Abstract
The sophistication and success of recently reported microfabricated organs-on-chips and human organ constructs have made it possible to design scaled and interconnected organ systems that may significantly augment the current drug development pipeline and lead to advances in systems biology. Physiologically realistic live microHuman (µHu) and milliHuman (mHu) systems operating for weeks to months present exciting and important engineering challenges such as determining the appropriate size for each organ to ensure appropriate relative organ functional activity, achieving appropriate cell density, providing the requisite universal perfusion media, sensing the breadth of physiological responses, and maintaining stable control of the entire system, while maintaining fluid scaling that consists of ~5 mL for the mHu and ~5 µL for the µHu. We believe that successful mHu and µHu systems for drug development and systems biology will require low-volume microdevices that support chemical signaling, micro-fabricated pumps, valves and microformulators, automated optical microscopy, electrochemical sensors for rapid metabolic assessment, ion mobility-mass spectrometry for real-time molecular analysis, advanced bioinformatics, and machine learning algorithms for automated model inference and integrated electronic control. Toward this goal, we are building functional prototype components and are working toward top-down system integration.
Keywords: Artificial biological organs, biological systems, biotechnology, systems biology
I. Introduction
INTEREST is rapidly growing in using microfabricated human organs-on-chips (OoCs) and tissue-engineered human organ constructs (HoCs) to complement the cell-culture/animal/ human tests comprising the current drug development pipeline [1]. While there are a number of individual OoCs/HoCs as described in recent reviews [2]–[4], only limited reports describe coupled organs [5], [6]. The study of how multiple organs affect drug efficacy and toxicity requires the interconnection of many OoCs/HoCs to create physiologically realistic microHuman (µHu) and milliHuman (mHu) systems that can operate stably for weeks (see Fig. 1). This presents a remarkable series of engineering challenges, which if met could revolutionize drug discovery and systems biology.
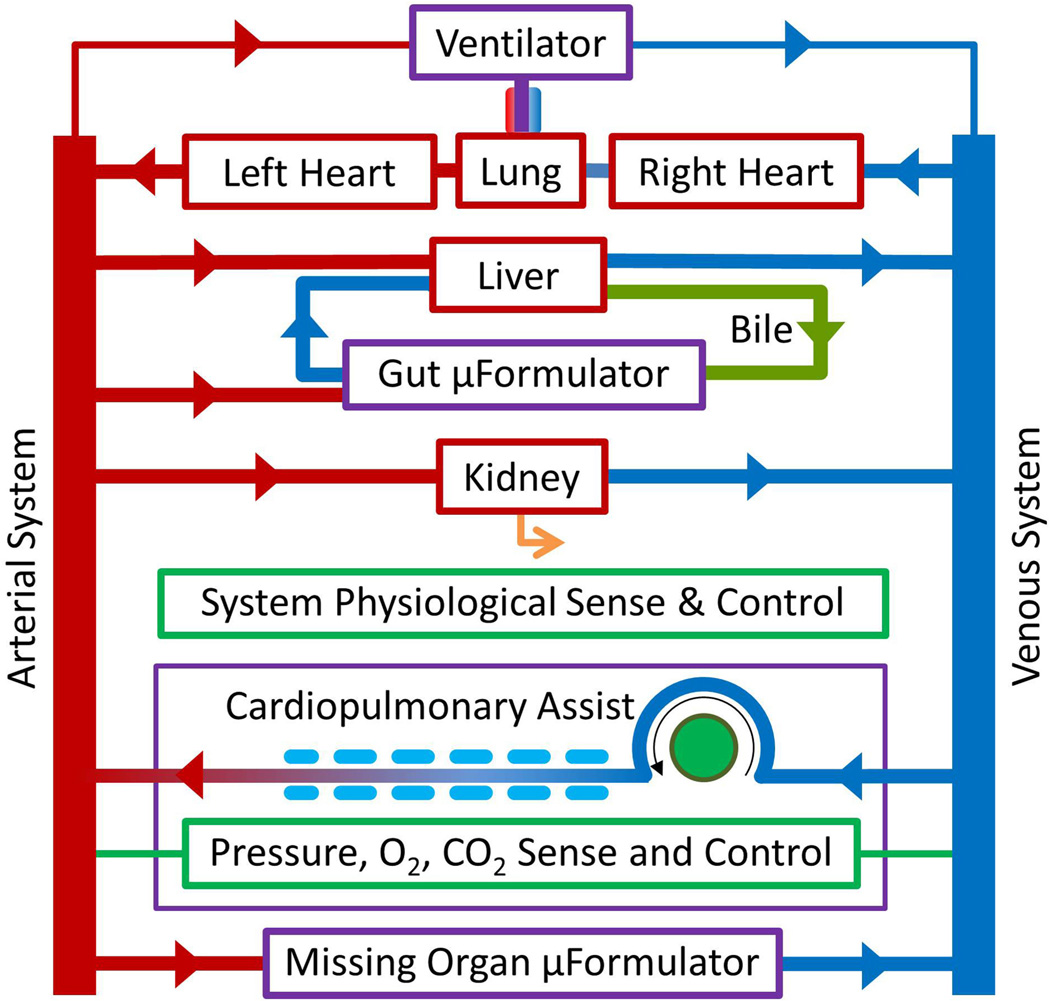
Fig. 1.
Schematic representation of how multiple OoC devices might be interconnected to form an instrumented mHu system.
While there are numerous individual challenges associated with the development of individual organs (e.g., bone marrow, brain, gastrointestinal system, heart, kidney, liver, lung, lymph node, pancreas, skeletal muscle, skin, testes, ovary, uterus, etc.), it is important to recognize that there are two complementary approaches when developing OoC systems. Bottom-up begins with a detailed specification of each organ, progresses to combine them into coupled systems (e.g., heart–lung and intestine–liver), and continues adding organs to create more complex models. Top–down considers the abstract, system-level architecture [7] of a human and then breaks the system down into the functionality of compositional organ systems (e.g., scaling, blood surrogate, interconnections, sensing, and control). The descriptive scale and interconnections are successively refined until the baseline specifications are defined. Both approaches are needed and ideally will be complementary. We now address these challenges from a top-down perspective.
II. Engineering Challenges
As summarized by the partial list in Table I, the challenges in developing useful coupled OoC/HoC systems span a breadth of engineering, biological, and medical disciplines. These challenges range from fundamental issues of organ scaling to obtaining analytical measurements in very small volumes, and include modeling, control, and cost.
TABLE I.
Engineering Challenges for Coupled OoCs
| Determining the proper size of each organ |
| Fluidic control of mL and µL volumes |
| Analytical chemistry in µL and nL volumes, including comprehensive molecular characterization in real time |
| Maintaining and controlling coupled organ systems |
| Vascularizing organs with appropriate surface-to-volume ratios |
| Developing a universal blood surrogate |
| Missing organs and the adjustment of blood surrogate |
| Modeling coupled organ systems |
| Characterization of organ health and disease |
| Minimizing organ cost to enable high-content screening |
A. Determining the Proper Size of Each Organ
The coupled OoC programs funded by the Food and Drug Administration (FDA) [8], the Defense Advanced Research Projects Agency (DARPA) [9], the National Institutes of Health (NIH) [10], and the Defense Threat Reduction Agency (DTRA) [11] aim to develop miniature, coupled organoid systems that utilize human cells to measure drug efficacy, toxicity, and interorgan interactions, and possibly study the effects of maliciously engineered pathogens. Hence, the success of these projects is contingent on constructing organ models that have the correct relative sizes and vascular volumes, lest drug metabolites from one organ are presented to other organs in unrealistically high or low concentrations. For instance, if a 0.1 µLung [12] were coupled to a mLiver [13], the mLiver might not respond significantly to drugs or toxins delivered through or metabolized by the µLung, e.g., the conversion of angiotensin I into angiotensin II. To design and achieve organ systems representing the appropriate fraction of an adult human is a major systems engineering challenge.
In contrast to the various animals used in conventional pharmacokinetics and pharmacodynamics (PKPD) studies, whose relative organ sizes have evolved in a self-consistent manner over eons, the relative size of organs for a coupled HoC/OoC system is at the discretion of the systems integrator. This flexibility has important implications for the assessment of the effects of drugs and toxins on system physiology, and may also provide an opportunity for creating model systems that are better matched to human physiology of various ages or disease states than are smaller mammals.
