Abstract
Estimates of foodborne illness are important for setting food safety priorities and making public health policies. The objective of this analysis is to estimate domestically acquired, foodborne illness in Canada, while identifying data gaps and areas for further research. Estimates of illness due to 30 pathogens and unspecified agents were based on data from the 2000–2010 time period from Canadian surveillance systems, relevant international literature, and the Canadian census population for 2006. The modeling approach required accounting for under-reporting and underdiagnosis and to estimate the proportion of illness domestically acquired and through foodborne transmission. To account for uncertainty, Monte Carlo simulations were performed to generate a mean estimate and 90% credible interval. It is estimated that each year there are 1.6 million (1.2–2.0 million) and 2.4 million (1.8–3.0 million) episodes of domestically acquired foodborne illness related to 30 known pathogens and unspecified agents, respectively, for a total estimate of 4.0 million (3.1–5.0 million) episodes of domestically acquired foodborne illness in Canada. Norovirus, Clostridium perfringens, Campylobacter spp., and nontyphoidal Salmonella spp. are the leading pathogens and account for approximately 90% of the pathogen-specific total. Approximately one in eight Canadians experience an episode of domestically acquired foodborne illness each year in Canada. These estimates cannot be compared with prior crude estimates in Canada to assess illness trends as different methodologies were used.
Introduction
Foodborne illness remains a global public health concern (Flint et al., 2005; Newell et al., 2010). Causes of foodborne illness include bacteria, parasites, viruses, toxins, metals, and prions. Symptoms can range from mild and self-limiting vomiting and diarrhea to severe and life-threatening neurological conditions. Estimates of foodborne illness are important for public health decision-makers. Determining these estimates can be challenging, given the large variability within foodborne illness in terms of pathogen, host, and environmental factors. Several attempts by different countries have been made to estimate foodborne illness at a national level (Mead et al., 1999; Adak et al., 2002; Adak et al., 2005; Hall et al., 2005; Vaillant et al., 2005; Cressey et al., 2011; Gkogka et al., 2011; Scallan et al., 2011a; Scallan et al., 2011b; Havelaar et al., 2012).
Cases of foodborne illness are underascertained by public health surveillance systems because of underdiagnosis (i.e., cases that do not seek medical care, are not tested or the laboratory test does not identify a causative agent) and under-reporting (i.e., positive laboratory test results are not reported to surveillance systems) (MacDougall et al., 2008). In order to be captured in a laboratory-based surveillance system in Canada, a sick individual must (1) seek care; (2) have a sample (stool, urine, or blood) requested; (3) submit a sample for testing; (4) the sample must be tested with a test capable of identifying the causative agent; and finally (5) this positive test result must be reported to the surveillance system (Thomas et al., 2008). In order to more accurately estimate the burden of foodborne illness, it is necessary to account for underascertainment and to estimate what proportion of cases result from foodborne transmission, as pathogens rarely are exclusive to one transmission route.
The Public Health Agency of Canada (the Agency) previously used values derived in part from earlier United States (U.S.) estimates (Mead et al., 1999) to generate a crude estimate of 11 million episodes of foodborne illness each year in Canada (Thomas et al., 2008). A more refined methodology and the most current available data have been used to generate updated estimates of foodborne illness domestically acquired in Canada from both known pathogens and unspecified agents. Similar methodology to that which was recently used by the U.S. Centers for Disease Control and Prevention (U.S.-CDC) was employed (Scallan et al., 2011a; Scallan et al., 2011b). The objective of this analysis is to provide a more accurate estimate of domestically acquired foodborne illness in Canada, while identifying data gaps and areas for further research.
Materials and Methods
The recent U.S.-CDC published list of 31 pathogens was used for the development of the Canadian estimates (Scallan et al., 2011b). Based on expert consultation, Mycobacterium bovis and Streptococcus Group A were excluded as they were not deemed to be foodborne pathogens for Canada; adenovirus was added to this list based on expert consultation and its potential to be foodborne (FAO/WHO, 2008; Hall et al., 2005), for a total of 30 pathogens included in the Canadian estimates of foodborne illness.
Analytical approach
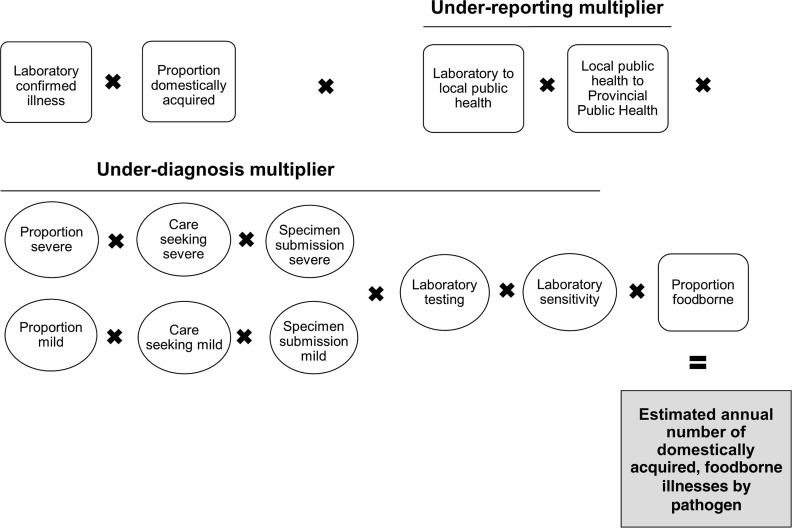
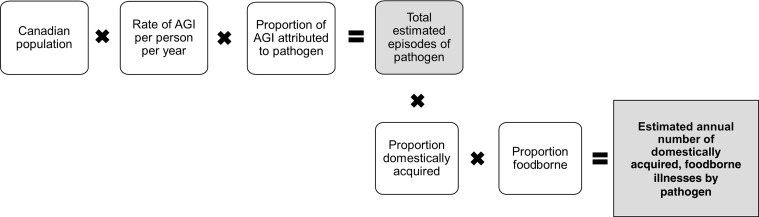
Estimates were developed from stochastic models to consider uncertainty of the input values. Two main modeling approaches were utilized: (1) laboratory-confirmed cases of pathogens scaled up to account for underascertainment due to underdiagnosis and under-reporting (Fig. 1) and (2) models that started with the total Canadian population and used incidence data to scale down the estimated number of illnesses (Fig. 2). Model outputs are described as mean values with 90% credible intervals (90%CrI).
FIG. 1.
Schematic of the general model used to estimate the number of cases of pathogens for which laboratory confirmed illnesses were scaled up
FIG. 2.
Schematic of general model used to estimate the number of cases of pathogens for which the Canadian population was scaled down
Data sources focused on the 2000–2010 time period, and all estimates were based on the approximate Canadian population in 2006 (32,500,000 people) as a midpoint of the selected time period (Statistics Canada, 2008). Details of the input and modeling approaches for each pathogen are provided (Supplementary Technical Appendix S1; Supplementary Data are available online at www.liebertpub.com/fpd). Preference was given to nationally representative data for Canada, followed by provincial data and Canadian-based published literature. If there was a gap in the Canadian-based literature, the values and references used by the U.S.-CDC estimates (Scallan et al., 2011b) were generally used as a proxy for a Canadian-based input.
