma membrane is represented by two dashed lines in A, and the location of a masked out neighboring receptor is indicated by a dashed outline in B. A variable number of rapsyn lobes is observed in the different class averages. When more than one lobe is present, the different lobes are labeled with roman numerals. The ratio r between the volume of each lobe and the theoretical volume of one rapsyn molecule is shown. Represented at the lower right of the figure are the surface contours of class averages C, D, and E, in a 1-voxel-thick slice located at the level of the rapsyn lobes. Position I is occupied in class averages C and E; position II in class average C, D, and E; and position III in class average D and E. The relative numbers of receptors sorted to the different classes are (from class A to E) 21.5%, 23.1%, 16.5%, 20.1%, and 18.8%.
A straightforward interpretation of these lobes is that they correspond to rapsyn molecules. Rapsyn is the only protein present in amounts as high as receptors in this experimental system (10, 20), and we demonstrated the submembrane localization of rapsyn by gold immunolabeling (Fig. S2). Moreover, the lobes all have the same shape and appear with strong densities, comparable with those of the receptor itself. This can only happen, as a result of reinforcement during averaging, if the attached proteins have similar shapes and equivalent orientations and are present in similar concentrations as the receptor itself.
We assume that class A corresponds to the case where there is only one rapsyn molecule present because the measured lobe volume nearly corresponds to the volume estimated for a protein of rapsyn mass (21) (ratio between the measured volume and the estimated volume; r = 1.3; Fig. 2A). The approximately two-times-larger volume in class B (r = 2.2; Fig. 2B) is consistent with a dimer of rapsyn molecules connecting the receptor with another receptor situated outside the mask (as suggested by the dotted line in Fig. 2B; see also Fig. S1). This agrees with the fact that rapsyn can self-associate (3, 4).
Class C (Fig. 2, cyan) shows two rapsyn lobes, which are separated by an angle of ∼72° (a fifth of 360°). This angle suggests that rapsyns are bound to homologous sites of two adjacent receptor subunits. Class D (Fig. 2, violet) also shows two rapsyn lobes but they are ∼180° apart, suggesting that in this alternative configuration the two rapsyn molecules are bound to nonhomologous locations on two receptor subunits. The Class D lobes may be responsible for the twofold symmetric structure that has been observed at the base of the receptor in tubular crystals (22). Class E (Fig. 2, pink) has three rapsyn lobes bound to the receptor that we identify as I, II, and III (Fig. 2E). The angle between I and II is ∼72°, whereas the angle between I and III is ∼180°. The two rapsyn lobes of class C match closely lobes I and II of class E (Fig. 2, Lower Right, cyan and pink contours). Similarly the two rapsyn lobes of class D match closely class E lobes II and III (Fig. 2, Lower Right, violet and pink contours). These correlations strongly suggest that the acetylcholine receptor possesses just three specific rapsyn-binding sites. Whenever present, lobe III appears to contact the receptor at a level closer to the plasma membrane than do lobes I and II. This further supports the hypothesis that rapsyns corresponding to lobes I and II bind to homologous sites of receptor adjacent subunits, whereas the rapsyn corresponding to lobe III binds to a nonhomologous site.