There is vast literature on allometric scaling of organs as a function of body mass from the shrew to the whale [14]–[18]. While an animal’s mass M increases as linear dimension L3, structural issues require that the cross-sectional area of the bones increase out of linear proportion [15], [16]. Quantities such as liver mass are found to be proportional to the body mass raised to a power, termed the allometric coefficient. Metabolic rates scale approximately as M0.75, blood circulation time scales as M0.25, and pulmonary and vascular networks exhibit M0.75 scaling. Different organ parameters scale with different allometric coefficients. For example, the allometric coefficients for hepatic mass, blood flow, blood volume, and oxygen consumption are 0.886, 0.91, 0.86, and 0.69, respectively [19]. Table III shows allometric scaling of organ mass with body mass using published primate scaling laws [14], where the organ weight Mo in grams is given by Mo = A × MbB, where Mb is the body weight in kilograms. Examination of the rightmost column, which shows the ratio of 1000 times the mHu organ mass, i.e., 1000×mHu Mo, to that of the adult human, 1.0×HuMo, demonstrates that because of the extrapolation over three orders of magnitude in mass and the differing allometric coefficients for each organ, it is not possible to use allometric scaling to construct a mHu that has the same relative proportions as an adult human—the discrepancy between organ sizes ranges from a factor of 0.4 to 10. The situation is even worse in scaling metabolic rates and blood circulation time, with allometric coefficients of 0.75 and 0.25, so that were one to use a simple allometric scaling approach in building a mHu, one might create a system that was representative of a mouse, with a heart rate of ~500 beats per minute (bpm) and the appropriate metabolic rates, rather than a mHu, with a heart rate of ~70 bpm. Furthermore, there is no universal agreement as to whether relative organ sizes should be normalized by mass, surface area, volumetric flow [20], or other principles [21], and one should not expect to find a consistent set of allometric coefficients that would allow one to predict the correct value of various organ parameters. As organs are made smaller, scaling will ultimately fail, since individual cells have a fixed size and appear at low density (three leukocytes per microliter of cerebral spinal fluid, and a ventricular myocardium must be at least one cell thick).
TABLE III.
Allometric Scaling of Organs for a Hu and mHu
| Organ | A | B | Hu Mb=60 kg |
mHu Mb =60 g |
1000 mH /H |
||
|---|---|---|---|---|---|---|---|
| Mo, g | % Mb | Mo, g | % Mb | ||||
| Liver | 33 | 0.93 | 1500 | 2.5% | 2.4 | 4.0% | 1.6 |
| Brain | 85 | 0.66 | 1300 | 2.1% | 13 | 22% | 10 |
| Lung | 9.7 | 0.94 | 4600 | 0.8% | 0.69 | 1.2% | 1.5 |
| Heart | 5.2 | 0.97 | 280 | 0.46% | 0.34 | 0.57% | 1.2 |
| Kidneys | 6.3 | 0.87 | 2200 | 0.37% | 0.54 | 0.91% | 2.4 |
| Pancreas | 2 | 0.91 | 83 | 0.14% | 0.15 | 0.26% | 1.9 |
| Spleen | 1.5 | 0.85 | 49 | 0.081% | 0.14 | 0.23% | 2.8 |
| Thyroid | 0.15 | 1.1 | 15 | 0.025% | 0.01 | 0.01% | 0.44 |
| Adrenals | 0.53 | 0.7 | 9.3 | 0.016% | 0.07 | 0.12% | 7.9 |
| Pituitary | 0.03 | 0.68 | 0.49 | 0.001% | 0.00 | 0.01% | 9.1 |
The pharmaceutical industry has found it more appropriate to use physiology-based scaling rather than simple allometric scaling [22]–[24] as it extrapolates data from well-plates and cell-culture dishes in the calculation, for example, of the first dose to primates or humans [25]. Simple scaling of drug doses by body weight alone is convenient but can be misleading, particularly if clearance, volume of distribution, and enzyme kinetics are not considered properly in the extrapolation [26]–[29]. It will be necessary to ensure that the mHu-to-Hu extrapolation includes differences in protein binding, drug transport, metabolism, and biliary and urinary excretion between the engineered mHu and adult Hu tissues [30], and also how these factors are affected by missing organs. The delivery of some drugs may be permeability limited, whereas for others flow may be the limiting factor, such that both the circulatory and permeability properties [31] of the mHu HoC/OoC system must be considered relative to the intact Hu.
Hence, we believe that the correct approach for determining the size of an organ is to choose the size that provides the appropriate relative organ functional activity, e.g., heart: volume pumped; lung: gas exchanged; liver: metabolism; kidney: molecular filtering and transport; brain: blood-brain barrier function and synapse formation. It is critical to specify the relevant functions that are to be modeled, determine the efficiency with which each function can be realized in an OoC/HoC, and then construct OoCs/HoCs with the physical dimensions that support the desired functional performance. An OoC/HoC system should reflect a small fraction of an adult human, and, in the words of D. E. Ingber (personal communication), “create living histological sections of an adult human.” A combination of device design, attention to physiological details, measured enzyme kinetics, computational models, and confirmatory experiments will be required to specify the physical dimensions of OoCs/HoCs that will accomplish this.
B. Fluidic Control of Milliliter and Microliter Volumes
Just as the organs need to be scaled functionally, so must be the blood volume. Given that the average blood volume of an adult human (1 Hu) is ~5 L, the blood volume of a mHu and µHu should be ~5 mL and ~5 µL, respectively. The volume of circulating blood surrogate must match organ size to avoid unphysiological dilution of metabolites, hormones, and paracrine signals to the point that each organ operates independent of the other organs. This presents a variety of challenges, in that the small volumes required for mHu and µHu systems for drug development and systems biology place severe constraints on many systems components. Highly instrumented bioreactors will need to support tissue-density cells [32] in a manner that enables paracrine and endocrine signaling [33]. This in turn requires low-volume pumps [34] and miniature valves, all at low cost, and compact and affordable support hardware to allow massively parallel experiments over weeks to months.
These strict requirements place severe fabrication constraints on not only the organs but the larger systems that interconnect, sense, and control their function, as illustrated in Fig. 2. Conventional commercially available syringe and peristaltic pumps have dead volumes that are orders of magnitude larger than the total blood volume of 1.0 µHu or even 10.0 µHu. Hence, there are significant challenges for microfluidic engineers to develop pumps, valves, bubble traps, and interconnects, as well as pressure, flow, and osmolarity sensors with small dead volume, all appropriately scaled to coupled OoCs/HoCs, that do not bind drugs, and that are cheap enough for massive parallelization and high-content screening (HCS). There is progress in meeting these challenges [34] but there is much work to be done in this remarkable creative opportunity.
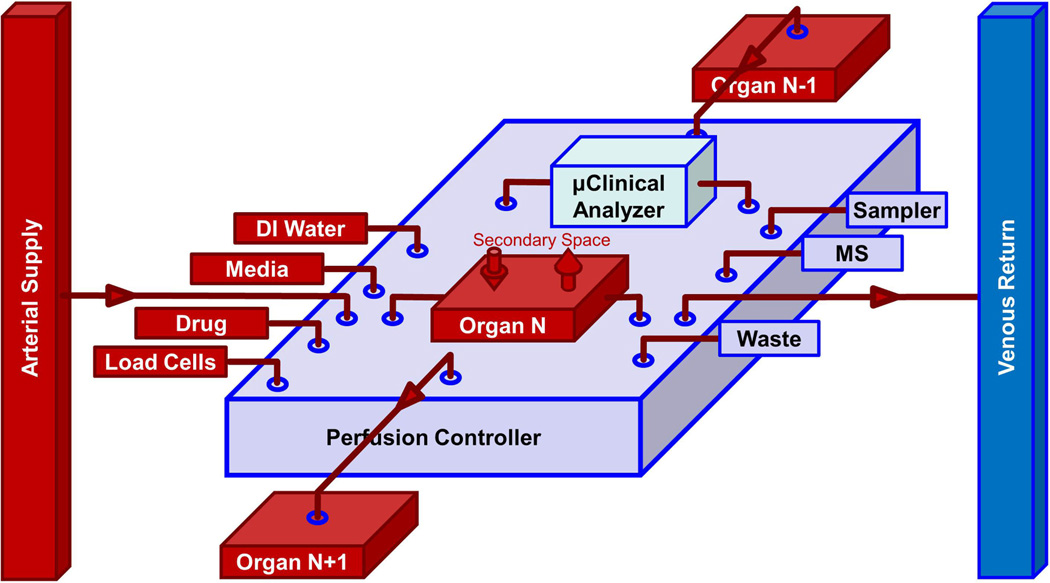
Fig. 2.
Conceptual drawing of an organ-chip cartridge that includes a PC and µCA for both the microvascular and interstitial compartments of a single OoC or HoC. On the left are the inputs to the organ, and on the right are outputs, including an online MS if desired. The vertical arrows access the secondary space of the organ (interstitial, bronchial, biliary, urinary, etc.). Interconnected cartridges would comprise an instrumented mHu or µHu system, either in series as shown for selected organs or in parallel as in Fig. 1.
C. Analytical Chemistry in Microliter and Nanoliter Volumes and Comprehensive Molecular Characterization in Real Time
The study of cellular and system-level physiology is an advanced science that benefits from an armamentarium of analytical instruments, techniques, and measurement protocols. While it may be possible to construct mHu and µHu systems with minimal instrumentation, the need to quantify the PKPD of the integrated OoCs/HoCs with the aforementioned volume constraint severely affects the design of the analytical approaches to characterize organ function and drug response. In contrast to a typical hypothesis-testing biological experiment where a small number of variables are measured dynamically and only a single experimental parameter is varied at a time, the monitoring of an OoC/HoC system would benefit from the dynamic measurement of a large number of physiological variables and the adjustment and even dynamic control of a large number of experimental parameters [35]. Furthermore, binary live/dead assays common in several bioactivity strategies are insufficient to perform complex failure analyses in the integration of multiple artificial organs.