As per the U.S.-CDC estimates (Scallan et al., 2011a), we generated an estimate for unspecified agents that cause domestically acquired foodborne illness. The estimate of unspecified agents includes illnesses not related to the 30 specified pathogens. Unspecified agents were defined using the same definition as the U.S.-CDC estimates: “Known agents with insufficient data for estimating agent-specific episodes of illness; known agents not yet recognized as causing food-borne illness; microbes, chemicals or other substances known to be in food but for which pathogenicity is unproven; and agents not yet described” (Scallan et al., 2011a).
Illnesses
Laboratory-based surveillance data were available for 18 pathogens (Table 1). These data were adjusted for underdiagnosis and under-reporting using pathogen-specific multipliers. Laboratory-based surveillance data at the national level were obtained from two surveillance systems: (1) Canadian Notifiable Disease Surveillance System (CNDSS) (Government of Canada, 2005; Government of Canada, 2012b), and (2) National Enteric Surveillance Program (NESP) (Government of Canada, 2012c); CNDSS data were used when available (with the exception of Escherichia coli O157, which relied on NESP data) followed by NESP data. Additional laboratory-based surveillance occurs at the provincial level for select pathogens.
Table 1.
Modeling Approaches Used to Estimate the Total Number of Illnesses for Different Types of Data, Canada
|
Pathogens for which laboratory-confirmed illnesses were scaled up |
|
|
|
|---|---|---|---|
| National reportable disease data | Provincial reportable disease data | Pathogens for which Canadian population scaled down | Other methods |
| Brucella spp. | Trichinella spp. | Adenovirus | Escherichia coli, other diarrheagenic |
| Campylobacter spp. | Listeria monocytogenes | Astrovirus | ETEC |
| Clostridium botulinum | Vibrio parahaemolyticus | Norovirus | VTEC non-O157 |
| Cryptosporidium spp. | Yersinia enterocolitica | Rotavirus | Bacillus cereus |
| Cyclospora cayetanensis | Sapovirus | Staphylococcus aureus | |
| VTEC O157 | Toxoplasma gondii | ||
| Giardia sp. | Clostridium perfringens | ||
| Hepatitis A | |||
| Salmonella spp., nontyphoidal | |||
| Salmonella Typhi | |||
| Shigella spp. | |||
| Vibrio cholerae | |||
| Vibrio spp., othera | |||
| Vibrio vulnificusa | |||
These pathogens are reported to the laboratory-based system of NESP and are not considered to be reportable to CNDSS.
VTEC, verotoxigenic Escherichia coli; ETEC, enterotoxigenic Escherichia coli.
To adjust for underdiagnosis due to medical care seeking behavior, specimen request and submission, laboratory testing, and test sensitivity, pathogen-specific multipliers were generated. Results from the National Studies on Acute Gastrointestinal Illness (NSAGI) population surveys completed in 2001–2002, 2002–2003, and 2005–2006 were combined and used to develop multiplier estimates for: medical care seeking; stool sample requests; and stool submission behaviors (Majowicz et al., 2004; Thomas et al., 2006; Sargeant et al., 2008). This information was stratified based on symptoms to account for differences in practices when symptoms are severe or mild. We defined individuals with acute diarrheal illness to be those who experienced ≥3 loose stools in 24 h with duration lasting >1 day and whose symptoms did not result from pregnancy, medication, food allergy, and/or medical conditions previously diagnosed by a doctor (e.g., colitis, diverticulitis, Crohn's disease, irritable bowel syndrome). Those who met the case definition for acute diarrheal illness and had bloody diarrhea or diarrhea that lasted greater than 7 days were considered to be severe, and proportions of those seeking medical care and stool sample submission were 44% and 27%, respectively. Those who met the case definition of acute diarrheal illness but did not experience bloody diarrhea and whose duration of illness was ≤7 days were considered to be mild, and proportions of those seeking medical care and stool sample submission were 14% and 20%, respectively.
For each pathogen, the proportion of laboratory-confirmed illnesses with severe and mild illness was estimated. Data from C-EnterNet (Canada's integrated enteric disease surveillance system based on sentinel sites) (Government of Canada, 2012a) were used to estimate the proportion of cases that experience bloody diarrhea or duration >7 days (i.e., severe) and those that did not experience bloody diarrhea and had a duration of ≤7 days (i.e., mild) for most pathogens captured in this surveillance system. For those pathogens where C-EnterNet data were not available or appropriate due to small case counts, values from the U.S. estimates (Scallan et al., 2011b) were used as a proxy for proportion severe. For certain pathogens, bloody diarrhea or long duration may not be typical (e.g., Vibrio parahaemolyticus, Brucella spp.); however, the care-seeking and sample submission behaviors were still based on these symptoms as a proxy.
Due to the severe nature of certain pathogens, the proportions of seeking medical care and sample submission for severe cases were altered to be 90% and 80%, respectively. This applied to Vibrio vulnificus, Clostridium botulinum, Listeria monocytogenes and hepatitis A (Scallan et al., 2011b).
To account for clinical laboratory testing practices and test sensitivity, data from the NSAGI Laboratory Survey (Flint, 2002; Government of Canada, 2002), available literature, and consultation with the Canadian Public Health Laboratory Network (CPHLN) directors were used to determine input values (Supplementary Technical Appendix S2).
To account for under-reporting from a clinical laboratory to a provincial public health authority, we used data from the NSAGI Laboratory and Public Health Reporting surveys (Flint, 2002; Government of Canada, 2002; Flint et al., 2004). For pathogens not included in these surveys, we used the under-reporting multiplier for nontyphoidal Salmonella spp. as a proxy. It was assumed that there was no under-reporting from provincial to national public health reporting systems.
Alternative approaches were used for the 12 pathogens not reported by routine surveillance or where data were not considered to be complete. Adenovirus, astrovirus, norovirus, rotavirus, sapovirus, and C. perfringens were estimated using incidence estimates from a community-based study completed in the United Kingdom (UK) 2008–2009 (Tam et al., 2011; Tam et al., 2012). They were applied to estimates of acute gastrointestinal illness (AGI) from NSAGI population surveys in Canada. Provincial reported disease data from two provinces for Bacillus cereus, Staphylococcus aureus and C. perfringens were extrapolated to estimate a national annual case count for each pathogen. The ratios of the mean extrapolated national case counts of B. cereus and S. aureus to C. perfringens were applied to the population-level estimate of C. perfringens (based on the UK community-based study [Tam et al., 2011; Tam et al., 2012]) to generate an estimate of total illnesses related to Bacillus cereus and Staphylococcus aureus. Verotoxigenic E. coli (VTEC) non-O157, enterotoxigenic E. coli, and other diarrheagenic E. coli non-VTEC and non-enterotoxigenic E. coli were all estimated relative to the estimate of domestic foodborne cases of VTEC O157 based on literature reports (Chui et al., 2011; Scallan et al., 2011b). Toxoplasma gondii was estimated based on the incidence estimated from nationally representative serologic data from the U.S. National Health and Nutrition Examination Survey (Jones et al., 2007; Scallan et al., 2011b) and then applied to the Canadian population. The proportion symptomatic was based on literature (WHO, 1969); however, as most symptomatic infections are thought to be mild, further estimates were made specific to the number of cases resulting in ocular disease and the number of congenital infections experiencing symptoms (Kemmeren et al., 2006; Havelaar et al., 2007).
