Abstract
Bovine peripheral lymph nodes (LNs), including subiliac LNs, have been identified as a potential source of human exposure to Salmonella enterica, when adipose trim containing these nodes is incorporated into ground beef. In order to gain a better understanding of the burden of S. enterica in peripheral LNs of feedlot and cull cattle, a cross-sectional study was undertaken in which 3327 subiliac LNs were collected from cattle at harvest in seven plants, located in three geographically distinct regions of the United States. Samples were collected in three seasons: Fall 2010, Winter/Spring 2011, and Summer/Fall 2011. A convenience sample of 76 LNs per day, 2 days per season (approximately 1 month apart), was collected per plant, from carcasses held in the cooler for no less than 24 h. Every 10th carcass half on a rail was sampled, in an attempt to avoid oversampling any single cohort of cattle. Median point estimates of S. enterica contamination were generally low (1.3%); however, median Salmonella prevalence was found to be greater in subiliac LNs of feedlot cattle (11.8%) compared to those of cull cattle (0.65%). Enumeration analysis of a subset of 618 feedlot cattle LNs showed that 67% of those harboring S. enterica (97 of 144) did so at concentrations ranging from <0.1 to 1.8 log10 CFU/g, while 33% carried a higher burden of S. enterica, with levels ranging from 1.9 to >3.8 log10 CFU/g. Serotyping of S. enterica isolated identified 24 serotypes, with the majority being Montevideo (44.0%) and Anatum (24.8%). Antimicrobial susceptibility phenotypes were determined for all isolates, and the majority (86.1%) were pansusceptible; however, multidrug-resistant isolates (8.3%) were also occasionally observed. As Salmonella contained within LNs are protected from carcass interventions, research is needed to define opportunities for mitigating the risk of Salmonella contamination in LNs of apparently healthy cattle.
Introduction
Nontyphoidal Salmonella enterica is a significant cause of morbidity and mortality in the United States and is estimated to cause over 1 million illnesses each year (Mead et al., 1999; Guo et al., 2011; Scallan et al., 2011). The vast majority of these cases are foodborne and while produce, poultry, and eggs account for most illnesses, beef has been identified as a vehicle of exposure, associated with sporadic cases and outbreaks (Guo et al., 2011). S. enterica (hereafter referred to as Salmonella) has been frequently recovered from the hides and feces of healthy cattle (Barkocy-Gallagher et al., 2003; Loneragan and Brashears, 2005; Brichta-Harhay et al., 2008; Kunze et al., 2008; Loneragan et al., 2012), and it is theorized that animal carriage of Salmonella ultimately contributes to ground beef contamination (Bosilevac et al., 2009).
During harvest, hides of cattle are likely the primary source of Salmonella contamination of carcass surfaces (Barkocy-Gallagher et al., 2001; Barkocy-Gallagher et al., 2003; Arthur et al., 2008; Brichta-Harhay et al., 2008). Accordingly, substantial effort is afforded to preventing contamination of carcasses and removal of contamination through strategies outlined in plant Hazard Analysis Critical Control Point plans. These strategies appear quite effective, as Salmonella prevalence after antimicrobial intervention application is typically undetectable or less than 1% (Barkocy-Gallagher et al., 2003; Rivera-Betancourt et al., 2004; Brichta-Harhay et al., 2008). However, despite successful control of surface contamination, it is still possible for Salmonella to be recovered from ground beef. In a study of commercial ground beef from seven regions of the United States (n=4136 samples collected over 2 years), Salmonella was recovered from 4.2% of ground beef samples (Bosilevac et al., 2009). Similarly, government testing of ground beef indicates that Salmonella contamination averages around 2.1%, and that little improvement in contamination has been achieved over the past decade (FSIS, 2011), even while the prevalence of Escherichia coli O157:H7 in ground beef has declined more than 70% (from 0.80% in 2001 to 0.23% in 2010 [FSIS, 2012]).
Research suggests that pathogen contamination of ground beef also can occur by means of contaminated lymph nodes (LNs) (Arthur et al., 2008). Cattle possess many LNs located within fatty tissues that are frequently incorporated into ground beef and thus have the potential to contaminate the final product. Contaminated LNs may explain the difference in Salmonella prevalence between postintervention carcasses or trim, and ground beef. When present in LNs, Salmonella are protected from chemical and thermal antimicrobial carcass interventions, and as a consequence sanitary harvest procedures may not address this potential source of contamination. Therefore, the objective of this cross-sectional study was to gain a better understanding of the burden of S. enterica in peripheral LNs of harvest-ready cattle, by examining the point prevalence, serotypes, and antimicrobial susceptibility phenotypes present in subiliac LNs of cull and fed cattle at harvest, in three regions of the United States.