There are several measurable parameters well suited to existing sensor technology that can characterize the dynamic state of OoCs/HoCs (see Table II), including optical imaging of fluorescence markers and reporters, electrochemical sensors for quantifying metabolites, fluorescence- or label-free assays to capture and quantify soluble factors, capillary electrophoresis, and mass spectrometry (MS). Each analytical method needs to be implemented with the appropriate frequency response, as well as small sensor volumes [36]. Given the small size of the OoCs/HoCs, one must address interferences between sensing modalities. Ideally, electrical and electrochemical sensors of interstitial parameters such as pH and glucose, lactate, and oxygen concentrations in microliter (see Fig. 3, [37]–[40]) and nanoliter bioreactors [41]–[43] would free up optical bandwidth for fluorescent measurement of intracellular fluorophores such as those for cytosolic calcium and surface or immunohistological markers of protein expression [33], [44], [45]. In recognition of the desire to use OoC/HoC systems for drug screening, it is inevitable that these systems will require capabilities in automated, high-content optical microscopy and image analysis that are far beyond standard high-throughput robotic well-plate screening.
TABLE II.
Sensors and Actuators for HoC/OoCs
| Parameters that can be measured (sensors) |
|---|
| Morphology: size, shape, optical density, motility, division, organelle configuration, non-translational motion, nuclear shape |
| Force: shear, tension, deformation |
| Intracellular signaling (optical, MALDI-IM-MS): GFP/luciferase reporters, [Ca]I, pHi, Vm, MMP, GFP, GFP-FRET |
| Extracellular electrolytes (electrochemical): [Na]e, [Ca]e, [K]e, [Mg]e, [PO4]e, [Cl]e, [HCO3]e |
| Neurotransmitters (electrochemical): serotonin, acetylcholine, GAB A, … |
| Extracellular metabolites (electrochemical): [glucose](t), [lactate](t), [pH](t), [O2](t), [NO](t), [H2O2](t),… |
| Extracellular metabolites (GC/UPLC-IM-MS): amino acids, carbohydrates, small metabolites, stable isotopic markers |
| Surface expression: specific affinity probes (dyes, Q-dots) |
| Soluble gene expression (Luminex, nESI-IM-MS): cytokines, growth factors, hormones, enzymes |
| Cytosolic proteins and membrane lipids (GFPs, MALDI-IM-MS) |
| Gene expression (mRNA arrays, RT-PCR) |
| Parameters that can be controlled (actuators) |
| Base formulation of universal perfusion medium (blood surrogate) |
| [glucose](t), [lactate](t), amino acids (~15), cytokines (N»1), growth factors (N»1), hormones (N»1), toxins and drugs (N»1) |
| [Na]e(t), [Ca]e(t), [K]e(t), [Mg]e(t), pH(t), [O2](t), [PO4]e(t) |
| Mechanical, electrical, optical, thermal, genetic (siRNA, optogenetic). |
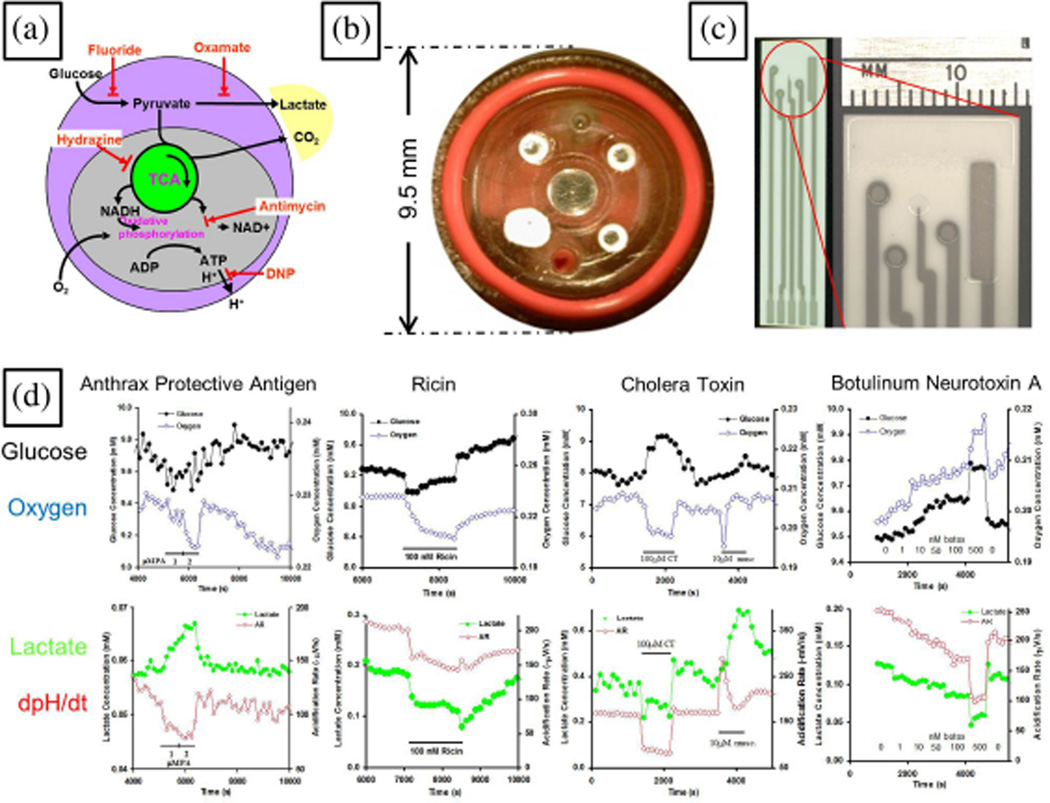
Fig. 3.
Dynamic electrochemical measurements in microbioreactors. (a) Core carbon metabolism and agents that affect it. (b) Modified Cytosensor™ sensor head for measuring glucose, lactate, and oxygen concentrations for ~105 cells in a ~3µL chamber [37]. (c) Screen-printed electrode for the same measurements. Adapted from [38]. (d) Demonstration of how dynamic metabolic measurements can discriminate between different biological toxins. Adapted from [39].
Optical methods alone will be insufficient for OoC/HoC systems. In classical cell culture and tissue engineering, the presence of a specific protein or biomarker is determined by classical western blot and antibody-based enzyme-linked immunosorbent assay, but these targeted assays typically require many micro-liters or even milliliters of supernatant or cytosol. There is great promise for microfluidic implementations of these approaches that can work in nanoliter volumes [46] and quantify specific target molecules.
Untargeted identification of biomolecules in a cell culture or bioreactor typically involves one of many implementations of MS, but used alone, this technique is compromised by the fact that many biomolecules can have similar or identical mass/charge ratios (m/z). Historically, these issues are ad-dressed by separating first by gas chromatography (GC) or high-performance liquid chromatography (HPLC). However, GC requires derivatization of metabolites [47] and hence may not be well suited for an untargeted search. A single HPLC separation requires minutes to hours, and multiple separations may be required. We believe that ion mobility-mass spectrometry (IM-MS) is the best approach to study the effluent of these OoCs/HoCs [48], [49]. In this approach, low-vacuum, gas-phase electrophoresis (IM) can measure molecular cross-sections in microseconds to milliseconds, followed by MS analysis in microseconds. The appeal of this approach is that high-sensitivity ionization techniques coupled to IM-MS can detect thousands of molecular species in a 100-nL sample (see Fig. 4). In contrast with HPLC-MS techniques, the IM-MS approach is nearly real time. The challenge is to apply this technique to analyze OoCs/HoCs without withdrawing volumes larger than a small fraction of the 5 mL and 5 µL volume budget for a mHu and µHu, respectively. A subset of this problem is to devise affinity or microdialysis techniques to remove the salts from the perfusion media without compromising sensitivity or temporal resolution, or exceeding the fluid-withdrawal budget [50]. The high levels of salts in biological media serve to attenuate overall signal response and must be removed rapidly prior to IM-MS analyses [50]. Obviously, given the ~$1 million cost of an IM-MS instrument, they are best suited for a discovery approach wherein unexpected metabolites or side effects are examined in detail to ascertain the underlying pharmacokinetics and metabolomics. Discovering important molecules in such a search will benefit from advanced bioinformatics techniques [51], [52]. IM-MS instruments are too expensive to be used to monitor every OoC/HoC in a large pharmacological study, but the data are exquisitely well suited for guiding the rapid translation of IM-MS discoveries from such untargeted searches into low-cost, low-volume, point-of-use assays that can track specific drug responses in an individual OoC/HoC. Because of the volume constraint, many existing assays are not suitable and new ones must be devised.