Domestically acquired foodborne illness
Data from C-EnterNet surveillance (2005–2010) and the British Columbia reportable disease systems (2008–2010) (Taylor et al., 2010; Government of Canada, 2012a) were used to estimate the proportion of travel-related cases by pathogen. This proportion was subtracted from the total laboratory-confirmed cases (i.e., prior to incorporating the under-reporting and underdiagnosis multipliers), assuming that the underascertainment of travel-related cases would be different from domestic cases (i.e., those with symptoms and who have traveled may be more likely to seek medical care, have a sample requested, and to submit a sample, thus altering the underdiagnosis multiplier). The proportion of cases attributed to food was determined based on a Canadian expert elicitation (Ravel et al., 2010) and other relevant literature (Supplementary Technical Appendix S3).
Unspecified agents
To estimate the number of cases of AGI attributed to unspecified agents, NSAGI population survey data from 2001 to 2002, 2002 to 2003, and 2005 to 2006 (Majowicz et al., 2004; Thomas et al., 2006; Sargeant et al., 2008) were used (Supplementary Technical Appendix S4). We estimated the proportion of individuals with AGI to be those who experienced ≥3 loose stools in 24 hours or any vomiting in the past 28 days, excluding those with chronic conditions, or concurrent symptoms of coughing, sneezing, sore throat, or runny nose. Data were standardized by gender and 5-year age categories to the 2006 Canadian census population (Statistics Canada, 2008), with an estimated rate of 0.630 (95% confidence intervals 0.574–0.689) episodes per person-year. This incidence was applied to the approximated 2006 Canadian population to estimate the total annual number of episodes of AGI.
The number of cases attributed to the 25 pathogens known to cause symptoms of vomiting or diarrhea was subtracted from the estimated total number of episodes of AGI to generate an estimate of the total number of cases of AGI related to unspecified agents. Five pathogens were not included since they do not typically cause symptoms of AGI (i.e., Toxoplasma gondii, hepatitis A, L. monocytogenes, Brucella spp., and C. botulinum). The estimated proportions domestically acquired and foodborne for the 25 known pathogens that cause symptoms of vomiting and diarrhea were used as a proxy for the proportion domestically acquired and proportion foodborne related to unspecified agents. These were applied to the estimate of the total number of cases of AGI related to unspecified agents to estimate the total number of domestically acquired, foodborne illnesses attributed to unspecified agents.
Uncertainty analysis
To capture the uncertainty associated with the estimates, inputs were described using probability distributions that captured the range (minimum, maximum) and most likely value. Empirical data were used to inform these distributions whenever possible; however, in situations where data were unavailable, expert opinion was used. In both situations, the minimum, maximum, and most likely values for the variable were developed (using the PERT distribution) (Supplementary Technical Appendix S1) (Vose, 2008). The final estimates (reported as mean with 90%CrI around the mean) were generated using Monte Carlo simulation (100,000 iterations using @Risk software, an add-in for Microsoft Excel).
Results
The combined estimate for 30 specified pathogens and unspecified agents is an annual estimate of 4.0 million (90%CrI: 3.1–5.0 million) episodes of domestically acquired, foodborne illness in Canada circa 2006. We estimate each year that there are 1.6 million (90%CrI: 1.2–2.0 million) episodes of domestically acquired foodborne illness related to 30 specified pathogens (Table 2). The pathogens that cause the greatest number of illnesses are norovirus (1 million), C. perfringens (177,000), Campylobacter spp. (145,000), and nontyphoidal Salmonella spp. (88,000) (Table 3). It is estimated that there are 9132 domestically acquired, foodborne toxoplasmosis cases annually in Canada, of which 293 (90%CrI: 203–388) present as ocular disease and 31 (90%CrI: 14–50) and 34 (90% CrI 14–58) present as symptomatic congenital infections in the first year of life and years 1–20, respectively. We estimate there are 2.4 million (90%CrI: 1.8–3.0 million) episodes of domestically acquired foodborne illness related to unspecified agents (Supplementary Technical Appendix S3).
Table 2.
Estimated Annual Number of Cases of Domestically Acquired Foodborne Illness for 30 Pathogens, Under-reporting and Underdiagnosis Multipliers, Canada Circa 2006
| |
|
|
|
Multipliers |
Domestically acquired |
|
Domestically acquired foodborne |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pathogen | Laboratory confirmed | Travel-related | Domestically acquired laboratory confirmed | Under-reporting | Underdiagnosis | Total | Mean | 90% CrI (low) | 90% CrI (high) | Foodborne | Mean | 90% CrI (low) | 90% CrI (high) |
| Bacteria | |||||||||||||
| Bacillus cereus | n/a | 0% | n/a | n/a | n/a | n/a | 36,275 | 19,520 | 55,381 | 100% | 36,269 | 19,517 | 55,371 |
| Brucella spp.a | 8 | 80% | 2 | 1.1 | 25.1 | 26.8 | 43 | 13 | 90 | 50% | 22 | 6 | 45 |
| Campylobacter spp. | 10,344 | 24% | 7861 | 1.1 | 25.2 | 27.2 | 213,749 | 144,288 | 308,837 | 68% | 145,350 | 95,686 | 212,971 |
| Clostridium botulinum | 5 | 0% | 5 | 1.1 | 2.5 | 2.7 | 14 | 0 | 27 | 100% | 14 | 0 | 27 |