Materials and Methods
Sample collection
Subiliac LNs were collected from carcasses of cull (cows/bulls and dairy) and feedlot (steers/heifers) cattle that had passed federal inspection at commercial abattoirs. Samples were collected during three time periods including September, October, and November 2010 (Fall 2010), February and March 2011 (Winter/Spring 2011), and July, August, and September 2011 (Summer/Fall 2011). A convenience sample of seven commercial processing plants consisting of three plants that primarily harvest feedlot cattle and four that primarily harvest cull cows was initially enrolled. An eighth plant, harvesting primarily feedlot cattle, contributed two sets of samples during the Summer/Fall 2011 sample period. Participating abattoirs were located in regions 2, 3, and 5 of the microbiological monitoring regions defined by the Beef Industry Food Safety Council (Fig. 1). A convenience sample of 76 LNs per day, 2 days per season (approximately 1 month apart), was collected per plant, from postintervention carcasses that had been held in the cooler for no less than 24 h. Approximately every 10th carcass half on a rail was sampled, resulting in sample collection from cattle harvested over 1–2 production days, in an attempt to avoid oversampling any single cohort of cattle. However, the possibility remains that multiple nodes from a single cohort reside in a given sample set. Samples were shipped to Texas Tech University in Lubbock, Texas or the U. S. Meat Animal Research Center in Clay Center, Nebraska for processing.
FIG. 1.
Map of the Beef Industry Food Safety Council microbiological monitoring regions from which fed and cull cattle lymph nodes were obtained.
LN sample processing and Salmonella detection
LNs were processed as previously described (Brichta-Harhay et al., 2012). Briefly, surrounding fat and fascia were trimmed from LN samples, which were weighed, surface sterilized by submersion in a boiling water bath, placed into individual filtered sample bags (Nasco, Atlanta, GA), pulverized using a rubber mallet, and then enriched in 80 mL of tryptic soy broth (Becton Dickinson, Sparks, MD) by incubating at 25°C for 2 h and then 42°C for 12 h. Enrichments were subjected to immunomagnetic separation using anti-Salmonella beads (Dynabeads; Invitrogen, Carlsbad, CA). Recovered beads were transferred to 3 mL of Rappaport-Vassiliadis (Remel, St. Louis, MO) broth, incubated at 42°C for 18–20 h, then streaked onto xylose lysine desoxycholate (XLD; Remel, St. Louis, MO) and brilliant green sulfa (Becton Dickinson, Franklin Lakes, NJ) agar plates prior to incubation at 37°C for 18–20 h.
Presumptive Salmonella isolates were confirmed using invA polymerase chain reaction (Rahn et al., 1992; Nucera et al., 2006). Isolates were subjected to molecular serotyping methods (Herrera-León et al., 2004). Resulting phenotypes were further confirmed by traditional slide agglutination (O typing) and tube agglutination (flagellar H typing) methods, using commercial antisera (Difco, BD Diagnostic Systems, Sparks, MD) following manufacturer's guidelines.
Antimicrobial susceptibility testing
Susceptibility to 15 antimicrobial agents was determined using broth microdilution (Sensititre CMV1AGNF test plates; TREK Diagnostic Systems, Inc., Cleveland, OH) according to manufacturer's guidelines. Isolates were classified as susceptible, intermediate, or resistant for each agent using breakpoints established by the Clinical and Laboratory Standards Institute (CLSI, 2010). Isolates resistant to two or more classes of antimicrobials, as defined by the National Antimicrobial Resistance Monitoring System (FDA, 2011), were considered multi-drug resistant (MDR).
Salmonella enumeration
Salmonella prevalence values observed in the Fall 2010 sample period for feedlot cattle LNs indicated that enumeration analysis of this population could yield data on levels of Salmonella present in contaminated tissues. Conversely, enumeration of cull cattle LNs was not attempted because low prevalence suggested the analysis would yield little data. For enumeration, all feedlot cattle LNs collected in the Summer/Fall 2011 sample period (n=618) were processed as described above and immediately following homogenization, 1 mL of tryptic soy broth/LN homogenate was plated onto Petrifilm™ Enterobacteriaceae Count Plates (EB; 3M Microbiology, St. Paul, MN) in quadruplicate, and incubated at 37°C for 22–26 h. Petrifilm plates were then held at 4°C until presumptive culture results were obtained. Colonies were counted and 10–100% (depending on number of colonies present) were streaked to XLD for further confirmation. Colonies indicative of Salmonella on XLD plates were counted and the proportion of Salmonella-positive colonies were applied to the EB Petrifilm counts, in order to estimate the Salmonella count for each enumerated sample. LNs observed to harbor Salmonella, but at concentrations below the limit of detection (∼1 CFU/g) were designated in the <0.1 log10 CFU/g category.