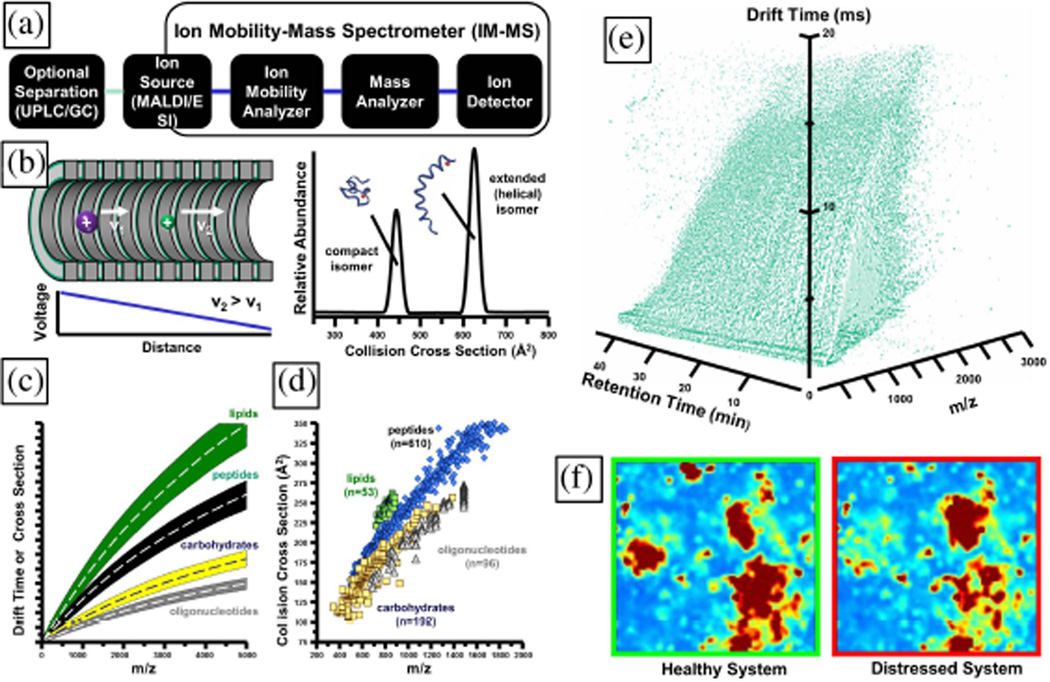
Fig. 4.
Ion mobility-mass spectrometry (IM-MS). (a) Configuration of an IM-MS with GC or ultraperformance liquid chromatography (UPLC) preseparation. (b) Use of gas-phase electrophoresis for rapid separation of molecules by their collision cross-section in low-pressure inert gas. (c) Conceptual ordering of molecules in 2-D IM-MS conformational space. The vertical axis is relative on drift time (µs) or collision cross-section (molecular surface area, Å2) and the horizontal axis is mass-to-charge ratio (m/z). (d) Empirical data demonstrating chemical class specific ordering by a single pass through an IM-MS. (e) 4-D dataset representing ca. 100 000 molecular signals. The entire dataset represents one time point sample from the microfluidic device. Each voxel encodes molecule-specific data of 1) UPLC retention time, 2) IM cross section, 3) MS mass-to-charge, and 4) relative abundance. These data occupy only 10−6 of the instrument phase space. (f) Self-organizing IM-MS heat maps of metabolites in biological system state space. This type of analysis is also ideal for time-series measurements. Panels (a) and (b) are adapted with kind permission from Springer Science+Business Media: Anal. Bioanal. Chem., Biomolecular structural separations by ion mobility-mass spectrometry, 391, 2008, 906, L. S. Fenn and J. A. McLean, Fig. 1(a) and (b). Panel (c) is adapted with kind permission from Springer Science+Business Media: Anal. Bioanal. Chem., Biomolecular structural separations by ion mobility-mass spectrometry, 391, 2008, 906, L. S. Fenn and J. A. McLean, Fig. 2(a). Panel (d) is adapted with kind permission from Springer Science+Business Media: Anal. Bioanal. Chem., Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples, 394, 2009, 235, L. S. Fenn, M. Kliman, A. Mahsut, S. R. Zhao, J. A. McLean, Fig. 1(a).
D. Maintaining and Controlling Coupled Organ Systems
The perfusion controller (PC) and microclincal analyzer (µCA) connections, sensors, instruments, and controls required for the single organ-chip cartridge in Fig. 2 might be implemented with existing pneumatic microfluidic pumps and valves [53], but a cost analysis would suggest that the need for an array of computer-controlled solenoid valves for binary control of multiple microfluidic valves and three solenoid valves for a peristaltic pump, at between $50 and $100 per solenoid, and the confustication of pneumatic tubing between the organ and its controller would limit either the sophistication of the OoC/HoC system or the number that could be implemented. The system in Fig. 2 could be implemented with a new class of compact, low-cost, on-chip rotary planar peristaltic micropump and rotary planar valve [54]. The required capabilities of the µCA/PC in Fig. 2 are illustrated in Figs. 5 and 6.
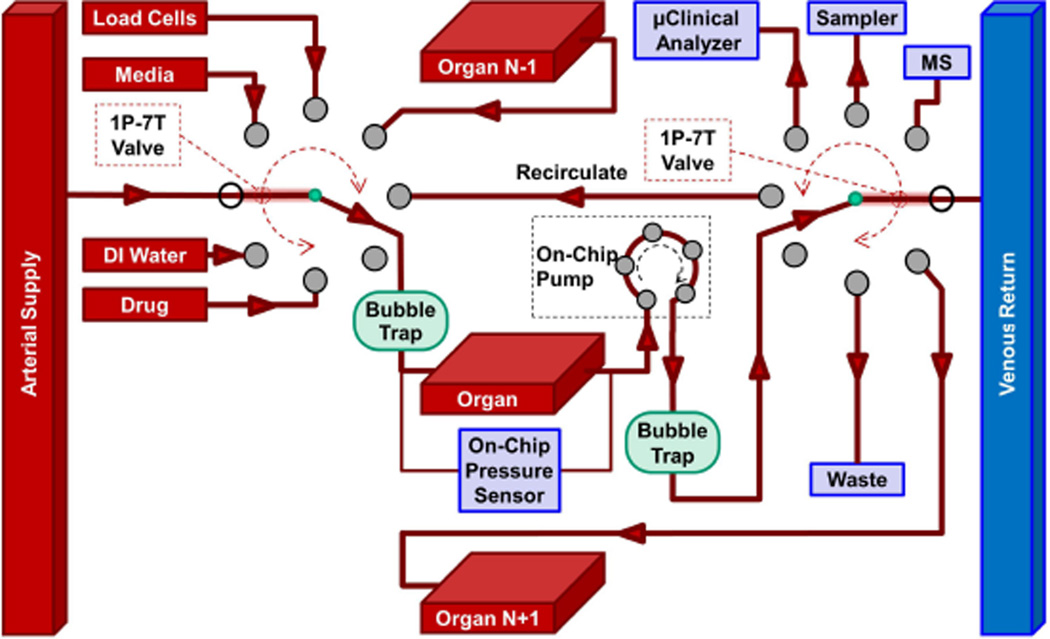
Fig. 5.
Conceptual drawing of how two single-pole, seven-throw (1P-7 T) valves and an on-chip peristaltic pump could provide the requisite PC functions in Fig. 2. The valve configuration shown has perfusate from the arterial supply perfusing Organ N and then exiting into the venous return. Other configurations of the valve on the left are used for adding media to make up any that is removed for sampling or flushing, loading cells (with the effluent going to waste), accepting fluid from an upstream organ, local recirculation, infusing a drug, dye, or marker into the single organ, or adding deionized (DI) water to adjust osmolarity after evaporation of water out of the device. The valve on the right is for venous return (as configured), sending fluid to a downstream organ, waste, recirculations, the µCA, an external autosampler, or an online MS. A second PC might be required to prepare cells in any secondary spaces, for example, to establish a confluent alveolar epithelium in a lung.
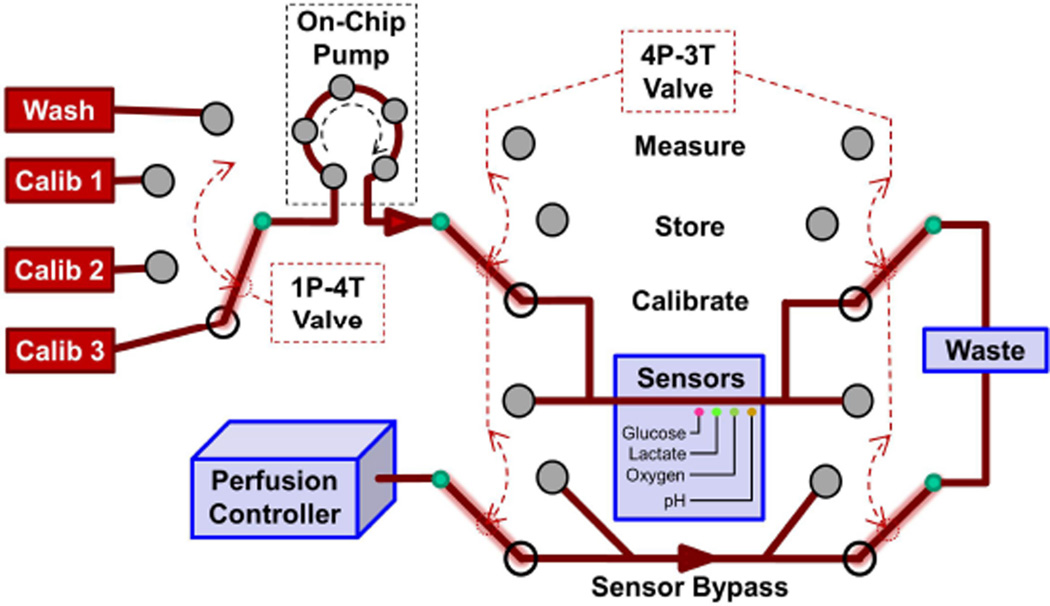
Fig. 6.