| Clostridium perfringens | n/a | 0% | n/a | n/a | n/a | n/a | 176,992 | 95,239 | 270,211 | 100% | 176,963 | 95,225 | 270,160 |
| VTEC O157 | 883 | 5% | 844 | 1.1 | 18.4 | 20.1 | 16,913 | 6968 | 29,668 | 76% | 12,827 | 5225 | 22,750 |
| VTEC, non-O157 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 20,523 | 8361 | 36,400 |
| ETEC | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 3848 | 1568 | 6825 |
| Escherichia coli, other diarrheagenic | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | 2565 | 1045 | 4550 |
| Listeria monocytogenes | 131 | 6% | 122 | 1.1 | 1.6 | 1.7 | 212 | 147 | 335 | 84% | 178 | 122 | 283 |
| Salmonella spp., nontyphoidal | 5676 | 26% | 4191 | 1.1 | 24.5 | 26.1 | 109,384 | 74,530 | 154,994 | 80% | 87,510 | 58,832 | 125,525 |
| Salmonella Typhi | 117 | 76% | 28 | 1.1 | 11.9 | 12.7 | 358 | 195 | 605 | 80% | 287 | 155 | 487 |
| Shigella spp. | 886 | 57% | 381 | 1.0 | 16.8 | 17.5 | 6675 | 3746 | 10,927 | 18% | 1202 | 561 | 2151 |
| Staphylococcus aureus | n/a | 0% | n/a | n/a | n/a | n/a | 25,114 | 13,514 | 38,341 | 100% | 25,110 | 13,512 | 38,334 |
| Vibrio cholerae, toxigenic | 5 | 99% | 0 | 1.0 | 11.9 | 11.9 | 0 | 0 | 1 | 100% | 0 | 0 | 1 |
| Vibrio vulnificus | 1 | 2% | 1 | 1.2 | 1.9 | 2.3 | 1 | 0 | 4 | 82% | 1 | 0 | 3 |
| Vibrio parahaemolyticus | 27 | 13% | 24 | 1.2 | 75.4 | 92.0 | 2193 | 1075 | 3995 | 82% | 1798 | 869 | 3305 |
| Vibrio spp., other | 17 | 11% | 15 | 1.2 | 75.4 | 92.0 | 1356 | 715 | 2308 | 82% | 1112 | 578 | 1911 |
| Yersinia enterocolitica | 975 | 16% | 824 | 1.1 | 36.4 | 39.3 | 32,394 | 19,788 | 49,597 | 80% | 25,915 | 15,344 | 40,457 |
| Subtotal | 621,675 | 541,495 | |||||||||||
| Parasites | |||||||||||||
| Cryptosporidium spp. | 746 | 30% | 522 | 1.3 | 36.9 | 48.5 | 25,318 | 14,331 | 45,955 | 9% | 2321 | 1012 | 4530 |
| Cyclospora cayetanensis | 159 | 71% | 46 | 1.4 | 36.9 | 53.5 | 2,475 | 1475 | 3867 | 99% | 2450 | 1460 | 3829 |
| Giardia sp. | 4374 | 39% | 2668 | 1.1 | 36.9 | 40.7 | 108,507 | 70,532 | 158,740 | 7% | 7776 | 4742 | 11,874 |
| Toxoplasma gondii | n/a | 3% | n/a | n/a | n/a | n/a | 18,265 | 14,418 | 22,566 | 50% | 9132 | 6953 | 11,628 |
| Trichinella spp.a | 5 | 4% | 5 | 1.1 | 11.7 | 12.5 | 63 | 0 | 235 | 100% | 63 | 0 | 235 |
| Subtotal | 154,627 | 21,743 | |||||||||||
| Viruses | |||||||||||||
| Adenovirus | n/a | 0% | n/a | n/a | n/a | n/a | 747,805 | 576,880 | 929,637 | 1% | 3739 | 1367 | 6398 |
| Astrovirus | n/a | 0% | n/a | n/a | n/a | n/a | 382,447 | 260,748 | 516,572 | 1% | 1912 | 671 | 3409 |
| Hepatitis A virus | 393 | 33% | 262 | 1.1 | 12.2 | 13.9 | 3640 | 1953 | 6053 | 7% | 271 | 115 | 516 |
| Norovirus | n/a | 0% | n/a | n/a | n/a | n/a | 3,379,990 | 3,002,927 | 3,778,461 | 31% | 1,047,733 | 679,576 | 1,434,048 |
| Rotavirus | n/a | 0% | n/a | n/a | n/a | n/a | 850,233 | 665,213 | 1,045,617 | 1% | 4252 | 1559 | 7206 |
| Sapovirus | n/a | 0% | n/a | n/a | n/a | n/a | 1,898,561 | 1,619,335 | 2,195,201 | 1% | 9491 | 3539 | 15,698 |
| Subtotal | 7,262,676 | 1,067,398 | |||||||||||
| Total mean and 90% CrI | 8,038,978 | 7,461,815 | 8,631,524 | 1,630,636 | 1,237,848 | 2,042,713 | |||||||
Pathogens for which Canadian foodborne transmission is thought to be from wild-game consumption due to surveillance and eradication programs in domestic food animals.
CrI, 90% credible intervals; n/a, not applicable; VTEC, verotoxigenic Escherichia coli; ETEC, enterotoxigenic Escherichia coli.
Table 3.
Total Estimated Domestically Acquired Foodborne Illness in Canada, Estimated Cases per 100,000 and Pathogen Rank
| Pathogen | Mean count | % of total (30 pathogens) | Estimated cases per 100,000 | Overall rank | Rank for pathogens based primarily on Canadian data | Rank for pathogens based on limited Canadian data |
|---|---|---|---|---|---|---|
| Norovirus | 1,047,733 | 65.12 | 3,223.79 | 1 | 1 | |
| Clostridium perfringens | 176,963 | 11.00 | 544.50 | 2 | 2 | |
| Campylobacter spp. | 145,350 | 8.42 | 447.23 | 3 | 1 | |
| Salmonella spp., nontyphoidal | 87,510 | 5.07 | 269.26 | 4 | 2 | |
| Bacillus cereus | 36,269 | 2.25 | 111.60 | 5 | 3 | |
| Yersinia enterocolitica | 25,915 | 1.49 | 79.74 | 6 | 3 | |
| Staphylococcus aureus | 25,110 | 1.56 | 77.26 | 7 | 4 | |
| VTEC non-O157 | 20,523 | 1.19 | 63.15 | 8 | 5 | |
| VTEC O157 | 12,827 | 0.75 | 39.47 | 9 | 4 | |
| Sapovirus | 9491 | 0.59 | 29.20 | 10 | 6 | |
| Toxoplasma gondii | 9132 | 0.57 | 28.10 | 11 | 7 | |
| Giardia spp. | 7776 | 0.45 | 23.93 | 12 | 5 | |
| Rotavirus | 4252 | 0.26 | 13.08 | 13 | 8 | |
| ETEC | 3848 | 0.22 | 11.84 | 14 | 9 | |
| Adenovirus | 3739 | 0.23 | 11.51 | 15 | 10 | |
| Escherichia coli, other diarrheagenic | 2565 | 0.15 | 7.89 | 16 | 11 | |
| Cyclospora cayetanensis | 2450 | 0.14 | 7.54 | 17 | 6 | |
| Cryptosporidium spp. | 2321 | 0.13 | 7.14 | 18 | 7 | |
| Astrovirus | 1912 | 0.12 | 5.88 | 19 | 12 | |
| Vibrio parahaemolyticus | 1798 | 0.10 | 5.53 | 20 | 8 | |
| Shigella spp. | 1202 | 0.07 | 3.70 | 21 | 9 | |
| Vibrio, other spp. | 1112 | 0.06 | 3.42 | 22 | 10 | |
| Salmonella Typhi | 287 | 0.02 | 0.88 | 23 | 11 | |
| Hepatitis A | 271 | 0.02 | 0.83 | 24 | 12 | |
| Listeria monocytogenes | 178 | 0.01 | 0.55 | 25 | 13 | |
| Trichinella spp. | 63 | <0.01 | 0.19 | 26 | 14 | |
| Brucella spp. | 22 | <0.01 | 0.07 | 27 | 15 | |
| Clostridium botulinum | 14 | <0.01 | 0.04 | 28 | 16 | |
| Vibrio vulnificus | 1 | <0.01 | 0.00 | 29 | 17 | |
| Vibrio cholerae, toxigenic | 0 | 0.0 | 0.00 | 30 | 18 |
VTEC, verotoxigenic Escherichia coli; ETEC, enterotoxigenic Escherichia coli.