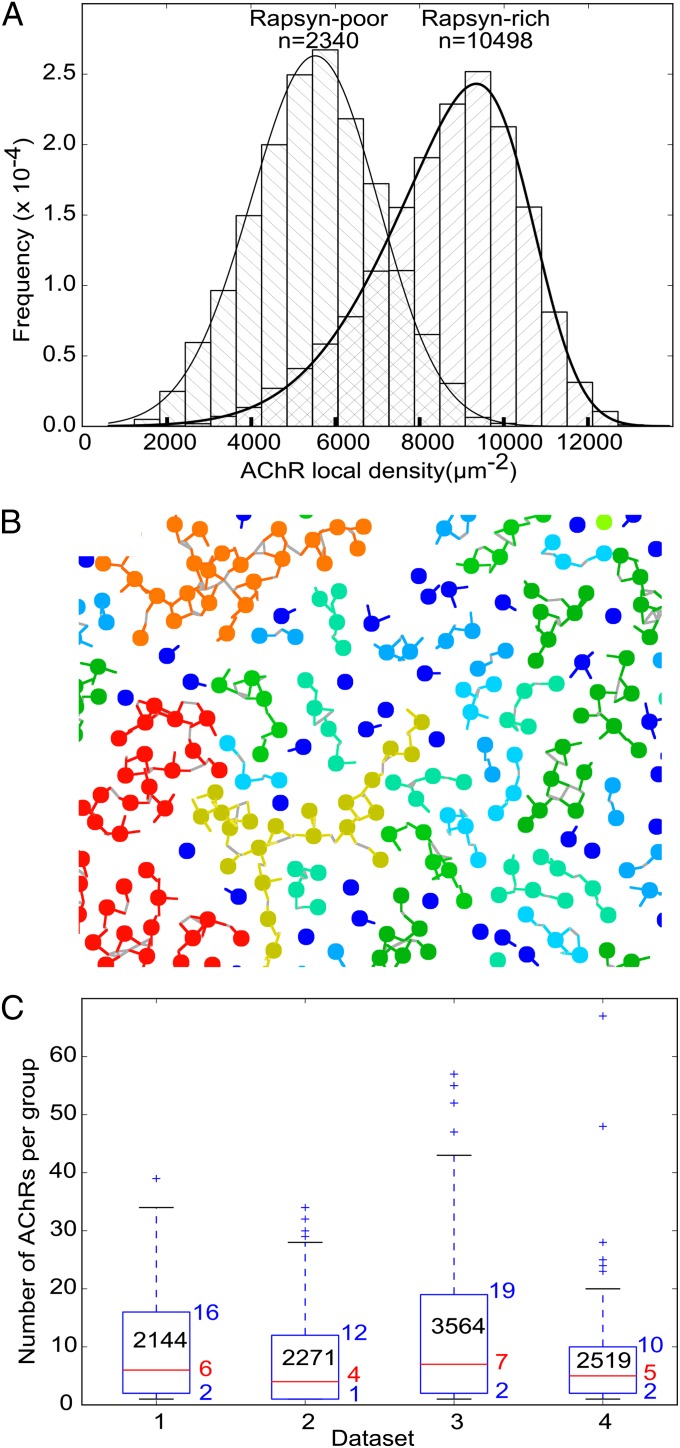
The results above were obtained from more than 3,000 receptor–rapsyn complexes extracted from a single tomogram. Additionally, we analyzed five other tomograms. Similar results were obtained from three of them. The remaining two tomograms showed considerably less filamentous structures, resulting in the absence of class averages with more than one rapsyn lobe. For each receptor in all six tomograms, we measured the local receptor density and plotted two histograms: one for rapsyn-rich and the other for rapsyn-deficient tomograms (Fig. 3A). The mean density was significantly higher in rapsyn-rich (8,708 ± 1,840 µm−2; fitted skew normal distribution maximum: 10,119 µm−2; n = 10,498) than in rapsyn-deficient tomograms (5,458 ± 1,582 µm−2; fitted skew normal distribution maximum: 5,522 µm−2; n = 2,340; P < 0.001 by Mann–Whitney test), confirming the importance of rapsyn for maintaining a high receptor concentration.
Fig. 3.
Large-scale organization of receptors and rapsyns. (A) Frequency histograms of the local concentration of receptors in rapsyn-rich and rapsyn-poor tomograms, respectively. (B) Detail of a receptor–rapsyn interaction network projected on a 2D plane. Receptors are represented as beads, and rapsyn lobes are represented as colored lines. Gray lines are drawn between contacting rapsyn lobes. The color of receptors and rapsyns is related to the size of the groups [from dark blue (single receptor) to red (>50 receptors)]. (C) Boxplots of the size of the group of rapsyn-connected receptors to which each receptor belonged. One boxplot is shown per rapsyn-rich dataset. The lower and upper edges of the boxes represent the 25th and 75th percentiles, respectively, whereas the red lines represent the median. The corresponding numerical values are written next to the boxes. The number of receptors in each area of investigated postsynaptic membrane is reported in the boxes. The whiskers extend to the most extreme data point comprised within ranges d1 = [p25 − 1.5 × (p75 – p25), p25] and d2 = [p75, p75 + 1.5 × (p75 – p25)], where p25 is the 25th percentile and p75 is the 75th percentile. More extreme values are shown as crosses.