Conceptual drawing of how 1P-4 T and 4P-3 T valves and another on-chip peristaltic pump could provide the requisite µCA functions in Fig. 2. The valve configuration shown would allow calibration of the electrochemical sensor array with one of three standard solutions (configuration shown) or sensor washing without affecting the organ. Other modes allow the organ effluent to bypass the washed sensor to minimize biofouling (Store), or the delivery of organ fluid to the calibrated electrochemical sensor array (Measure). In this design, all of the µCA output is sent to waste rather than to venous return.
Figs. 2 and 5 indicate that an organ may need to be connected to an upstream (N − 1) or downstream (N + 1) organ so that two organs can be connected in series, e.g., intestine or spleen with the liver by the portal vein, and the hypothalamus with the pituitary. The system in Fig. 5 does not show the secondary inputs and outputs to an organ, e.g., lung (bronchial tree), thymus and spleen (efferent lymphatic vessels), liver (bile duct), brain (CSF), and kidney (ureter). The interstitial space of organs could drain into a lymphatic system. Some secondary spaces may need independent initial or long-term perfusion control, as in a lung-on-a-chip where the alveolar endothelium is cultured under fluid flow prior to being exposed to air, and thereafter they are nourished by adjacent perfused, capillary endothelial cells [12], [55].
E. Controlling Coupled Organ Systems
It is na¨ıve to assume that a collection of OoCs will exist in a stable equilibrium. Excess metabolic activity of one region without concomitant increases in oxygenation and nutrients and removal of carbon dioxide will lead to acidification and/or unwanted downstream effects. In living systems, homeostasis is maintained by a plethora of chemical, neural, and biomechanical signals. It would be reasonable to assume that such a complex, nonlinear system would oscillate, but not necessarily the way humans do. An OoC system will require an equivalent regulatory system. An appropriate control algorithm [56] should be able to stabilize the entire organ system, as long as there are sufficient sensors and the required actuators.
F. Vascularizing Organs With Appropriate Surface-to-Volume Ratios
Ultimately, the size of tissue-engineered 3-D organ replicates may be limited by vascularization. Typically, diffusive transport of nutrients and metabolites to and from metabol-ically active cells is effective for up to ~100 to ~300 µm, although diffusion can be effective to 2 mm [57]. To support the high metabolic activity of mammalian hearts, each myocyte (~10 µm × ~10 µm × ~100µm) invivoisin direct contact with a microcapillary. In conventional planar tissue constructs with perfusion above a slab of cells, metabolically active cells at the base may become anoxic if the layer is thicker than ~100 µm, (possibly alleviated by having flowing media above the cells), which is one reason that the confluent monolayer is a staple in laboratory cell culture. In large-scale tissue culture, this has been addressed by the use of hollow-fiber bioreactors [58], which may prove useful in the development of HoCs such as the liver [13]. In polydimethylsiloxane (PDMS) microfluidic systems, long-term organotypic cultures can be stable as long as there is flow of media and the distances are short [59]. However, because of the difficulties in making and maintaining microfluidic channels whose dimensions are comparable to ~7 µm diameter of actual capillaries, microfluidic devices typically utilize channels with as much as 50 times the cross-sectional area. Hence, it is at present difficult to fabricate macroscopic microfluidic perfusion networks with the appropriate surface-to-volume ratio and the vascular-to-tissue volume ratio, which in turn will ultimately affect the PKPD scaling of HoCs/OoCs systems. A promising solution to this problem is to enable perfusable microvascular networks to self-assemble within the HoC/OoC bioreactor. The spontaneous formation of endothelial tubes is well documented [60]–[62], but the connection of them to an external perfusion system is a greater challenge. Recently, it has been shown that interstitial flow can guide the migration of endothelial cells into a fibrin-filled PDMS channel to create 50– 150-µm-diameter and 100–1600-µm-long capillaries [63] and, more exciting, lead to the self-assembly of complete, patent 2-D microvascular networks [64], [65]. This approach should have profound implications for the development of a number of different HoCs/OoCs.
G. Developing a Universal Blood Surrogate
The culture of primary human cells and the differentiation of stem-cell-derived ones is a developing art. Maintaining a dozen different cell types with a common universal media has yet to be done, but the isolation provided by a layer of endothelial cells between the tissue and the microvasculator may aid this goal [12]. If a funding program does not allow serum in the blood surrogate, transport proteins must be added, as well as large molecules to maintain osmolarity. Unless red blood cells, with a limited lifetime, are used, the scaling of blood velocity and capillary length and diameter may compromise OoC/HoC metabolism unless oxygen carriers such as perfluorocarbons or vesicular hemoglobin are used.
H. Missing Organs and the Adjustment of Blood Surrogate
As the OoC/HoC systems are brought on line, there will always be missing organs that either have yet to be implemented or are not desired for particular tests. It may be necessary, for example, to deliver insulin to an OoC/HoC system that has yet to integrate a pancreas, or to add distilled water to maintain osmolarity as water evaporates through the devices or is secreted in urine. This can be accomplished with a missing-organ micro-formulator that is comparable to smaller, proven microfluidic designs [66]. Such a device could also prove invaluable in the automated exploration of growth-factor cocktails for stem cell differentiation that may prove invaluable in human HoC/OoC systems. It may also be necessary to remove compounds that would otherwise be metabolized by missing organs.
I. Modeling Coupled Organ Systems
The Physiome project [67] is working to develop comprehensive, multiscale models of mammalian physiology. The OoC/HoC effort can learn much from this, but enjoys the extra degree of freedom of being able to adjust both the physical and computational models to optimize either experiments, computation, or both. The design of the instruments that maintain and control the OoCs/HoCs is in fact the design of active physiological components, which could clearly benefit from advanced multiphysics modeling [68], [69]. An even greater opportunity would be to utilize a fully automated OoC/HoC system coupled to machine-learning algorithms [70], [71] to automatically infer physiological or control models of the system. Such an approach may soon be applied to the optimal design of experiments to estimate pharmacokinetic compartments.
J. Characterization of Organ Health and Disease
In all likelihood, the OoC/HoC effort will develop models not only of healthy organs but diseased ones as well. Can we maintain cellular heterogeneity? Clearly, it may be relatively easy to make models of heart, liver, and kidney failure. It is reassuring that studies of a lung-on-a-chip were able to replicate by design the normal lung [12] and a diseased one [55]. That said, we will need to devise methods to determine the overall health of our mHu and µHu, and one might expect that this could parallel clinical diagnosis, which can be viewed as a probability-based decision tree [72]—hence the need for detailed physiological information about each OoC/HoC.
Given the fluid volume constraints, it could be nontrivial to devise means to fully evaluate the contractility of the heart, cardiac valves, and smooth and skeletal muscle, since one needs, for example, to control the preload and afterload of tissues while measuring the length and tension of the musculature [73]–[77]. However, it is important that highly instrumented OoCs/HoCs will provide information not available to the bedside diagnostician: detailed internal data, the possiblity of localized biochemical or mechanical challenges, and interpretation of the resulting changes in terms of a computational biology systems model. While a systems model of an entire human is a distant goal of the systems biology community, it seems reasonable that it may be easier to model the limited organ set of a simplified mHu and µHu than to model an entire person. Given the small size of organs and the small numbers of replicates in the early stages of this effort, it may also be necessary to develop noise-tolerant techniques for estimation of pharmacokinetic parameters for individual organ systems [78].
K. Minimizing Organ Cost to Enable HCS
For OoC/HoC systems to become widely used in the pharmaceutical industry, the cost of these systems must be low enough for them to be purchased and used in very large quantities. The cell-culture chip will need to be low-cost and disposable. The control system in Fig. 2, if dedicated to each chip, should also be inexpensive but not necessairly disposable. There are tradeoffs between size, reliability, and cost that can be readily optimized through manufacturing engineering while minimizing hardware overdesign. We envison a complete coupled HoC/OoC system consisting of a small number of long-life, capital-intensive components (computers, sensor electronics, and microscopes), a number of subsystems that can be reused a specified number of times (pumps, valves, chip-scale imagers, sensors, microcontrollers, mounting hardware, etc.), and a large number of single-use, disposable, sterilizable organ chips that are readily replaced after several weeks of cell culture.