Discussion
We estimate that in Canada there are 4.0 million episodes of domestically acquired foodborne illnesses each year, circa 2006, attributed to 30 known pathogens as well as unspecified agents that cause symptoms of AGI. The main pathogens associated with illnesses are norovirus, C. perfringens, Campylobacter spp., and nontyphoidal Salmonella spp., representing 90% of the total pathogen-specific illnesses. Unspecified agents are the main contributor to the total estimate of domestically acquired foodborne illnesses.
This estimate differs from the previous Canadian estimate of 11 million (Thomas et al., 2008), which used values from earlier U.S. estimates (Mead et al., 1999) applied to the total Canadian population and an incidence estimate of 1.3 episodes per person-year based on a sensitive AGI case definition of “any vomiting or diarrhea in the past 4 weeks” and did not exclude travel-related illnesses. The new approach uses a more specific case definition for AGI (that is more consistent with the U.S.-CDC estimates [Scallan et al., 2011b]) and thus the incidence of AGI is lower as well. This estimate cannot be compared with the estimate of 11 million for the purpose of assessing trends. The CNDSS (Government of Canada, 2012b) provides the best national data on trends over time.
Comparisons between different country estimates are difficult to make, given the inherent differences in methodological approaches and data sources. However, the pathogen-specific results presented here are similar to other country estimates (United States, Australia, the Netherlands, New Zealand, and France) that also found norovirus (Vaillant et al., 2005; Hall et al., 2005; Cressey et al., 2011; Scallan et al., 2011b; Havelaar et al., 2012) to be a main contributor and Campylobacter spp. and nontyphoidal Salmonella spp. (United States, Australia, New Zealand, France, the UK, and Greece) (Adak et al., 2002; Hall et al., 2005; Vaillant et al., 2005; Cressey et al., 2011; Gkogka et al., 2011; Scallan et al., 2011b) to be leading bacterial pathogens in national foodborne illness estimates. Food poisoning and foodborne toxin-producing pathogens (namely, B. cereus, C. perfringens, and S. aureus) are among the top 10 pathogens in recent national foodborne illness estimates (United States, Australia, the Netherlands, New Zealand, France, the UK and Greece) (Adak et al., 2002; Hall et al., 2005; Vaillant et al., 2005; Cressey et al., 2011; Gkogka et al., 2011; Scallan et al., 2011b; Havelaar et al., 2012); however, estimates for these pathogens are typically based on limited information, warranting future research.
Among the national published foodborne illness estimates, the proportion foodborne for norovirus ranges from around 11% in the United Kingdom (Adak et al., 2002) to 39% in New Zealand (Cressey et al., 2011). As identified in the U.S.-CDC estimates (Scallan et al., 2011b), the estimate of the proportion foodborne for norovirus can have a substantial impact on the overall estimate. Estimates of the incidence of foodborne illness due to nontyphoidal Salmonella spp. and Campylobacter spp. in Australia (Hall et al., 2005) and New Zealand (Cressey et al., 2011) are substantially greater than those in Canada and the U.S.-CDC estimates (Scallan et al., 2011b). This is likely due to the high laboratory-confirmed incidence rate of these pathogens in Australia and New Zealand, suggesting that although they are leading bacterial causes of foodborne illness in all four countries, the actual incidence of these pathogens is lower in North America compared with Australia and New Zealand.
Comparison between the Canadian and the U.S.-CDC estimates (Scallan et al., 2011b) shows that the top four pathogens are identical (i.e., norovirus, C. perfringens, Campylobacter spp., and nontyphoidal Salmonella spp.) and account for nearly 90% of the total pathogen-specific burden, though their relative order changes between the two estimates. The overall total estimate (specified pathogens and unspecified agents) for Canada is slightly less than the U.S.-CDC estimate; approximately one in eight Canadians compared to one in six Americans experience foodborne illness per year.
A key difference between the United States and Canadian approaches is the incorporation of duration of illness to define severity by pathogen. Duration and presence of bloody diarrhea has been used in Australia (Hall et al., 2008) to determine severity of illness by pathogen and has been shown to influence the likelihood of seeking care (MacDougall et al., 2008). The measure of severity is used to describe differences in behavior, which contribute to differences in the underdiagnosis of a pathogen. By incorporating duration with bloody diarrhea, the proportion of cases considered severe is higher, resulting in a lower estimate of underascertainment and the pathogen-specific total.
An additional difference between the United States and Canadian approaches was the decision to estimate rotavirus, astrovirus, and sapovirus for the total population, rather than only for those under the age of 5 years (Scallan et al., 2011b). Evidence suggests that illnesses associated with these viruses do occur in adults ( Svenungsson et al., 2000; de Wit et al., 2001; Anderson et al., 2004; Pang et al., 2009; Bresee et al., 2012; Anderson et al., 2012). This decision effectively reduced the number of cases estimated in the unspecified portion, as they are accounted for among these viruses. Since these viruses are not thought to be primarily foodborne, the final proportion of foodborne related to specified pathogens is less, which in turn reduces the estimated total for foodborne-unspecified agents. The inclusion of adenovirus in the Canadian estimate effectively reduces the unspecified agent estimate for Canada in comparison with that of the U.S.-CDC estimate.
Because toxoplasmosis is often mild with nonspecific symptoms (i.e., lymphadenopathy, fatigue, and general malaise), estimates of congenital toxoplasmosis (i.e., symptoms of chorioretinitis, intracranial calcifications and hydrocephalus, and central nervous system abnormalities in the first year of life and the development of chorioretinitis in the first 20 years of life) and acquired ocular toxoplasmosis were generated as a subset of the full estimate, to demonstrate specific severe outcomes (Kimball et al., 1971; Alford et al., 1974; Koppe et al., 1986; Guerina et al., 1994; Gratzl et al., 1998; Lopez et al., 2000; Binquet et al., 2003; Kemmeren et al., 2006; Havelaar et al., 2007). Primary infection during pregnancy can also result in spontaneous abortion and stillbirth; however these are not explicitly captured in the estimate of toxoplasmosis (Kemmeren et al., 2006; Havelaar et al., 2007).