Statistical analysis
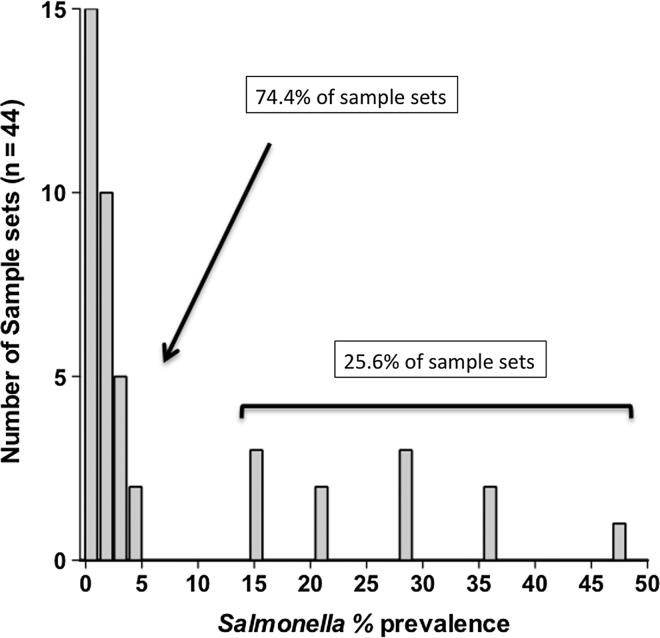
Salmonella point prevalence was calculated for each set of LNs collected (n=approximately 76 per set) in a given processing day (n=44 sample sets). Observed prevalence values were grouped by outcome and the frequency distribution was depicted as a histogram (Fig. 2). Median and mean point prevalence values were determined based on animal type, season, and region, and univariate analyses of results were performed using a one-way analysis of variance and Bonferroni's multiple comparison post-test, the Kruskal-Wallis test for nonparametric data and Dunn's multiple comparison post-test, or the Mann–Whitney t-test for nonparametric data, as indicated in the footnotes of Table 1. Enumeration data also were plotted as total estimated CFU/LN versus LN weight in grams (Fig. 3), and the geometric mean log10 CFU/g for enumerable samples (114 of 618) was determined. Comparisons of prevalence estimates and construction of data plots were performed using Prism 5.0d, GraphPad Software, Inc. (www.graphpad.com, San Diego, CA) and p values<0.05 were considered significantly different.
FIG. 2.
Histogram of Salmonella prevalence outcomes for the 44 lymph node (LN) sample sets (n=approximately 76 LN per set) collected in this study.
Table 1.
Salmonella Percent (%) Prevalence, and Standard Error (SE) in Subiliac Lymph Nodes (LNs) of Fed and Cull Cattle at Harvest, by Region and Season
| |
Cull cattle subiliac lymph nodesa |
|
Fed cattle subiliac lymph nodesa |
|
||||
|---|---|---|---|---|---|---|---|---|
| Sample period | Region 2 | Region 3 | Region 5 | All Cull | Region 3 | Region 5 | All Fed | Overall by season |
| Fall 2010 | ||||||||
| Sample sets collectedb | 2 | 4 | 2 | 8 | 4 | 2 | 6 | 14 |
| Number LNs tested | 152 | 304 | 152 | 608 | 279 | 152 | 431 | 1039 |
| Mean % (SE) | 0.65 (0.65) | 0.97 (0.62) | 0 | 0.65 (0.35) | 29.6 (1.5) | 0 | 19.7EF (6.3) | 8.8 (3.7) |
| Median % | 0.65 | 0.65 | 0 | 0D | 28.3 | 0 | 27.6 | 0.65 |
| Minimum % | 0 | 0 | 0 | 0 | 27.6 | 0 | 0 | 0 |
| Maximum % | 1.3 | 2.6 | 0 | 2.6 | 34.1 | 0 | 34.1 | 34.1 |
| Winter/Spring 2011 | ||||||||
| Sample sets collectedb | 2 | 4 | 2 | 8 | 4 | 2 | 6 | 14 |
| Number LNs tested | 151 | 305 | 152 | 608 | 305 | 147 | 452 | 1060 |
| Mean % (SE) | 3.3 (0.7) | 0.32 (0.32) | 0 | 0.98 (0.54) | 2.3 (0.34) | 0.7 (0.7) | 1.8E (0.44) | 1.3 (0.37) |
| Median % | 3.3 | 0 | 0 | 0D | 2.6 | 0.7 | 1.9 | 1.3 |
| Minimum % | 2.6 | 0 | 0 | 0 | 1.3 | 0 | 0 | 0 |
| Maximum % | 4.0 | 1.3 | 0 | 4 | 2.8 | 1.4 | 2.8 | 4.0 |
| Summer/Fall 2011 | ||||||||
| Sample sets collectedb | 2 | 4 | 2 | 8 | 6 | 2 | 8 | 16 |
| Number LNs tested | 152 | 306 | 152 | 610 | 466 | 152 | 618 | 1228 |
| Mean % (SE) | 1.3 (0) | 0.65 (0.38) | 12.5 (8.6) | 3.8 (2.5) | 24.7 (8.0) | 13.2 (1.4) | 21.4F (5.9) | 12.0 (3.8) |
| Median % | 1.3 | 0.65 | 12.5 | 1.3D | 20.0 | 13.2 | 17.7 | 3.9 |
| Minimum % | 1.3 | 0 | 3.9 | 0 | 1.3 | 11.8 | 1.3 | 0 |
| Maximum % | 1.3 | 1.3 | 21.1 | 21.1 | 47.4 | 14.5 | 47.4 | 47.4 |
| Overall by region or type | Lymph nodes overall | |||||||
| Sample sets collectedb | 6 | 12 | 6 | 24 | 14 | 6 | 20 | 44 |
| Number LNs tested | 455 | 915 | 456 | 1826 | 1050 | 451 | 1501 | 3327 |
| Mean % (SE) | 1.75 (0.56) | 0.65 (0.25) | 4.2 (3.4) | 1.8 (0.88) | 19.3 (4.4) | 4.6 (2.7) | 14.7 (3.5) | 7.5 (1.9) |
| Median % | 1.3A | 0A | 0A | 0.65G | 20.0B | 0.7C | 11.8H | 1.3 |
| Minimum % | 0 | 0 | 0 | 0 | 1.3 | 0 | 0 | 0 |
| Maximum % | 4.0 | 2.6 | 21.1 | 21.1 | 47.4 | 14.5 | 47.4 | 47.4 |
Common uppercase superscripts indicate values that are not significantly different (p>0.05).