L. Determining the Required Accuracy of Organ Systems
Just as systems biology needs to be aware of the risks of striving for totally realistic models of biology, the OoC community needs to be conscious of the risks of attempting to create a perfectly realistic milli- or micromodel of a human. This is placed in perspective by a fictional account of ancient cartography [79]:
From Travels of Praiseworthy Men (1658) by J. A. Suarez Miranda: (…)In that Empire, the craft of Cartography attained such Perfection that the Map of a Single province covered the space of an entire City, and the Map of the Empire itself an entire Province. In the course of Time, these Extensive maps were found somehow wanting, and so the College of Cartographers evolved a Map of the Empire that was of the same Scale as the Empire and that coincided with it point for point. Less attentive to the Study of Cartography, succeeding Generations came to judge a map of such Magnitude cumbersome, and, not without Irreverence, they abandoned it to the Rigours of sun and Rain. In the western Deserts, tattered Fragments of the Map are still to be found, Sheltering an occasional Beast or beggar; in the whole Nation, no other relic is left of the Discipline of Geography.
In the same vein, one recognizes that “the best model for a cat is another, or preferably the same cat” [80]. A mHu or µHu will never be a perfect model of a human. The issue is how does one abstract the complexity of biology to obtain a meaningful model that will be useful for studying the properties of the entire system [7]. Paraphrasing Albert Einstein, one should make one’s OoCs systems simple enough, but not too simple.
III. CONCLUSION
We present a daunting list of challenges that confront the human OoC efforts, but we do so with the conviction that each of these challenges can be met by the engineering and biology community. Given the breadth and magnitude of the funding by the FDA, DARPA, NIH, and DTRA, and the caliber of the investigators working in this area, we anticipate that each of these limitations can be addressed successfully in a timely manner. The community is eager to uncover and resolve as yet unrecognized challenges—this is the heart of engineering and the physical sciences. We expect that the OoC/HoC effort will lead to exciting new advances in drug discovery, identification of the effects of emerging and maliciously engineered pathogens, and fundamental advances in the study of pharmaceutical modes of action, organ–organ interactions, physiological regulation, and systems biology.
Acknowledgment
The authors would like to thank E. Curtis, Z. Eagleton, B. Evans, L. Hofmeister, A. Kole, and W. Matloff for their detailed analyses of organ-on-chip scaling; G. Hamilton, D. Ingber, D. Levner, and K. K. Parker for extensive conversations and discussions related to precursors to Figs. 2, 5, and Fig. 6 and collaboration that led to successful funding of DARPA W911NF-12-2-0036; R. Iyer and K. Pant for participating in discussions during the creation of Fig. 1 and the writing of DTRA proposals that led to funding of DTRA100271 A-5196; A. Przekwas for many discussions on modeling and PKPD; F. E. Block, Jr., for suggestions regarding the application of anesthesiology principles and techniques to organ-on-a-chip systems; and A. Price and D. Berry for their editorial assistance.
Footnotes
This work was supported in part by the Defense Threat Reduction Agency under Grants HDTRA1-09-1-00-13 and DTRA100271 A-5196, the National Institutes of Health under Grants R01GM092218, RC2DA028981, and, through the NIH Common Fund, 1UH2-TR000491-01, the Defense Advanced Research Projects Agency under Grant W911NF-12-2-0036, a Vanderbilt University Discovery Grant, and the Vanderbilt Institute for Integrative Biosystems Research and Education.
Contributor Information
John P. Wikswo, Email: john.wikswo@vanderbilt.edu, Departments of Biomedical Engineering, Molecular Physiology & Biophysics, and Physics, and Astronomy, Vanderbilt University, Nashville, TN 37235-1807 USA.
Frank E. Block, III, Email: frank.block@vanderbilt.edu, Department of Biomedical Engineering, Vanderbilt University, Nashville, TN 37235-1631 USA.
David E. Cliffel, Email: d.cliffel@vanderbilt.edu, Department of Chemistry, Vanderbilt University, Nashville, TN 37235-1822 USA.
Cody R. Goodwin, Email: cody.r.goodwin@vanderbilt.edu, Department of Chemistry, Vanderbilt University, Nashville, TN 37235-1822 USA.
Christina C. Marasco, Email: chrissy.marasco@vanderbilt.edu, Department of Biomedical Engineering, Vanderbilt University, Nashville, TN 37235-1631 USA.
Dmitry A. Markov, Email: d.markov@vanderbilt.edu, Department of Cancer Biology, Vanderbilt University, Nashville, TN 37232-6840 USA.
David L. McLean, Email: david.l.mclean@vanderbilt.edu, Department of Physics & Astronomy, Vanderbilt University, Nashville, TN 37235-1807 USA.
John A. McLean, Email: john.a.mclean@vanderbilt.edu, Department of Chemistry, Vanderbilt University, Nashville, TN 37235-1822 USA.
Jennifer R. McKenzie, Email: jennifer.mckenzie@vanderbilt.edu, Department of Chemistry, Vanderbilt University, Nashville, TN 37235-1822 USA.
Ronald S. Reiserer, Email: ron.reiserer@vanderbilt.edu, Department of Physics & Astronomy, Vanderbilt University, Nashville, TN 37235-1807 USA.
Philip C. Samson, Email: philip.samson@vanderbilt.edu, Department of Physics & Astronomy, Vanderbilt University, Nashville, TN 37235-1807 USA.
David K. Schaffer, Email: d.schaffer@vanderbilt.edu, Department of Physics & Astronomy, Vanderbilt University, Nashville, TN 37235-1807 USA.
Kevin T. Seale, Email: kevin.seale@vanderbilt.edu, Department of Biomedical Engineering, Vanderbilt University, Nashville, TN 37235-1631 USA.
Stacy D. Sherrod, Email: stacy.d.sherrod@vanderbilt.edu, Department of Physics & Astronomy, Vanderbilt University, Nashville, TN 37235-1807 USA.
References
- 1.Kola I. The state of innovation in drug development. Clin. Pharmacol. Ther. 2008;vol. 83(no. 2):227–230. doi: 10.1038/sj.clpt.6100479. [DOI] [PubMed] [Google Scholar]
- 2.Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: Organs-on-chips. Lab Chip. 2012;vol. 12(no. 12):2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 3.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Organs-on-a-Chip: A focus on compartmentalized microdevices. Ann. Biomed. Eng. 2012;vol. 40(no. 6):1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 4.van der Meer AD, van den Berg A. Organs-on-chips: Breaking the in vitro impasse. Integr. Biol. 2012;vol. 4:461–470. doi: 10.1039/c2ib00176d. [DOI] [PubMed] [Google Scholar]
- 5.Esch MB, King TL, Shuler ML. The role of body-on-a-chip devices in drug and toxicity studies. Annu. Rev. Biomed. Engr. 2011;vol. 13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 6.Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;vol. 10(no. 4):446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- 7.Huang S, Wikswo J. Dimensions of systems biology. In: Amara SG, Bamberg E, Gudermann T, Hebert SC, Jahn R, Lederer WJ, Lill R, Miyajima A, Offermanns S, editors. Reviews of Physiology, Biochemistry and Pharmacology. 157th ed. Berlin, Heidelberg: Springer; 2007. pp. 81–104. [DOI] [PubMed] [Google Scholar]
- 8.Advancing Regulatory Science through Novel Research and Science-Based Technologies (U01) 2010 Feb 24; RFA No. RFA-RM-10–006, NIH [Online] Available: http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-10-006.html.
- 9.Microphysiological Systems, Solicitation no. DARPA-BAA-11–73. 2011 Sep 11; [Online] https://www.fbo.gov/index?s=opportunity&mode=form&id=956b160c42aaa386cf5762f12c21be9f&tab=core&cview=0.
- 10.Integrated Microphysiological Systems for Drug Efficacy and Toxicity Testing in Human Health and Disease (UH2/UH3) 2011 Nov 11; FOA number: RFA-RM-11–022, NIH [Online]. Available: http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-11-022.html.
- 11.Chemical and Biological Defense Innovations and Technologies. 2011 Dec 12; [Online] Available: http://www.grants.gov/search/search.do?mode=VIEW&oppId=133833.