Finally, we examined how receptor–rapsyn complexes are organized on a larger scale. For that purpose, the coordinates of the center of each rapsyn density and of the cytoplasmic region of the receptor in the class averages were transposed into the coordinate system of the original tomogram. This was done by applying the inverse rotation and translation operations found during subtomogram alignment. Two receptors were considered connected if the centers of their respective rapsyn densities were separated by less than twice the rapsyn density radius (Fig. 3B). For each receptor, the size of the group that it belonged to was then obtained. In the four rapsyn-rich tomograms, groups of up to 67 connected receptors were found. Overall, 75% receptors were connected to at least another receptor, 50% belonged to groups of 5 receptors or more, and 25% belonged to groups of 14 or more (Fig. 3C).
Discussion
In this study, we have examined the fine structure of the network formed by the nicotinic acetylcholine receptor and rapsyn at the Torpedo electric organ synapse, a model system for the neuromuscular junction. In the early 1980s, Sealock demonstrated that rapsyn appears as a thin line in the cytoplasm underlying the receptors in electron micrographs of Torpedo cross-sectioned postsynaptic membranes (17). The author also showed that the very same structure was present in mouse neuromuscular junctions (18). At the same time, Burden et al. showed that rapsyn and the receptor β-subunit are closely associated (10). This led to the hypothesis that rapsyn forms a subcortical scaffold for receptors. Later on, other subunits of the receptor were found to interact with rapsyn by colocalization studies in heterologous cell systems of coexpressed rapsyn and either individual full-length subunits of the receptor or chimeric membrane proteins containing cytoplasmic loops of individual receptor subunits (6, 11). However, only the maximum number of rapsyn molecules potentially interacting with each receptor could be inferred from these fluorescence microscopy studies. Also, because of the artificial systems used, it cannot be excluded that these results are artifactual.
Our direct structural approach, investigating the receptor–rapsyn complexes in a native membrane system, avoids the ambiguities that arise from studies with heterologous cell systems, enabling a clear description of the rapsyn–receptor network as it exists in situ. The single average of all receptors showed three cytoplasmic associated densities, identified as rapsyn molecules (Fig. 1 B and C). Subtomogram classification reveals that the receptor can have one, two, or three rapsyns in positions denoted I, II, and III (Fig. 2E) and that there are two alternative configurations of rapsyn–receptor complexes involving two rapsyns (Fig. 2 C and D). The precise arrangement of the binding sites with respect to receptor subunits could not be determined in the present study because the resolution (∼4 nm; Fig. S3) was not sufficient to identify individual receptor subunits. Nevertheless, binding sites I and II seem to be homologous and located on two adjacent subunits, because they are separated by an angle of ∼72° and positioned at a similar level along the receptor long axis. On the other hand, site III seems to be in a nonhomologous position, because it is located closer to the membrane and separated from sites I and II by ∼108° and ∼180°, respectively. The absence in Fig. 2 of a class where only the positions I and III would be occupied indicates that the affinity for rapsyn is stronger at site II than at the two other sites. It has been reported that the β-subunit strongly binds rapsyn (10, 11). Therefore, it seems likely that site II is located either fully on the β-subunit or at the interface between the β-subunit and another subunit.
Our comparison of tomograms comprising different amounts of rapsyn reveals that acetylcholine receptors are more densely packed when rapsyn is more concentrated (Fig. 3A), in agreement with reports of tighter packing and stability of the receptor following increase in rapsyn expression during development or rapsyn overexpression (23). We did not observe a very large reduction in the concentration of acetylcholine receptors that might have been expected to occur over time in the rapsyn-depleted membranes. This is because receptors could not diffuse far away because of the limited size of isolated membrane patches (Methods).