Approximately 76 LNs collected per set, two sets collected per plant, approximately 1 month apart in each season.
Median cull cattle prevalence by region was not significantly different (p=0.2356) one-way analysis of variance (ANOVA) Kruskal–Wallis nonparametric test; Dunn's multiple comparison post-test.
Median fed cattle prevalence by region was significantly different (p=0.0198) Mann–Whitney nonparametric two-tailed t-test.
Median cull cattle prevalence by season was not significantly different (p=0.3007) one-way ANOVA Kruskal–Wallis nonparametric test; Dunn's multiple comparison post-test.
Mean fed cattle prevalence by season was significantly different (p=0.0304) one-way ANOVA and Bonferroni's post-test.
Median lymph node prevalence overall for cull and fed cattle was significantly different (p=0.0006) Mann–Whitney nonparametric two-tailed t-test.
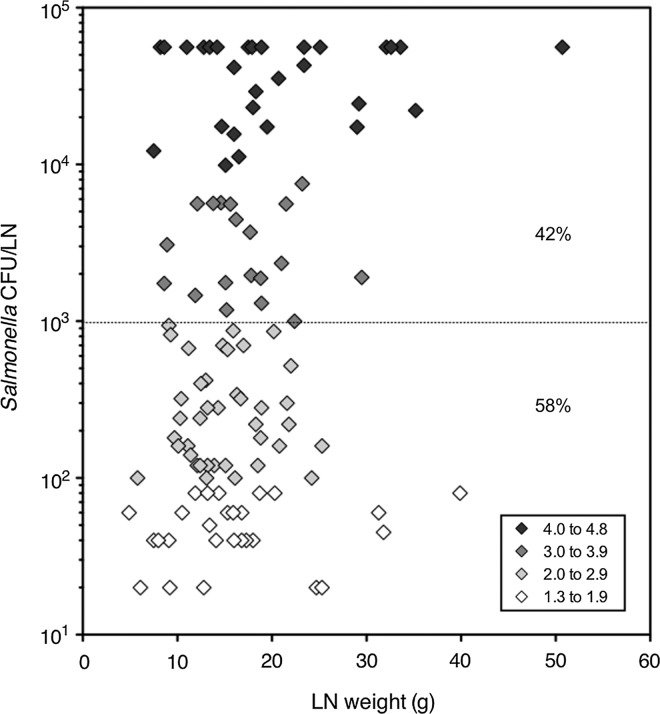
FIG. 3.
Concentration of Salmonella in contaminated lymph nodes (LN) of feedlot cattle at harvest. Enumeration data were collected from 618 LN from carcasses of feedlot cattle in Summer/Fall 2011, using the methods described. Of these, 144 LN were positive for Salmonella contamination and 114 LN contained Salmonella at enumerable levels. Total estimated Salmonella CFU/LN was plotted versus LN weight (grams). Estimated total levels of Salmonella (log10 CFU/LN) are indicated by symbol color intensity (as depicted in the key), with white being the lowest (1.3–1.9 log10 CFU/LN) and dark gray being the highest (4.0–4.8 CFU/LN).
Results
Salmonella point prevalence and enumeration
In this cross-sectional study, a total of 3327 subiliac LNs were collected from fed and cull cattle carcasses, in three regions of the United States, over 12 months (Table 1). Examination of the frequency distribution of observed prevalence values showed that the majority of sample sets (74.4%) had few-to-no Salmonella positives (Fig. 2). As a consequence, the median point estimate for Salmonella contamination was found to be 1.3%. However, some sample sets (25.6%) yielded considerably higher prevalence values. These sample sets skewed the resulting distribution such that arithmetic mean prevalence of Salmonella contamination was 7.5%. Salmonella point prevalence in feedlot cattle LNs was significantly greater than that observed in cull cattle (p=0.0006) and appeared to be affected by region and season (Table 1), with levels significantly higher in region 3 as compared with region 5 (p=0.0198), and in Summer/Fall 2011 as compared with Winter/Spring 2011 (p=0.0304), for samples collected in region 3. Conversely, Salmonella prevalence in cull cattle LNs did not appear to be affected by region or season, as levels in this population tended to be low with a median overall point prevalence of 0.65%, although an exception to that trend was observed in region 5 in the Summer/Fall 2011 sample period (Table 1). It should be noted however, that further investigation revealed that the cull cows contributing to this outlier data point originated in region 3 but were transported to region 5 for harvest.