- 12.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010 Jun;vol. 328(no. 5986):1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeilinger K, Schreiter T, Darnell M, Soderdahl T, Lubberstedt M, Dillner B, Knobeloch D, Nussler AK, Gerlach JC, Andersson TB. Scaling down of a clinical three-dimensional perfusion multicompartment hollow fiber liver bioreactor developed for extra-corporeal liver support to an analytical scale device useful for hepatic pharmacological in vitro studies. Tissue Eng. Part C Methods. 2011;vol. 17(no. 5):549–556. doi: 10.1089/ten.TEC.2010.0580. [DOI] [PubMed] [Google Scholar]
- 14.Stahl WR. Organ weights in primates and other mammals. Science. 1965 Nov;vol. 150(no. 3699):1039–1042. doi: 10.1126/science.150.3699.1039. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt-Nielsen K. How Animals Work. Cambridge, U.K: Cambridge Univ. Press; 1972. [Google Scholar]
- 16.Schmidt-Nielsen K. Scaling: Why is Animal Size so Important? New York, NY: USA Cambridge Univ. Press; 1984. Organ size and tissue metabolism,” in; pp. 90–98. [Google Scholar]
- 17.West GB, Brown JH, Enquist BJ. A general model for the origin of allometric scaling laws in biology. Science. 1997;vol. 276(no. 5309):122–126. doi: 10.1126/science.276.5309.122. [DOI] [PubMed] [Google Scholar]
- 18.West GB, Brown JH. The origin of allometric scaling laws in biology from genomes to ecosystems: towards a quantitative unifying theory of biological structure and organization. J. Exp. Biol. 2005;vol. 208(no. 9):1575–1592. doi: 10.1242/jeb.01589. [DOI] [PubMed] [Google Scholar]
- 19.Prothero JW. Organ scaling in mammals: The liver. Comp. Biochem. Physiol. A, Physiol. 1982;vol. 71(no. 4):567–577. doi: 10.1016/0300-9629(82)90205-5. [DOI] [PubMed] [Google Scholar]
- 20.Anderson BJ, Mckee AD, Holford NHG. Size, myths and the clinical pharmacokinetics of analgesia in paediatric patients. Clin. Pharmacokinet. 1997;vol. 33(no. 5):313–327. doi: 10.2165/00003088-199733050-00001. [DOI] [PubMed] [Google Scholar]
- 21.Bejan A. Design in Nature: How the Constructal Law Governs Evolution in Biology, Physics, Technology, and Social Organization. New York, NY, USA: Doubleday; 2012. [Google Scholar]
- 22.Johnson TN, Rostami-Hodjegan A, Tucker GT. Prediction of the clearance of eleven drugs and associated variability in neonates, infants and children. Clin. Pharmacokinet. 2006;vol. 45(no. 9):931–956. doi: 10.2165/00003088-200645090-00005. [DOI] [PubMed] [Google Scholar]
- 23.Danhof M, de Jongh J, De Lange ECM, Della Pasqua O, Ploeger BA, Voskuyl RA. Mechanism-based pharmacokinetic-pharmacodynamic modeling: Biophase distribution, receptor theory, and dynamical systems analysis. Annu. Rev. Pharmacol. Toxicol. 2007 Jan;vol. 47(no. 1):357–400. doi: 10.1146/annurev.pharmtox.47.120505.105154. [DOI] [PubMed] [Google Scholar]
- 24.Leung HW. General, Applied and Systems Toxicology. New York, NY, USA: Wiley; 2009. Physiologically based pharmacokinetic modelling. [Google Scholar]
- 25.Boxenbaum H, Dilea C. First-time-in-human dose selection—Allometric thoughts and perspectives. J. Clin. Pharmacol. 1995;vol. 35(no. 10):957–966. doi: 10.1002/j.1552-4604.1995.tb04011.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahmood I, Balian JD. Interspecies scaling: predicting clearance of drugs in humans. Three different approaches. Xenobiotica. 1996 Jan;vol. 26(no. 9):887–895. doi: 10.3109/00498259609052491. [DOI] [PubMed] [Google Scholar]
- 27.Mordenti J, Chen SA, Moore JA, Ferraiolo BL, Green JD. Interspecies scaling of clearance and volume of distribution data for 5 therapeutic proteins. Pharm. Res. 1991;vol. 8(no. 11):1351–1359. doi: 10.1023/a:1015836720294. [DOI] [PubMed] [Google Scholar]
- 28.Ward KW, Smith BR. A comprehensive quantitative and qualitative evaluation of extrapolation of intravenous pharmacokinetic parameters from rat, dog, and monkey to humans. I. Clearance. Drug Metab. Dispos. 2004;vol. 32(no. 6):603–611. doi: 10.1124/dmd.32.6.603. [DOI] [PubMed] [Google Scholar]
- 29.Ito K, Houston JB. Prediction of human drug clearance from in vitro and preclinical data using physiologically based and empirical approaches. Pharm. Res. 2005;vol. 22(no. 1):103–112. doi: 10.1007/s11095-004-9015-1. [DOI] [PubMed] [Google Scholar]
- 30.Sharma V, McNeill JH. To scale or not to scale: The principles of dose extrapolation. Brit. J. Pharmacol. 2009;vol. 157(no. 6):907–921. doi: 10.1111/j.1476-5381.2009.00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson MD, Beard DA. Development of appropriate equations for physiologically based pharmacokinetic modeling of permeability-limited and flow-limited transport. J. Pharmacokinet. Pharmacodyn. 2011;vol. 38(no. 4):405–421. doi: 10.1007/s10928-011-9200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wikswo JP, Prokop A, Baudenbacher F, Cliffel D, Csukas B, Velkovsky M. Engineering challenges of BioNEMS: The integration of microfluidics, and micro- and nanodevices, models, and external control for systems biology. IEE Proc.—Nanobiotechnol. 2006;vol. 153(no. 4):81–101. doi: 10.1049/ip-nbt:20050045. [DOI] [PubMed] [Google Scholar]
- 33.Faley S, Seale K, Hughey J, Schaffer DK, VanCompernolle S, McKinney B, Baudenbacher F, Unutmaz D, Wikswo JP. Microfluidic platform for real-time signaling analysis of multiple single T cells in parallel. Lab Chip. 2008;vol. 8(no. 10):1700–1712. doi: 10.1039/b719799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darby S, Moore M, Wikswo JP, Reiserer R, Friedlander T, Schaffer DK, Seale KT. A metering rotary nanopump for microfluidic systems. Lab Chip. 2010;vol. 10(no. 23):3218–3226. doi: 10.1039/c0lc00087f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miesenbock G. The optogenetic catechism. Science. 2009 Oct;vol. 326(no. 5951):395–399. doi: 10.1126/science.1174520. [DOI] [PubMed] [Google Scholar]
- 36.Snider RM, McKenzie JR, Kraft L, Kozlov E, Wikswo JP, Cliffel DE. The effects of Cholera Toxin on cellular energy metabolism. Toxins. 2010;vol. 2(no. 4):632–648. doi: 10.3390/toxins2040632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eklund SE, Cliffel DE, Kozlov E, Prokop A, Wikswo JP, Jr., Baudenbacher FJ. Modification of the cytosensor ™ microphysiometer to simultaneously measure extracellular acidification and oxygen consumption rates. Anal. Chim. Acta. 2003;vol. 496(no. 1–2):93–101. [Google Scholar]
- 38.Hiatt LA, McKenzie JR, Deravi LF, Harry RS, Wright DW, Cliffel DE. A printed superoxide dismutase coated electrode for the study of macrophage oxidative burst. Biosens. Bioelectron. 2012;vol. 33:128–133. doi: 10.1016/j.bios.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eklund SE, Taylor D, Kozlov E, Prokop A, Cliffel DE. A microphysiometer for simultaneous measurement of changes in extracellular glucose, lactate, oxygen, and acidification rate. Anal. Chem. 2004;vol. 76(no. 3):519–527. doi: 10.1021/ac034641z. [DOI] [PubMed] [Google Scholar]
- 40.Eklund SE, Thompson RG, Snider RM, Carney CK, Wright DW, Wikswo J, Cliffel DE. Metabolic discrimination of select list agents by monitoring cellular responses in a multianalyte microphysiometer. Sensors. 2009;vol. 9(no. 3):2117–2133. doi: 10.3390/s90302117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ges IA, Currie KP, Baudenbacher F. Electrochemical detection of catecholamine release using planar iridium oxide electrodes in nanoliter microfluidic cell culture volumes. Biosens. Bioelectron. 2012;vol. 34(no. 1):30–36. doi: 10.1016/j.bios.2011.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ges IA, Baudenbacher F. Enzyme-coated microelectrodes to monitor lactate production in a nanoliter microfluidic cell culture device. Biosens. Bioelectron. 2010 Oct;vol. 26(no. 2):828–833. doi: 10.1016/j.bios.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ges IA, Baudenbacher F. Enzyme electrodes to monitor glucose consumption of single cardiac myocytes in sub-nanoliter volumes. Biosens. Bioelectron. 2010 Jan;vol. 25(no. 5):1019–1024. doi: 10.1016/j.bios.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warnement MR, Faley SL, Wikswo JP, Rosenthal SJ. Quantum dot probes for monitoring dynamic cellular response: Reporters of T cell activation. IEEE Trans. NanoBiosci. 2006;vol. 5(no. 4):268–272. doi: 10.1109/tnb.2006.886573. [DOI] [PubMed] [Google Scholar]
- 45.Faley SL, Copland M, Wlodkowic D, Kolch W, Seale KT, Wikswo JP, Cooper JM. Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab Chip. 2009;vol. 9(no. 18):2659–2664. doi: 10.1039/b902083g. [DOI] [PubMed] [Google Scholar]