To better understand the topology of the rapsyn–receptor network, we replaced each receptor analyzed by subtomogram averaging by its corresponding class average and studied the connectivity of the receptors resulting from rapsyn links. Our analysis showed that most receptors belong to groups of two to 14 interconnected receptors (Fig. 3 B and C). Typically, 25% of the receptors present in a cluster are not connected to any other receptors. We consider two possible explanations. (i) Rapsyn–receptor interaction is dynamic, and receptors are transiently unbridged but not long enough to allow them to escape their cluster. (ii) The unbridged receptors cannot diffuse away from their receptor cluster because they are trapped between other receptors that are immobilized through rapsyn bridges. The latter hypothesis is in good agreement with a recent report of single-molecule tracking experiments in cultured myoblasts where 20% of the receptors showed restricted diffusion in small domains of ∼50 nm, whereas the majority of the receptors were immobile (24).
Confocal microscopy of overexpressed rapsyn in nonmuscle cells suggested that rapsyn self-assembles into a submembrane network and that acetylcholine receptors can subsequently dock onto this network (6). However, this hypothesis was brought into question by experiments in C2 myotubes showing that rapsyn does not form a network in the absence of acetylcholine receptors (8, 9). Our results are consistent with the latter observations. Indeed, we did not observe only one large mesh of rapsyn per postsynaptic membrane but, instead, a collection of small bridges connecting pairs of receptors.
Because most receptors possess two or more bridges, cross-linked groups of more than two receptors develop. Our observation that the receptor contains three binding sites for rapsyn molecules seems important for the formation of 2D networks. This number is the minimum needed because two binding sites will only lead to linear or circular arrays. On the other hand, with more binding sites, as proposed on the basis of single receptor subunit overexpression studies (6, 11), the connectivity (and, hence, stability) of the network might be too high to facilitate acetylcholine receptor turnover (25).
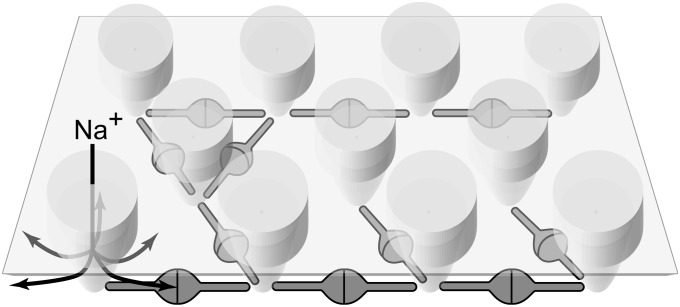
Viewed from the side, the main mass of the network lies well below the plasma membrane (Figs. 1C and 2), leaving ∼30 Å of space underneath the membrane for ions to diffuse and providing them with unrestricted access to the lateral openings of the acetylcholine-gated channels (22) (Fig. 4). Cytoskeletal elements such as actin filaments were not observed in any of the analyzed tomograms. The cross-linking of receptors by rapsyn, therefore, seems to be sufficient to reduce receptor diffusion and, thereby, maintain an elevated receptor concentration, in agreement with recent single-molecule tracking experiments (24). It is, nonetheless, likely that interactions between rapsyn and the actin cytoskeleton via the dystroglycan complex further stabilize receptor clusters (26).
Fig. 4.
Model of the nicotinic acetylcholine receptor–rapsyn network. Receptors are tightly packed and are interconnected by rapsyn dimers. Up to three rapsyn dimers contact each receptor in specific locations at its base. The interconnecting arms of individual rapsyn molecules lie below the intracellular entrances of the receptor channels (arrows), leaving unrestricted submembrane space within which ions can freely diffuse.
In summary, we have described the architecture of the native Torpedo postsynaptic membrane, showing that the majority of acetylcholine receptors interact with rapsyn. The receptor possesses three binding sites for rapsyn, the minimum number required to form a 2D network. This explains how acetylcholine receptors are maintained in high concentrations at the synapse. Other pentameric neurotransmitter-gated ion channels such as GABAA receptors, glycine receptors, or type 3 serotonin receptors might interact with their clustering partners by similar principles.
Methods
Postsynaptic Membrane Preparation.