Enumeration analysis of 618 feedlot cattle LNs collected in Summer/Fall 2011 showed that 23.3% (n=144) harbored Salmonella and that 18.4% (n=114) contained levels detectable with the enumeration methods employed (limit of detection ∼1 CFU/g). The geometric mean concentration of Salmonella for enumerable samples was 1.75 log10 CFU/g; however, as shown in Figure 3, a wide range of values were observed. While the majority of quantifiable nodes (58.8%; 67 of 114) contained Salmonella at concentrations ranging from 0.1 to 1.8 Log10 CFU/g, 41.2% (47 of 114) carried higher levels, ranging from 1.9 to 3.8 log10 CFU/g, or greater (Fig. 3).
Salmonella serotypes and antimicrobial susceptibility phenotypes
Twenty-four serotypes were identified, with the majority being either Montevideo (44.0%) or Anatum (24.8%; Table 2). Eighteen serotypes were identified among the 33 positive LNs from cull cattle, whereas 14 serotypes were observed among the 233 positive LNs from feedlot cattle. At least two colonies were serotyped for each positive LN, resulting in the isolation of multiple Salmonella serotypes from 3.8% (n=10) of positive samples. Multiple serotypes were only observed in feedlot cattle LNs, and for prevalence estimates only one of the two serotypes was counted. Serotype combinations in these samples included Montevideo and Anatum (n=7), Montevideo and Infantis (n=1), Montevideo and Muenchen (n=1), and Anatum and Kentucky (n=1).
Table 2.
Salmonella Serotypes Recovered from Subiliac Lymph Nodes (LNs) of Feedlot Cattle and Cull Cows
| Serotype | All LNs(n=266)overall % | Fed cattleLNs (n=233)relative % | Cull cattleLNs (n=33)relative % |
|---|---|---|---|
| Montevideo | 44.0 | 48.5 | 12.1 |
| Anatum | 24.8 | 27.5 | 6.1 |
| Reading | 4.9 | 5.2 | 3.0 |
| Thompsona | 3.8 | 3.9 | 3.0 |
| Meleagridis | 3.0 | 3.4 | |
| Kentucky | 3.0 | 1.7 | 12.1 |
| C07 NT | 2.3 | 2.6 | |
| Mbandaka | 2.3 | 1.3 | 9.1 |
| Muenchena | 1.5 | 1.7 | |
| Bredeney | 1.1 | 9.1 | |
| Infantis | 1.1 | 1.3 | |
| Newport | 1.1 | 0.9 | 3.0 |
| Braenderup | 0.8 | 6.1 | |
| Brandenburg | 0.8 | 6.1 | |
| Cerro | 0.8 | 0.9 | |
| Dublin | 0.8 | 6.1 | |
| Muenster | 0.8 | 0.9 | |
| Panamaa | 0.8 | 6.1 | |
| Saint Paul | 0.8 | 0.4 | 3.0 |
| Cubana | 0.4 | 3.0 | |
| Give | 0.4 | 3.0 | |
| Kiambua | 0.4 | 3.0 | |
| Typhimurium | 0.4 | 3.0 | |
| Uganda | 0.4 | 3.0 |
Indicates predicted serotype—serotype data incomplete due to the presence of R H antigens that do not react with H antisera. Isolates designated as Thompsona may be Thompson (6,7,14: k: 1,5) or Ohio (6,7,14: b: l,w); isolates designated as Panamaa may be Panama (1,9,12: l,v: 1,5) or Javiana (1,9,12: l,z28: 1,5). CO7 NT – Nontypeable Salmonella that reacts with O-group 6,7 antisera, but H-antigens are nonreactive.
The majority of Salmonella isolates (229 of 266) were susceptible to all antimicrobial agents tested. Multidrug resistance (MDR, defined as resistance to two or more antimicrobial drug classes; [FDA, 2011]) was observed in 8.3% (n=266) of isolates (Table 3). Seventeen isolates (6.4%) exhibited the MDR-AmpC phenotype (co-resistance to at least ampicillin, chloramphenicol, streptomycin, sulfisoxazole, tetracycline, amoxicillin/clavulanic acid, ceftiofur, and ceftriaxone) (Gupta et al., 2003; Kunze et al., 2008). Serotypes demonstrating MDR-AmpC resistance included Reading (n=13), Newport (n=3), and Typhimurium (n=1), although it should be noted that 11 of the 13 Reading isolated were from a single set of 76 LNs collected from feedlot cattle in the Summer/Fall 2011 sample period, and likely represent a cluster originating from a single lot of cattle at harvest (Table 3).
Table 3.