- 46.Diercks AH, Ozinsky A, Hansen CL, Spotts JM, Rodriguez DJ, Aderem A. A microfluidic device for multiplexed protein detection in nano-liter volumes. Anal. Biochem. 2009 Mar;vol. 386(no. 1):30–35. doi: 10.1016/j.ab.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noguchi Y, Young JD, Aleman JO, Hansen ME, Kelleher JK, Stephanopoulos G. Tracking cellular metabolomicsinlipoapoptosis- and steatosis-developing liver cells. Mol. Biosyst. 2011;vol. 7(no. 5):1409–1419. doi: 10.1039/c0mb00309c. [DOI] [PubMed] [Google Scholar]
- 48.Enders JR, Goodwin CR, Marasco CC, Seale KT, Wikswo JP, Mclean JA. Advanced structural mass spectrometry for systems biology: Pulling the needles from haystacks. Spectrosc. Supp. Curr. Trends Mass Spectrom. 2011 Jul;:18–23. [Google Scholar]
- 49.Enders JR, Marasco CC, Kole A, Nguyen B, Sundarapandian S, Seale KT, Wikswo JP, Mclean JA. Towards monitoring realtime cellular response using an integrated microfluidics-MALDI/NESI-ion mobility-mass spectrometry platform. IET Syst. Biol. 2010;vol. 4(no. 6):416–427. doi: 10.1049/iet-syb.2010.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enders JR, Marasco CC, Wikswo JP, Mclean JA. A Dual-column solid phase extraction strategy for online collection and preparation of continuously flowing effluent streams for mass spectrometry. Anal. Chem. 2012;vol. 84(no. 20):8467–8474. doi: 10.1021/ac3021032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eichler GS, Huang S, Ingber DE. Gene expression dynamics inspector (GEDI): For integrative analysis of expression profiles. Bioinformatics. 2003 Nov;vol. 19(no. 17):2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 52.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010 Apr;vol. 26(no. 7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;vol. 288(no. 5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 54.Gould PA, Hoang LT, Scherrer JR, Matloff WJ, Seale KT, Curtis EL, Schaffer DK, Hall DJ, Kole A, Reiserer RS, Tidwell H, Samson PC, Wikswo JP. Peristaltic micropump and related systems and methods. Pat. Appl. No. 2012 Apr 12; PCT/US2011/055432. [Google Scholar]
- 55.Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA, Thorneloe KS, Mcalexander MA, Ingber DE. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;vol. 4(no. 159):159–147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LeDuc PR, Messner WC, Wikswo JP. How do control-based approaches enter into biology? Annu. Rev. Biomed. Eng. 2011;vol. 13:369–396. doi: 10.1146/annurev-bioeng-071910-124651. [DOI] [PubMed] [Google Scholar]
- 57.Griffith CK, Miller C, Sainson RCA, Calvert JW, Jeon NL, Hughes CCW, George SC. Diffusion limits of an in vitro thick prevascularized tissue. Tissue Eng. 2005;vol. 11(no. 1/2):257–266. doi: 10.1089/ten.2005.11.257. [DOI] [PubMed] [Google Scholar]
- 58.Yan X, Bergstrom DJ, Chen XB. Modeling of cell cultures in perfusion bioreactors. IEEE Trans. Biomed. Eng. 2012 Sep;vol. 59(no. 9):2568–2575. doi: 10.1109/TBME.2012.2206077. [DOI] [PubMed] [Google Scholar]
- 59.Markov DA, Lu JQ, Samson PC, Wikswo JP, McCawley LJ. Thick-tissue bioreactor as a platform for long-term organotypic culture and drug delivery. Lab Chip. 2012;vol. 12(no. 21):4560–4568. doi: 10.1039/c2lc40304h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martins-Green M, Li QJ, Yao M. A new generation organ culture arising from cross-talk between multiple primary human cell types. FASEBJ. 2005;vol. 18(no. 15):222–224. doi: 10.1096/fj.04-1725fje. [DOI] [PubMed] [Google Scholar]
- 61.Wood LB, Das A, Kamm RD, Asada HH. A stochastic broadcast feedback approach to regulating cell population morphology for microfluidic angiogenesis platforms. IEEE Trans. Biomed. Eng. 2009 Sep;vol. 56(no. 9):2299–2303. doi: 10.1109/TBME.2009.2026732. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Markov D, Wikswo J, McCawley L. Microfabricated scaffold-guided endothelial morphogenesis in three-dimensional culture. Biomed. Microdevices. 2011 Jun;vol. 13(no. 5):837–846. doi: 10.1007/s10544-011-9554-2. [DOI] [PubMed] [Google Scholar]
- 63.Yeon JH, Ryu HR, Chung M, Hu QP, Jeon NL. In vitro formation and characterization of a perfusable three-dimensional tubular capillary network in microfluidic devices. Lab Chip. 2012;vol. 12(no. 16):2815–2822. doi: 10.1039/c2lc40131b. [DOI] [PubMed] [Google Scholar]
- 64.Hsu YH, Moya ML, Abiri P, Hughes CCW, George SC, Lee AP. Full range physiological mass transport control in 3-D tissue cultures. Lab Chip. 2013;vol. 13(no. 1):81–89. doi: 10.1039/c2lc40787f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moya ML, Hsu Y-H, Lee AP, Hughes CCW, George SC. In vitro perfused human capillary networks. Tissue Eng. Part C. 2013 Feb 21;19(9) doi: 10.1089/ten.tec.2012.0430. [Online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hansen CL, Sommer MOA, Quake SR. Systematic investigation of protein phase behavior with a microfluidic formulator. Proc. Nat. Acad. Sci. USA. 2004;vol. 101(no. 40):14431–14436. doi: 10.1073/pnas.0405847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hunter P, Burrowes K, Fernandez PN, Smith N, Tawhai M. The IUPS physiome project: Progress and plans. In: Kriete A, Eils R, editors. Computational Systems Biology. Amsterdam, The Netherlands: Elsevier; 2006. pp. 383–393. [Google Scholar]
- 68.Kannan R, Przekwas A. A computational model to detect and quantify a primary blast lung injury using near-infrared optical tomography. Int. J. Numer. Methods Biomed. Eng. 2011;vol. 27(no. 1):13–28. [Google Scholar]
- 69.Technologies for Future Micro-Nano Manufacturing, The Manufacturing Technologies 2011. Workshop, Napa, CA: 2011. [Google Scholar]
- 70.King RD, Rowland J, Oliver SG, Young M, Aubrey W, Byrne E, Liakata M, Markham M, Pir P, Soldatova LN, Sparkes A, Whelan KE, Clare A. The automation of science. Science. 2009 Apr;vol. 324(no. 5923):85–89. doi: 10.1126/science.1165620. [DOI] [PubMed] [Google Scholar]
- 71.Schmidt MD, Vallabhajosyula RR, Jenkins JW, Hood JE, Soni AS, Wikswo JP, Lipson H. Automated refinement and inference of analytical models for metabolic networks. Phys. Biol. 2011;vol. 8(no. 5):055011. doi: 10.1088/1478-3975/8/5/055011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller RA. Medical diagnostic decision-support systems—past, present, and future: A threaded bibliography and brief commentary. J. Amer. Med. Inform. Assoc. 1994;vol. 1(no. 1):8–27. doi: 10.1136/jamia.1994.95236141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balachandran K, Alford PW, Wylie-Sears J, Goss JA, Grosberg A, Bischoff J, Aikawa E, Levine RA, Parker KK. Cyclic strain induces dual-mode endothelial-mesenchymal transformation of the cardiac valve. Proc. Nat. Acad. Sci. USA. 2011;vol. 108(no. 50):19943–19948. doi: 10.1073/pnas.1106954108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grosberg A, Alford PW, McCain ML, Parker KK. Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip. Lab Chip. 2011;vol. 11(no. 24):4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alford PW, Feinberg AW, Sheehy SP, Parker KK. Biohybrid thin films for measuring contractility in engineered cardiovascular muscle. Biomaterials. 2010 May;vol. 31(no. 13):3613–3621. doi: 10.1016/j.biomaterials.2010.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mathews G, Sondergaard C, Jeffreys A, Childs W, Le BL, Sahota A, Najibi S, Nolta J, Si MS. Computational analysis of contractility in engineered heart tissue. IEEE Trans. Biomed. Eng. 2012 May;vol. 59(no. 5):1429–1435. doi: 10.1109/TBME.2012.2187899. [DOI] [PubMed] [Google Scholar]
- 77.Sondergaard CS, Mathews G, Wang L, Jeffreys A, Sahota A, Wood M, Ripplinger CM, Si MS. Contractile and electrophysiologic characterization of optimized self-organizing engineered heart tissue. Ann. Thorac. Surg. 2012;vol. 94(no. 4):1241–1249. doi: 10.1016/j.athoracsur.2012.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seng KY, Nestorov I, Vicini P. Fuzzy least squares for identification of individual pharmacokinetic parameters. IEEE Trans. Biomed. Eng. 2009 Dec;vol. 56(no. 12):2796–2805. doi: 10.1109/TBME.2009.2029083. [DOI] [PubMed] [Google Scholar]
- 79.Borges JL, tr- Di Giovanni NT. A Universal History of Infamy. London, U.K: Penguin; 1975. Of exactitude in science. [Google Scholar]
- 80.Rosenblueth A, Wiener N. The role of models in science. Philosophy Sci. 1945 Oct;vol. 12(no. 4):316–321. [Google Scholar]