The approach for the bacterial toxins (B. cereus, C. perfringens, and S. aureus) was also different from the U.S. approach. Though Canadian data to estimate an outbreak to sporadic ratio were explored, they were deemed not suitable for these estimates. Instead, the ratio of these pathogens from the reported provincial data was used in addition to the estimate obtained through the population incidence of C. perfringens from the U.K. study (Tam et al., 2011; Tam et al., 2012).
There are many limitations in the approach used to estimate total domestically acquired foodborne illness. The proportion severe for many pathogens is based on information from the C-EnterNet surveillance program (Government of Canada, 2012a); however, these data are based on only one sentinel site and may not reflect the Canadian population.
Data on frequency of laboratory testing were based in part on a 2001 survey of Canadian Laboratories (Flint, 2002; Government of Canada, 2002) and vetted by members of the Canadian Public Health Laboratory Network; however, it is likely that laboratory practices have changed since 2001. Furthermore, there was little information in the literature to determine laboratory test sensitivity, warranting future research. Though not incorporated in these estimates or other national foodborne illness estimates, test specificity (false positives) should be explored.
The use of population surveys on self-reported illness and care-seeking behaviors related to AGI is a potential limitation. Recall bias and case definitions can alter the results (Majowicz et al., 2008; Cantwell et al., 2010; Thomas et al., 2011). Noninfectious causes of vomiting and diarrhea and illness associated with respiratory symptoms were excluded. A highly specific case definition was used (Hall et al., 2010; Majowicz et al., 2004; Sargeant et al., 2008; Thomas et al., 2006), yet cases may have been misclassified. The exclusion of cases with symptoms of runny nose or sore throat may have resulted in an overly conservative case definition, which may be a limitation.
For the estimation of norovirus, rotavirus, astrovirus, sapovirus, adenovirus; and C. perfringens, data from the U.K. cohort study were used (Tam et al., 2011; Tam et al., 2012). The UK study population differs somewhat from the Canadian population, with a higher proportion of participants in the 55 years and greater age group and fewer in the 15–44 years age group (compared with the 2006 Canadian census population). Also, the incidence of these pathogens in the United Kingdom may differ from that of Canada; however, at the time of this work, no suitable Canadian data were available.
Data on the proportion of travel-related cases is based on two Canadian geographical areas, which may not represent the rest of Canada. The case definition for travel-related episodes varied between the two surveillance databases used.
The proportion of illnesses transmitted by food greatly affects the estimates, but there is a paucity of data on which to base these values. One expert elicitation (Ravel et al., 2010) was used for many proportion estimates, but may be prone to bias. For pathogens not included in the expert elicitation, the U.S.-CDC estimates were used. This may not accurately reflect the Canadian situation. With full national implementation of Canadian Outbreak Summaries (currently implemented in some provinces only) (Savelli et al., 2009), future estimates could incorporate information from outbreak-related databases.
The estimate of domestically acquired foodborne illness related to unspecified agents is highly dependent on the accuracy of the estimates related to the 25 known AGI-causing pathogens. As the number of cases attributed to the 25 known pathogens increases, the number of cases attributed to the unspecified agents decreases. Furthermore, the proportion domestic and foodborne based on 25 known pathogens may not be the same for unspecified agents. This approach is consistent with the U.S. estimates (Scallan et al., 2011a; Scallan et al., 2011b) and thus allows for comparison.
To estimate the number of cases related to unspecified agents, it was necessary to subtract the total number of illnesses related to 25 known AGI pathogens. For simplicity and consistency with the U.S. methods (Scallan et al., 2011a; Scallan et al., 2011b), the same multiplier was applied to travel-related cases to subtract a total pathogen-specific estimate from the total estimate of AGI. Future work will be necessary to develop an estimate of travel-related foodborne illness in Canada.
With full implementation of surveillance programs like C-EnterNet (Government of Canada, 2012a) and Outbreak Summaries for Canada (Savelli et al., 2009), critical information will be collected to inform a reassessment of the annual burden of foodborne illness in Canada. A survey of Canadian clinical laboratories would refine our understanding of current laboratory practices and test sensitivities. Test specificity should be explored in future. As noted in Table 3, estimates of certain pathogens relied more heavily on non-Canadian data sources, indicating a Canadian data gap and that these estimates should be interpreted carefully.
Conclusions
As incidence of pathogens, availability of data, and laboratory practices change with time, re-estimating the number of episodes of foodborne illness in the future will be important to determine what pathogens are contributing to the overall burden, while assessing how identified data gaps have been addressed. Currently, efforts are focusing on estimating the number of hospitalizations and deaths related to foodborne illness, estimating costs and related Disability Adjusted Life Years, and estimating attribution of foodborne illness.
These revised estimates of domestically acquired foodborne illness for Canada represent the most accurate and current values and utilized rigorous and robust methodology. This information will be useful for policy-makers to direct prevention and control activities to those pathogens that cause the most illness. The estimates could inform education campaigns for consumers, provide information to industry and academics for research programs, and be incorporated in food-safety risk assessments.
Supplementary Material
Acknowledgments
We thank the members of the Canadian Public Health Laboratory Network and our partners at Provincial Public Health Authorities for providing data and expert advice for these estimates. We also thank the experts (from federal and provincial government and academic institutions) who provided their time to peer-review the approach taken to develop these estimates.
Disclosure Statement
No competing financial interests exist.