Salmonella Serotypes and Observed Antimicrobial Susceptibility Phenotypes Recovered from Subiliac Lymph Nodes of Feedlot Cattle and Cull Cows
| Animal type | Serotype | Antimicrobial resistance phenotypea | Number of lymph nodes |
|---|---|---|---|
| Cull | Mbandaka | S | 1 |
| Kentucky | S | 1 | |
| Dublin | Ap, C, K, G, Su | 1 | |
| Dublin | Ap, C, K, G, Su, Te | 1 | |
| Newport | Am, Ap, F, T, C, S, Su, Te | 1 | |
| Reading | Am, Ap, F, T, Ax, C, S, Su, Te | 1 | |
| Typhimurium | Am, Ap, F, T, Ax, C, S, Su, Te | 1 | |
| Fed | Montevideo | Te | 11 |
| Thompson | Te | 1 | |
| Montevideo | S | 1 | |
| Kentucky | Su, Te | 1 | |
| C O7 NTb | Su, TS | 2 | |
| Reading | Am, Ap, F, T, C, S, Su, Te | 11c | |
| Newport | Am, Ap, F, T, Ax, C, S, Su, Te | 2 | |
| Reading | Am, Ap, F, T, Ax, C, S, Su, Te | 1 |
Am, amoxicillin-clavulanic acid; Ap, ampicillin; F, cefoxitin; K, Kanamycin; G, Gentamicin; T, ceftiofur; Ax, ceftriaxone; C, chloramphenicol; S, streptomycin; Su, sulfisoxazole; Te, tetracycline; TS, trimethoprim/sulphamethoxazole.
CO7 NT, nontypeable Salmonella that reacts with O-group 6,7 antisera, but H-antigens nonreactive.
Detected as a cluster of isolates originating from one set of 76 lymph nodes and thus may originate from a single lot of cattle at harvest.
Discussion
Salmonella are versatile enteric pathogens noted for their ability to invade and survive within host lymphoid tissues (Stevens et al., 2009). Previous research has shown that cattle peripheral LNs can serve as a vehicle for Salmonella contamination, if fat trim containing these nodes is incorporated into ground beef (Arthur et al., 2008; Samuel et al., 1980). In this cross-sectional study, we also observed that Salmonella could be recovered from subiliac LNs, but additionally found that point estimates of prevalence in feedlot cattle populations appear to be greater than those in cull cattle populations, and that Salmonella harborage may be affected by season and region. While further study is needed to confirm the observed trends, these data nevertheless raise intriguing questions regarding the mechanism of Salmonella entry into bovine peripheral LNs, and the factors influencing this phenomenon. Possible explanations for the differences observed include diet effects and animal age, as well as management of cattle prior to harvest and differences in the prevalence of Salmonella in cattle environments.
Numerous studies have described a seasonal effect on Salmonella prevalence in cattle environments, with prevalence peaking in warmer months (summer and fall), and dipping in colder months (winter and spring) (APHIS, 2001; Barkocy-Gallagher et al., 2003; Edrington et al., 2004). Furthermore, mounting evidence from surveys of cattle feces, hides, and environments suggests a regional variation in Salmonella prevalence in North America, where the burden of Salmonella broadly increases across a southerly gradient. In a large study that included fecal samples of cattle housed in Canadian feedlots, Salmonella was recovered from 0.2% of cattle ready for harvest (Sorensen et al., 2002). In the Midwest, Barkocy-Gallagher et al. (Barkocy-Gallagher et al., 2003) reported peak fecal prevalence of 9.1% during the summer and fall months, while in Texas, Salmonella was recovered from 32.0% and 25.5% of fecal samples collected from healthy cattle housed in six feedlots and 22 dairies, respectively (Kunze et al., 2008; Farrow et al., 2009). The observed similarity in seasonal and regional prevalence between Salmonella in LNs and in cattle environments suggests the potential for an environmental component to the mechanism of how Salmonella gains entry to peripheral nodes. It is known that subiliac LNs receive afferent lymph from the skin of the abdominal wall, pelvis, prepuce, and hind limbs; thus, it is possible that Salmonella recovered from subiliac LNs may have entered via a transdermal route, through abrasions or biting insects. This idea has been suggested previously (Samuel et al., 1980), and given that cattle hides are a common reservoir for Salmonella (Loneragan and Brasheears, 2005; Brichta-Harhay et al., 2008; Kunze et al., 2008), the observed correlation between Salmonella prevalence on cattle hides, in cattle environments, and in peripheral LNs is perhaps not surprising. The observed difference in prevalence between feedlot and cull cattle in region 3 was unexpected, however, and may reflect differences in hygiene or mitigation practices (i.e., Salmonella vaccine use or differences in pest management) because of a greater perceived risk of Salmonella as an animal health issue in dairy cattle populations.
Having confirmed that subiliac LNs may be a source of Salmonella contamination in ground beef, it was important to examine the serotypes and antimicrobial susceptibility profiles of Salmonella occupying this niche, because depending on host status, transmission vehicle, and inoculum level, certain Salmonella serotypes appear to be more relevant to causing human disease (Jones et al., 2008). Results showed a diverse set of serotypes were isolated; however, two serotypes—Montevideo (44%) and Anatum (24.8%)—represented the majority of isolates (Table 2). These serotypes are frequently isolated from feces and hides of healthy feedlot cattle, especially in region 3 (Fluckey et al., 2007; Kunze et al., 2008), and as the majority of positive LN samples (79.7%) were collected from feedlot cattle in this region, their predominance is not unexpected. It is noteworthy, however, that Montevideo and Anatum are also the most commonly recovered Salmonella serotypes from ground beef in both federal testing programs (FSIS, 2011) and national surveys (Bosilevac et al., 2009). Considering the enumeration data presented here, demonstrating that 33% of contaminated LNs tested (47 of 144) harbored Salmonella at levels in the range of 1.9 to >3.8 log10 CFU/g, it is tempting to suggest that these data identify the mechanism by which the majority of Salmonella may be entering ground beef. To our knowledge, this is the first report to document the range in Salmonella contamination present in cattle peripheral LNs.