Postsynaptic membrane isolation was modified from a well-established protocol (27). All steps were performed at 4 °C; 100 g of T. marmorata electric organ was homogenized in 40 mL of isolation buffer [400 mM NaCl, 20 mM phosphate buffer, 0.3 mg/L leupeptin , 1 mg/L pepstatin (pH 7.4)]. During this process, cell membranes break up in patches only slightly larger than the areas containing the receptor clusters, which are mechanically distinct. The homogenate was centrifuged at 5,000 rpm in an SLA-1500 rotor (Sorvall) for 10 min. The supernatant was kept for later use; the pellet was resuspended in 40 mL of isolation buffer, homogenized, and centrifuged as before. The supernatant was recovered, mixed with the supernatant of the first centrifugation step, and centrifuged at 8,000 rpm for 1 h. The pellet was resuspended in 41.5% (wt/wt) sucrose solution and was homogenized with six strokes in a glass/Teflon tissue homogenizer. It was then poured over a 37%/41.5% sucrose gradient (15 mL each); 10 mL of 30% sucrose solution was poured on top. All sucrose solutions contained 0.02% Na azide. The sucrose gradient was centrifuged for 2.5 h at 45,000 rpm in a 45Ti rotor (Beckman Coulter). The band between 37% and 41.5% was collected and kept at 4 °C.
Electron Microscopy.
Postsynaptic membranes from the sucrose gradient were diluted 30× in isolation buffer; 10-nm BSA-gold beads (BBI) were mixed as fiducial markers to the postsynaptic membranes, and 4 µL of this mix was transferred on one side of lacy carbon 300 mesh copper grids (Agar Sci) that were glow-discharged previously in the presence of n-amylamine. The excess fluid was absorbed with a blotting paper applied on the other side of the grids, and the grids were plunge-frozen in liquid ethane. Imaging was carried out at a temperature of ∼80 K and at 300-kV acceleration voltage on a Polara electron microscope (FEI) equipped with an energy filter and a 4,096 × 4,096 pixels CCD camera (Gatan). Typically, single- and dual-axis tilt series were recorded with SerialEM (28) over a tilt range from −70° to 70° at a magnification of 69,000× using a camera binning factor 2 (calibrated binned pixel size: 3.74 Å), a defocus of 3–6 µm, and a total radiation of 40 to 100 electrons per square angstrom. Alignment parameters were determined using fiducial markers in IMOD (29). Images were binned by another factor 2 (pixel size: 7.48 Å), and tomograms were reconstructed by the simultaneous iterative reconstruction technique (SIRT) in IVE PRIISM (30). For dual-axis tilt series, the two SIRT tomograms were aligned and merged in IMOD (31). For visualization, tomograms were denoised by nonlinear anisotropic diffusion implemented in IMOD (32).
Subtomogram Averaging and Classification.
Subtomogram positions were manually determined in denoised tomograms, where receptor extracellular parts could be unequivocally identified. The orientation of the long axis of each receptor was estimated with custom python scripts using the SciPy module by fitting a plane to the positions of all surrounding receptors within a radius of 20 nm. Subtomograms were extracted and normalized (mean, 0; SD, 1) using Bsoft (33). An initial model was generated by averaging the extracted subtomograms after rotating them to bring their long receptor axis to a common register. Subtomogram averaging was performed using Xmipp ml_tomo (34) with a 17-nm-diameter spherical mask and correcting for the missing data because of incomplete angular sampling. Euler angle phi and rot were scanned in a range of ±20° around the receptor long axis estimate. Psi angle search (i.e., rotation around the receptor long axis) was not restricted.
For subtomogram classification, a mask was applied to keep only the central receptor and its potentially associated cytoplasmic densities (Fig. S1). Prior to application, the mask was rotated according to the predicted orientation of the receptor long axis. The excluded area was filled with Gaussian noise (mean, 0; SD, 0.9). Ten initial models were generated by randomly assigning each individual subtomogram to 1 of 10 classes after rotating them according to the orientation of their receptor computed long axis using Xmipp ml_tomo. Subtomograms were then iteratively aligned, classified, and averaged with a resolution cutoff at 2.5 nm. Out of the 10 class averages, several looked very similar. The class averages that appeared most different from one another were selected for further refinement against all subtomograms. The class exhibiting a single associated lobe was further separated using Xmipp ml_tomo-focused classification into the two classes shown in Fig. 2 A and B.
Volume Measurements.