References
- Adak GK. Long SM. O'Brien SJ. Trends in indigenous foodborne disease and deaths, England and Wales: 1992 to 2000. Gut. 2002;51:832–841. doi: 10.1136/gut.51.6.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adak GK. Meakins SM. Yip H. Lopman BA. O'Brien SJ. Disease risks from foods, England and Wales, 1996–2000. Emerg Infect Dis. 2005;11:365–372. doi: 10.3201/eid1103.040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford CA., Jr Stagno S. Reynolds DW. Congenital toxoplasmosis: Clinical, laboratory, and therapeutic considerations, with special reference to subclinical disease. Bull N Y Acad Med. 1974;50:160–181. [PMC free article] [PubMed] [Google Scholar]
- Anderson EJ. Weber SG. Rotavirus infection in adults. Lancet Infect Dis. 2004;4:91–99. doi: 10.1016/S1473-3099(04)00928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EJ. Katz BZ. Polin JA. Reddy S. Weinrobe MH. Noskin GA. Rotavirus in adults requiring hospitalization. J Infect. 2012;64:89–95. doi: 10.1016/j.jinf.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Binquet C. Wallon M. Quantin C. Kodjikian L. Garweg J. Fleury J. Peyron F. Abrahamowicz M. Prognostic factors for the long-term development of ocular lesions in 327 children with congenital toxoplasmosis. Epidemiol Infect. 2003;131:1157–1168. doi: 10.1017/s0950268803001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresee JS. Marcus R. Venezia RA. Keene WE. Morse D. Thanassi M. Brunett P. Bulens S. Beard RS. Dauphin LA. Slutsker L. Bopp C. Eberhard M. Hall A. Vinje J. Monroe SS. Glass RI US Acute Gastroenteritis Etiology Study Team. The etiology of severe acute gastroenteritis among adults visiting emergency departments in the United States. J Infect Dis. 2012;205:1374–1381. doi: 10.1093/infdis/jis206. [DOI] [PubMed] [Google Scholar]
- Cantwell LB. Henao OL. Hoekstra RM. Scallan E. The effect of different recall periods on estimates of acute gastroenteritis in the United States, FoodNet Population Survey 2006–2007. Foodborne Pathog Dis. 2010;7:1225–1228. doi: 10.1089/fpd.2010.0567. [DOI] [PubMed] [Google Scholar]
- Chui L. Lee MC. Malejczyk K. Lim L. Fok D. Kwong P. Prevalence of shiga toxin-producing Escherichia coli as detected by enzyme-linked immunoassays and real-time PCR during the summer months in northern Alberta, Canada. J Clin Microbiol. 2011;49:4307–4310. doi: 10.1128/JCM.05211-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressey P. Lake R. Report—New Zealand Government; Christ Church, New Zealand: 2011. Estimated incidence of foodborne illness in New Zealand: Application of overseas models and multipliers. [Google Scholar]
- de Wit MA. Koopmans MP. Kortbeek LM. Wannet WJ. Vinje J. van Leusden F. Bartelds AI. van Duynhoven YT. Sensor, a population-based cohort study on gastroenteritis in the Netherlands: Incidence and etiology. Am J Epidemiol. 2001;154:666–674. doi: 10.1093/aje/154.7.666. [DOI] [PubMed] [Google Scholar]
- [FAO/WHO] Food and Agriculture Organization of the United Nations/World Health Organization; FAO/WHO. Viruses in food: Scientific advice to support risk management activities. Meeting report. Microbiological Risk Assessment Series No. 13. Rome and Geneva; 2008. [Google Scholar]
- Flint J. Guelph: Government of Canada; 2002. Report of the 2001 Canadian Laboratory Study—National Studies on Acute Gastrointestinal Illness. [Google Scholar]
- Flint JA. Dore K. Majowicz SE. Edge VL. Sockett P. From stool to statistics: Reporting of acute gastrointestinal illnesses in Canada. Can J Public Health. 2004;95:309–313. doi: 10.1007/BF03405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint JA. Van Duynhoven YT. Angulo FJ. DeLong SM. Braun P. Kirk M. Scallan E. Fitzgerald M. Adak GK. Sockett P. Ellis A. Hall G. Gargouri N. Walke H. Braam P. Estimating the burden of acute gastroenteritis, foodborne disease, and pathogens commonly transmitted by food: An international review. Clin Infect Dis. 2005;41:698–704. doi: 10.1086/432064. [DOI] [PubMed] [Google Scholar]
- Gkogka E. Reij MW. Havelaar AH. Zwietering MH. Gorris LG. Risk-based estimate of effect of foodborne diseases on public health, Greece. Emerg Infect Dis. 2011;17:1581–1590. doi: 10.3201/eid1709.101766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Government of Canada. Guelph: ON: Public Health Agency of Canada; 2002. Report of the 2001 Canadian Laboratory Survey: National Studies on Acute Gastrointestinal Illness (NSAGI) [Google Scholar]
- Government of Canada. National Notifiable Diseases On-Line: Public Health Agency of Canada. 2005. http://dsol-smed.phac-aspc.gc.ca/dsol-smed/ndis/index-eng.php. [Oct 4;2012 ]. http://dsol-smed.phac-aspc.gc.ca/dsol-smed/ndis/index-eng.php
- Government of Canada. Guelph: ON: Public Health Agency of Canada, (2005–2010); 2012a. Canadian National Enteric Pathogen Surveillance System (C-EnterNet) [Google Scholar]
- Government of Canada. Public Health Agency of Canada; Ottawa: 2012b. Canadian Notifiable Disease Surveillance System National Report (2005–2008) [Google Scholar]
- Government of Canada. Public Health Agency of Canada; Guelph: 2012c. National Enteric Surveillance Program (2000–2010) [Google Scholar]
- Gratzl R. Hayde M. Kohlhauser C. Hermon M. Burda G. Strobl W. Pollak A. Follow-up of infants with congenital toxoplasmosis detected by polymerase chain reaction analysis of amniotic fluid. Eur J Clin Microbiol Infect Dis. 1998;17:853–858. doi: 10.1007/s100960050206. [DOI] [PubMed] [Google Scholar]
- Guerina NG. Hsu HW. Meissner HC. Maguire JH. Lynfield R. Stechenberg B. Abroms I. Pasternack MS. Hoff R. Eaton RB. Neonatal serologic screening and early treatment for congenital Toxoplasma gondii infection. The New England Regional Toxoplasma Working Group. N Engl J Med. 1994;330:1858–1863. doi: 10.1056/NEJM199406303302604. [DOI] [PubMed] [Google Scholar]
- Hall G. Kirk MD. Becker N. Gregory JE. Unicomb L. Millard G. Stafford R. Lalor K. OzFoodNet Working Group. Estimating foodborne gastroenteritis, Australia. Emerg Infect Dis. 2005;11:1257–1264. doi: 10.3201/eid1108.041367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall G. Yohannes K. Raupach J. Becker N. Kirk M. Estimating community incidence of Salmonella, Campylobacter, and Shiga toxin-producing Escherichia coli infections, Australia. Emerg Infect Dis. 2008;14:1601–1609. doi: 10.3201/eid1410.071042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall G. McDonald L. Majowicz SE. Scallan E. Kirk M. Sockett P. Angulo FJ. Respiratory symptoms and the case definition of gastroenteritis: An international analysis of the potential impact on burden estimates. Epidemiol Infect. 2010;138:117–124. doi: 10.1017/S0950268809990112. [DOI] [PubMed] [Google Scholar]
- Havelaar AH. Kemmeren JM. Kortbeek LM. Disease burden of congenital toxoplasmosis. Clin Infect Dis. 2007;44:1467–1474. doi: 10.1086/517511. [DOI] [PubMed] [Google Scholar]