Antimicrobial susceptibility phenotyping showed that the majority of Salmonella isolated (86%) were susceptible to all antimicrobials tested; however, MDR Salmonella (8.3%) were observed (Table 3). Notably, 15.2% of isolates from cull cattle LNs (n=33) were MDR while 7.3% of isolates from fed cattle (n=233) had MDR phenotypes. When considering Salmonella that are potentially more relevant to human disease, Typhimurium and Newport are two of the leading serotypes isolated in cases of human illness in the United States and have been associated with outbreaks attributed to contaminated ground beef (Gupta et al., 2003). Conversely, serotypes Montevideo and Anatum have been implicated in fewer laboratory-confirmed human salmonellosis cases (CDC, 2011), especially from ground beef sources, and lack medical relevance in comparison. These observations highlight the need for investigation into the virulence factors, or adaptive mechanisms that may be associated with increased human illness among medically relevant serotypes. In this study, serotypes Typhimurium and Newport were observed in 6.1% and 0.9% of cull and fed cattle LNs, respectively (Table 3). Quantification of pathogen load in peripheral LNs containing these medically relevant serotypes will aid in modeling the potential quantitative risk imposed by the addition of contaminated LNs to ground beef.
Conclusions
The data presented show that subiliac LNs can be a significant source of Salmonella, if incorporated in ground beef, and that prevalence appears to be affected by season, region, and animal type. Furthermore, we show that contaminated nodes can carry substantial levels of Salmonella (1.9 to >3.8 log10 CFU/g). As LN harborage protects Salmonella from carcass interventions, research is needed to define opportunities for mitigating the risk of Salmonella contamination in LNs of apparently healthy cattle.
Acknowledgments
Funding for this research was provided in part by the Beef Checkoff Program. The authors gratefully acknowledge the assistance of the cooperating abattoirs. We also thank Kim Kucera, Frank Reno, Bruce Jasch, Julie Dyer, Sydney Brodrick, Greg Smith, Rebecca McCarthy, Amy Parks, Max Wolf, Kathleen Fermin, Tanya Jackson, Martha Maradiaga, Ashley Hartzog-Hawkins, and Alexandra Calle for their excellent technical support, and Jan Watts and Jody Gallagher for administrative assistance.
Disclosure Statement
No competing financial interests exist.
References
- APHIS. Salmonella in United States feedlots. 2001. http://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot99/Feedlot99_is_Salmonella.pdf. [Mar 14;2012 ]. http://www.aphis.usda.gov/animal_health/nahms/feedlot/downloads/feedlot99/Feedlot99_is_Salmonella.pdf
- Arthur TM. Brichta-Harhay DM. Bosilevac JM, et al. Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J Food Prot. 2008;71:1685–1688. doi: 10.4315/0362-028x-71.8.1685. [DOI] [PubMed] [Google Scholar]
- Barkocy-Gallagher GA. Arthur TM. Rivera-Betancourt M, et al. Seasonal prevalence of shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. J Food Prot. 2003;66:1978–1986. doi: 10.4315/0362-028x-66.11.1978. [DOI] [PubMed] [Google Scholar]
- Barkocy-Gallagher GA. Arthur TM. Siragusa GR, et al. Genotypic analyses of Escherichia coli O157:H7 and O157 nonmotile isolates recovered from beef cattle and carcasses at processing plants in the midwestern states of the United States. Appl Environ Microbiol. 2001;67:3810–3818. doi: 10.1128/AEM.67.9.3810-3818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosilevac JM. Guerini MN. Kalchayanand N. Koohmaraie M. Prevalence and characterization of Salmonella in commercial ground beef in the United States. Appl Environ Microbiol. 2009;75:1892–1900. doi: 10.1128/AEM.02530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichta-Harhay DM. Arthur TM. Bosilevac JM, et al. Microbiological analysis of bovine lymph nodes for the detection of Salmonella enterica. J Food Prot. 2012;75:854–858. doi: 10.4315/0362-028X.JFP-11-434. [DOI] [PubMed] [Google Scholar]
- Brichta-Harhay DM. Guerini MN. Arthur TM, et al. Salmonella and Escherichia coli O157:H7 contamination on hides and carcasses of cull cattle presented for slaughter in the United States: An evaluation of prevalence and bacterial loads by immunomagnetic separation and direct plating methods. Appl Environ Microbiol. 2008;74:6289–6297. doi: 10.1128/AEM.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Atlanta, GA: Centers for Disease Control and Prevention; 2009. 2011. National Salmonella surveillance annual data summary. [Google Scholar]
- [CLSI] Clinical and Laboratory Standards Institute. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement. CLSI document M100-S20. [Google Scholar]