To measure the volume occupied by rapsyn densities, an isosurface threshold was selected to tightly encompass the acetylcholine receptor. Rapsyn densities were then manually contoured and their volume was computed in IMOD. A conversion factor of 1.21 nm3/kDa was used to estimate the corresponding protein mass. It was assumed from its amino acid sequence that one rapsyn molecule has a mass of 46 kDa (21).
Surface Concentration Measurements.
For each receptor, the local receptor surface density was computed as follows: the number of receptors(j) included in a sphere of 22.5-nm radius centered on receptor(i) was counted. This count was divided by the surface of a 22.5-nm radius disk. Because the membranes were essentially planar, this procedure yielded an accurate estimate of the local surface density. To exclude from the computation receptors(j) located on the opposite part of the flattened postsynaptic membrane vesicle, the angle formed between the long axes of receptor(i) and each receptor(j) was computed. If it was more than 90°, then receptor(j) was excluded from the computation of the density at receptor(i). A histogram of the local surface concentration was plotted for the histograms corresponding to rapsyn-rich and rapsyn-poor datasets. A skew normal distribution was fitted to rapsyn-rich dataset and rapsyn-poor dataset. The statistical significance of the difference between those datasets was assessed by the Mann–Whitney test.
Network Analysis.
The coordinates of the centers of mass of each rapsyn lobe and of the cytoplasmic end of the receptor were measured for each class average. For each subtomogram, the coordinates of the corresponding class average were transposed to the tomogram coordinate system by applying the opposite rotation and translation to those obtained during subtomogram alignment. This and the following steps were performed with a collection of custom python scripts. For each rapsyn center of mass, the distances to the centers of mass associated with different receptors were computed. If these were smaller than 5 nm (the extent of a rapsyn lobe as in Fig. 2 B–E), the two lobes were considered connected. For each receptor, the size of the group of connected receptors to which it belonged was measured and a boxplot analysis was performed.
Gold Immunolabeling.
Whenever buffer exchange was necessary, postsynaptic membranes were pelleted by centrifugation at 14,000 × g for 10 min. Before every step of the labeling procedure, the postsynaptic membranes (which form closed vesicles) were pipetted up and down 10 times in a volume of 20 µL with a 10-µL glass micropipette (Hamilton) to open them by shear force and make the cytoplasmic side accessible to buffer. All steps were performed at room temperature. The membranes were incubated for 1.5 h in 140 µL of PBS containing 0.5% BSA (0.5% BSA-PBS) and anti-rapsyn monoclonal antibody (R2029; Sigma-Aldrich) diluted 50-fold. They were washed three times for 10 min with 1 mL of PBS and then incubated with an anti-mouse antibody coupled to a 5-nm gold bead (G7527; Sigma-Aldrich) diluted fivefold in 140 µL 0.5% BSA-PBS for 1.5 h. The membranes were washed three times for 10 min with 1 mL of PBS. They were resuspended in 100 µL of isolation buffer and were immediately plunge-frozen. Gold beads coupled to secondary antibody served as fiducial marker for tilt series alignment.
Analysis of Gold-Immunolabeled Membrane Tomograms.
Tomograms were obtained as described above except that before reconstruction, the densities corresponding to gold beads in tilt series images were replaced by a disk containing noise with the same mean density and SD as the surrounding area (“gold erasure”). This was done so that gold density would not influence subsequent subtomogram alignment. Most receptors were picked, and a single average was obtained as described in Subtomogram Averaging and Classification with the use of a mask to focus alignment to the central receptor and its associated cytoplasmic densities. The positions of the gold beads in the tomograms were computed with IMOD. For each of them, the closest receptor was determined, and the position of the gold was reported in the coordinate system of the average structure using custom python scripts.
Graphical Representation.
Tomogram slice rendering was carried out with 3dmod (29). Isosurface rendering, automated fitting of receptor atomic model 2BG9, and receptor–rapsyn interaction-network representation were performed with UCSF Chimera (35). All plots were generated with the python plotting library Matplotlib (36).