- Havelaar AH. Haagsma JA. Mangen MJ. Kemmeren JM. Verhoef LP. Vijgen SM. Wilson M. Friesema IH. Kortbeek LM. van Duynhoven YT. van Pelt W. Disease burden of foodborne pathogens in the Netherlands, 2009. Int J Food Microbiol. 2012;156:231–238. doi: 10.1016/j.ijfoodmicro.2012.03.029. [DOI] [PubMed] [Google Scholar]
- Jones JL. Kruszon-Moran D. Sanders-Lewis K. Wilson M. Toxoplasma gondii infection in the United States, 1999–2004, decline from the prior decade. Am J Trop Med Hyg. 2007;77:405–410. [PubMed] [Google Scholar]
- Kemmeren J. Mangen M. van Duynhove Y. Havelaarm A. Priority setting of foodborne pathogens. Rijksinstituut voor Volksgezondheid en Milieu (RIVM)—Report. 2006 [Google Scholar]
- Kimball AC. Kean BH. Fuchs F. Congenital toxoplasmosis: A prospective study of 4,048 obstetric patients. Am J Obstet Gynecol. 1971;111:211–218. doi: 10.1016/0002-9378(71)90892-1. [DOI] [PubMed] [Google Scholar]
- Koppe JG. Loewer-Sieger DH. de Roever-Bonnet H. Results of 20-year follow-up of congenital toxoplasmosis. Lancet. 1986;1:254–256. doi: 10.1016/s0140-6736(86)90785-3. [DOI] [PubMed] [Google Scholar]
- Lopez A. Dietz VJ. Wilson M. Navin TR. Jones JL. Preventing congenital toxoplasmosis. MMWR Morb Mortal Wkly Rep. 2000;49:59–68. [PubMed] [Google Scholar]
- MacDougall L. Majowicz S. Doré K. Flint J. Thomas K. Kovacs S. Sockett P. Under-reporting of infectious gastrointestinal illness in British Columbia, Canada: Who is counted in provincial communicable disease statistics? Epidemiol Infect. 2008;136:248–256. doi: 10.1017/S0950268807008461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majowicz SE. Dore K. Flint JA. Edge VL. Read S. Buffett MC. McEwen S. McNab WB. Stacey D. Sockett P. Wilson JB. Magnitude and distribution of acute, self-reported gastrointestinal illness in a Canadian community. Epidemiol Infect. 2004;132:607–617. doi: 10.1017/s0950268804002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majowicz SE. Hall G. Scallan E. Adak GK. Gauci C. Jones TF. O'Brien S. Henao O. Sockett PN. A common, symptom-based case definition for gastroenteritis. Epidemiol Infect. 2008;136:886–894. doi: 10.1017/S0950268807009375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS. Slutsker L. Dietz V. McCaig LF. Bresee JS. Shapiro C. Griffin PM. Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell DG. Koopmans M. Verhoef L. Duizer E. Aidara-Kane A. Sprong H. Opsteegh M. Langelaar M. Threfall J. Scheutz F. van der Giessen J. Kruse H. Food-borne diseases—The challenges of 20 years ago still persist while new ones continue to emerge. Int J Food Microbiol. 2010;139(suppl 1):S3–S15. doi: 10.1016/j.ijfoodmicro.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang XL. Lee BE. Tyrrell GJ. Preiksaitis JK. Epidemiology and genotype analysis of sapovirus associated with gastroenteritis outbreaks in Alberta, Canada: 2004–2007. J Infect Dis. 2009;199:547–551. doi: 10.1086/596210. [DOI] [PubMed] [Google Scholar]
- Ravel A. Davidson VJ. Ruzante JM. Fazil A. Foodborne proportion of gastrointestinal illness: Estimates from a Canadian expert elicitation survey. Foodborne Pathog Dis. 2010;7:1463–1472. doi: 10.1089/fpd.2010.0582. [DOI] [PubMed] [Google Scholar]
- Sargeant JM. Majowicz SE. Snelgrove J. The burden of acute gastrointestinal illness in Ontario, Canada, 2005–2006. Epidemiol Infect. 2008;136:451–460. doi: 10.1017/S0950268807008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelli C. Public Health Agency of Canada. Implementation of a National Web-Based Outbreak Summary Reporting Application. Canadian Public Health Association Annual Conference; 2009. [Google Scholar]
- Scallan E. Griffin PM. Angulo FJ. Tauxe RV. Hoekstra RM. Foodborne illness acquired in the United States—Unspecified agents. Emerg Infect Dis. 2011a;17:16–22. doi: 10.3201/eid1701.P21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E. Hoekstra RM. Angulo FJ. Tauxe RV. Widdowson MA. Roy SL. Jones JL. Griffin PM. Foodborne illness acquired in the United States—Major pathogens. Emerg Infect Dis. 2011b;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistics Canada; Catalogue no. 94-581-XCB2006001. Profile for Canada, Provinces, Territories, Census Divisions and Census Subdivisions, 2006 Census. 2008.
- Svenungsson B. Lagergren A. Ekwall E. Evengard B. Hedlund KO. Karnell A. Lofdahl S. Svensson L. Weintraub A. Enteropathogens in adult patients with diarrhea and healthy control subjects: A 1-year prospective study in a Swedish clinic for infectious diseases. Clin Infect Dis. 2000;30:770–778. doi: 10.1086/313770. [DOI] [PubMed] [Google Scholar]
- Tam C. Viviani L. Adak B. Bolton E. Dodds J. Cowden J. Evans M. Gray J. Hunter P. Jackson K. Letley L. Neal K. Rait G. Smith G. Smyth B. Tompkins D. van der Es M. Rodrigues L. O'Brien S. The Second Study of Infectious Intestinal Disease in the Community (IID2 Study)—Report. 2011 [Google Scholar]
- Tam CC. Rodrigues LC. Viviani L. Dodds JP. Evans MR. Hunter PR. Gray JJ. Letley LH. Rait G. Tompkins DS. O'Brien SJ. IID2 Study Executive Committee. Longitudinal study of infectious intestinal disease in the UK (IID2 study): Incidence in the community and presenting to general practice. Gut. 2012;61:69–77. doi: 10.1136/gut.2011.238386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. MacDougall L. Li M. Galanis E. The impact of international travel on the epidemiology of enteric infections, British Columbia, 2008. Can J Public Health. 2010;101:332–336. doi: 10.1007/BF03405297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MK. Majowicz SE. MacDougall L. Sockett PN. Kovacs SJ. Fyfe M. Edge VL. Dore K. Flint JA. Henson S. Jones AQ. Population distribution and burden of acute gastrointestinal illness in British Columbia, Canada. BMC Public Health. 2006;6:307. doi: 10.1186/1471-2458-6-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MK. Majowicz SE. Pollari F. Sockett PN. Burden of acute gastrointestinal illness in Canada, 1999–2007: interim summary of NSAGI activities. Can Commun Dis Rep. 2008;34:8–15. [PubMed] [Google Scholar]
- Thomas MK. Perez E. Majowicz SE. Reid-Smith R. Olea A. Diaz J. Solari V. McEwen SA. Burden of acute gastrointestinal illness in the Metropolitan region, Chile, 2008. Epidemiol Infect. 2011;139:560–571. doi: 10.1017/S0950268810001160. [DOI] [PubMed] [Google Scholar]
- Vaillant V. de Valk H. Baron E. Ancelle T. Colin P. Delmas MC. Dufour B. Pouillot R. Le Strat Y. Weinbreck P. Jougla E. Desenclos JC. Foodborne infections in France. Foodborne Pathog Dis. 2005;2:221–232. doi: 10.1089/fpd.2005.2.221. [DOI] [PubMed] [Google Scholar]
- Vose D. England: John Wiley & Sons, Ltd.; 2008. Risk Analysis: A Quantitative Guide. [Google Scholar]
- [WHO] World Health Organization. Toxoplasmosis. In: Technical Report Series. Geneva: WHO; 1969. , No. 431. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.