- Edrington TS. Hume ME. Looper ML, et al. Variation in the faecal shedding of Salmonella and E. coli O157:H7 in lactating dairy cattle and examination of Salmonella genotypes using pulsed-field gel electrophoresis. Lett Appl Microbiol. 2004;38:366–372. doi: 10.1111/j.1472-765X.2004.01495.x. [DOI] [PubMed] [Google Scholar]
- Farrow RL. Loneragan GH. Edrington TS, et al. Quantitative herd-level evaluation of the potential for a commercially available vaccine to effectively control Salmonella in dairy cattle. Annual Meeting of the Conference of Research Workers in Animal Diseases; Chicago, IL. 2009. [Google Scholar]
- [FDA] Food and Drug Administration. Rockville, MD: U.S. Department of Health and Human Services, Food and Drug Administration; 2011. National Antimicrobial Resistance Monitoring System–Enteric Bacteria (NARMS): 2009 Executive Report. [Google Scholar]
- Fluckey WM. Loneragan GH. Warner R. Brashears MM. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J Food Prot. 2007;70:551–556. doi: 10.4315/0362-028x-70.3.551. [DOI] [PubMed] [Google Scholar]
- [FSIS] Food Safety and Inspection Service. Serotypes profile of Salmonella isolates from meat and poultry products: January 1998 through December 2010. 2011. http://www.fsis.usda.gov/PDF/Serotypes_Profile_Salmonella_2010.pdf. [Nov 10;2011 ]. http://www.fsis.usda.gov/PDF/Serotypes_Profile_Salmonella_2010.pdf
- FSIS. Microbiological results of raw ground beef products analyzed for Escherichia coli O157:H7, summarized by calendar year. 2012. http://www.fsis.usda.gov/science/Ground_Beef_E.Coli_Testing_Results/index.asp. [Feb 14;2012 ]. http://www.fsis.usda.gov/science/Ground_Beef_E.Coli_Testing_Results/index.asp (Online)
- Guo C. Hoekstra RM. Schroeder CM, et al. Application of bayesian techniques to model the burden of human salmonellosis attributable to U.S. food commodities at the point of processing: Adaptation of a Danish model. Foodborne Pathog Dis. 2011;8:509–516. doi: 10.1089/fpd.2010.0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. Fontana J. Crowe C, et al. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J Infect Dis. 2003;188:1707–1716. doi: 10.1086/379668. [DOI] [PubMed] [Google Scholar]
- Herrera-León S. McQuiston JR. Usera MA. Fields PI. Garaizar J. Echeita MA. Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J Clin Microbiol. 2004;42:2581–2586. doi: 10.1128/JCM.42.6.2581-2586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TF. Ingram LA. Cieslak PR, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. 2008;198:109–114. doi: 10.1086/588823. [DOI] [PubMed] [Google Scholar]
- Kunze DJ. Loneragan GH. Platt TM, et al. Salmonella enterica burden in harvest-ready cattle populations from the southern high plains of the United States. Appl Environ Microbiol. 2008;74:345–351. doi: 10.1128/AEM.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loneragan GH. Brashears MM. Effects of using retention-pond water for dust abatement on performance of feedlot steers and carriage of Escherichia coli O157 and Salmonella spp. J Am Vet Med Assoc. 2005;226:1378–1383. doi: 10.2460/javma.2005.226.1378. [DOI] [PubMed] [Google Scholar]
- Loneragan GH. Thomson DU. McCarthy RM, et al. Salmonella diversity and burden in cows on and culled from dairy farms in the Texas high plains. Foodborne Pathog Dis. 2012;9:1–7. doi: 10.1089/fpd.2011.1069. [DOI] [PubMed] [Google Scholar]
- Mead PS. Slutsker L. Dietz V, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucera DM. Maddox CW. Hoien-Dalen P. Weigel RM. Comparison of API 20E and invA PCR for identification of Salmonella enterica isolates from swine production units. J Clin Microbiol. 2006;44:3388–3390. doi: 10.1128/JCM.00972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn K. De Grandis SA. Clarke RC, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- Rivera-Betancourt M. Shackelford SD. Arthur TM, et al. Prevalence of Escherichia coli O157:H7, Listeria monocytogenes, and Salmonella in two geographically distant commercial beef processing plants in the United States. J Food Prot. 2004;67:295–302. doi: 10.4315/0362-028x-67.2.295. [DOI] [PubMed] [Google Scholar]
- Samuel JL. O'Boyle DA. Mathers WJ. Frost AJ. Isolation of Salmonella from mesenteric lymph nodes of healthy cattle at slaughter. Res Vet Sci. 1980;28:238–241. [PubMed] [Google Scholar]
- Scallan E. Hoekstra RM. Angulo FJ, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen O. Van Donkersgoed J. McFall M. Manninen K. Gensler G. Ollis G. Salmonella spp. shedding by Alberta beef cattle and the detection of Salmonella spp. in ground beef. J Food Prot. 2002;65:484–491. doi: 10.4315/0362-028x-65.3.484. [DOI] [PubMed] [Google Scholar]
- Stevens MP. Humphrey TJ. Maskell DJ. Molecular insights into farm animal and zoonotic Salmonella infections. Philos Trans R Soc Lond B Biol Sci. 2009;364:2709–2723. doi: 10.1098/rstb.2009.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]