Supplementary Material
Acknowledgments
We thank our colleagues at the Medical Research Council (MRC) Laboratory of Molecular Biology for fruitful discussions and Jake Grimmett, Shaoxia Chen, and Sjors Scheres for support with the high-performance computer cluster, the cryoelectron microscopes, and Xmipp ml_tomo, respectively. We thank members of the staff at the Station Biologique du Centre National de la Recherche Scientifique et de l'Université Pierre et Marie Curie, Roscoff, France for supplying T. marmorata. We thank Paul Margiotta for his contribution to Fig. 4. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco [supported by National Institutes of Health (NIH) Grant P41 RR001081]. This work was supported by a European Molecular Biology Organization long-term fellowship (to B.Z.), Swiss National Science Foundation Grant PP00P3_139098/1 (to B.Z.), NIH Grant GM61941 (to N.U.), and the MRC (U105184294).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The density maps reported in this paper have been deposited in the EMDataBank (http://www.emdatabank.org/; accession nos. EMD-2376, EMD-2377, EMD-2378, EMD-2381, EMD-2382, and EMD-2383) and the fitted atomic models have been deposited in the Protein Data Bank in Europe (http://www.ebi.ac.uk/pdbe/; accession nos. 4bog, 4boi, 4bon, 4boo, 4bor, and 4bot).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301277110/-/DCSupplemental.
References
- 1.Gautam M, et al. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature. 1995;377(6546):232–236. doi: 10.1038/377232a0. [DOI] [PubMed] [Google Scholar]
- 2.Engel AG. Current status of the congenital myasthenic syndromes. Neuromuscul Disord. 2012;22(2):99–111. doi: 10.1016/j.nmd.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramarao MK, Bianchetta MJ, Lanken J, Cohen JB. Role of rapsyn tetratricopeptide repeat and coiled-coil domains in self-association and nicotinic acetylcholine receptor clustering. J Biol Chem. 2001;276(10):7475–7483. doi: 10.1074/jbc.M009888200. [DOI] [PubMed] [Google Scholar]
- 4.Eckler SA, Kuehn R, Gautam M. Deletion of N-terminal rapsyn domains disrupts clustering and has dominant negative effects on clustering of full-length rapsyn. Neuroscience. 2005;131(3):661–670. doi: 10.1016/j.neuroscience.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 5.Bartoli M, Ramarao MK, Cohen JB. Interactions of the rapsyn RING-H2 domain with dystroglycan. J Biol Chem. 2001;276(27):24911–24917. doi: 10.1074/jbc.M103258200. [DOI] [PubMed] [Google Scholar]
- 6.Huebsch KA, Maimone MM. Rapsyn-mediated clustering of acetylcholine receptor subunits requires the major cytoplasmic loop of the receptor subunits. J Neurobiol. 2003;54(3):486–501. doi: 10.1002/neu.10177. [DOI] [PubMed] [Google Scholar]
- 7.Marchand S, Bignami F, Stetzkowski-Marden F, Cartaud J. The myristoylated protein rapsyn is cotargeted with the nicotinic acetylcholine receptor to the postsynaptic membrane via the exocytic pathway. J Neurosci. 2000;20(2):521–528. doi: 10.1523/JNEUROSCI.20-02-00521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marangi PA, et al. Acetylcholine receptors are required for agrin-induced clustering of postsynaptic proteins. EMBO J. 2001;20(24):7060–7073. doi: 10.1093/emboj/20.24.7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruneau EG, Brenner DS, Kuwada JY, Akaaboune M. Acetylcholine receptor clustering is required for the accumulation and maintenance of scaffolding proteins. Curr Biol. 2008;18(2):109–115. doi: 10.1016/j.cub.2007.12.029. [DOI] [PubMed] [Google Scholar]
- 10.Burden SJ, DePalma RL, Gottesman GS. Crosslinking of proteins in acetylcholine receptor-rich membranes: Association between the beta-subunit and the 43 kd subsynaptic protein. Cell. 1983;35(3 Pt 2):687–692. doi: 10.1016/0092-8674(83)90101-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Rudell J, Ferns M. Rapsyn interacts with the muscle acetylcholine receptor via alpha-helical domains in the alpha, beta, and epsilon subunit intracellular loops. Neuroscience. 2009;163(1):222–232. doi: 10.1016/j.neuroscience.2009.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cartaud J, et al. The Torpedo electrocyte: A model system to study membrane-cytoskeleton interactions at the postsynaptic membrane. Microsc Res Tech. 2000;49(1):73–83. doi: 10.1002/(SICI)1097-0029(20000401)49:1<73::AID-JEMT8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 13.Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2(11):791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- 14.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346(4):967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Heuser JE, Salpeter SR. Organization of acetylcholine receptors in quick-frozen, deep-etched, and rotary-replicated Torpedo postsynaptic membrane. J Cell Biol. 1979;82(1):150–173. doi: 10.1083/jcb.82.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brisson A, Unwin PN. Tubular crystals of acetylcholine receptor. J Cell Biol. 1984;99(4 Pt 1):1202–1211. doi: 10.1083/jcb.99.4.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sealock R. Cytoplasmic surface structure in postsynaptic membranes from electric tissue visualized by tannic-acid-mediated negative contrasting. J Cell Biol. 1982;92(2):514–522. doi: 10.1083/jcb.92.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sealock R. Visualization at the mouse neuromuscular junction of a submembrane structure in common with Torpedo postsynaptic membranes. J Neurosci. 1982;2(7):918–923. doi: 10.1523/JNEUROSCI.02-07-00918.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gendrel M, Rapti G, Richmond JE, Bessereau JL. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 2009;461(7266):992–996. doi: 10.1038/nature08430. [DOI] [PubMed] [Google Scholar]
- 20.Bridgman PC, Carr C, Pedersen SE, Cohen JB. Visualization of the cytoplasmic surface of Torpedo postsynaptic membranes by freeze-etch and immunoelectron microscopy. J Cell Biol. 1987;105(4):1829–1846. doi: 10.1083/jcb.105.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carr C, McCourt D, Cohen JB. The 43-kilodalton protein of Torpedo nicotinic postsynaptic membranes: Purification and determination of primary structure. Biochemistry. 1987;26(22):7090–7102. doi: 10.1021/bi00396a034. [DOI] [PubMed] [Google Scholar]
- 22.Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. Nicotinic acetylcholine receptor at 4.6 A resolution: Transverse tunnels in the channel wall. J Mol Biol. 1999;288(4):765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- 23.Gervásio OL, Armson PF, Phillips WD. Developmental increase in the amount of rapsyn per acetylcholine receptor promotes postsynaptic receptor packing and stability. Dev Biol. 2007;305(1):262–275. doi: 10.1016/j.ydbio.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Piguet J, Schreiter C, Segura JM, Vogel H, Hovius R. Acetylcholine receptor organization in membrane domains in muscle cells: Evidence for rapsyn-independent and rapsyn-dependent mechanisms. J Biol Chem. 2011;286(1):363–369. doi: 10.1074/jbc.M110.139782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akaaboune M, Grady RM, Turney S, Sanes JR, Lichtman JW. Neurotransmitter receptor dynamics studied in vivo by reversible photo-unbinding of fluorescent ligands. Neuron. 2002;34(6):865–876. doi: 10.1016/s0896-6273(02)00739-0. [DOI] [PubMed] [Google Scholar]
- 26.Pilgram GS, Potikanond S, Baines RA, Fradkin LG, Noordermeer JN. The roles of the dystrophin-associated glycoprotein complex at the synapse. Mol Neurobiol. 2010;41(1):1–21. doi: 10.1007/s12035-009-8089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobel A, Weber M, Changeux JP. Large-scale purification of the acetylcholine-receptor protein in its membrane-bound and detergent-extracted forms from Torpedo marmorata electric organ. Eur J Biochem. 1977;80(1):215–224. doi: 10.1111/j.1432-1033.1977.tb11874.x. [DOI] [PubMed] [Google Scholar]
- 28.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): A software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116(1):56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 31.Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol. 1997;120(3):343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 32.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135(3):239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 33.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157(1):3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Scheres SH, Melero R, Valle M, Carazo JM. Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure. 2009;17(12):1563–1572. doi: 10.1016/j.str.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Hunter J. Matplotlib: A 2D Graphics Environment. Comput Sci Eng. 2007;9(3):90–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.