Abstract
TRIM5α is a factor contributing to intracellular defense mechanisms against retrovirus infection. Rhesus and cynomolgus monkey TRIM5αs potently restrict HIV-1, whereas human TRIM5α shows weak effects against HIV-1. We investigated the association between a single nucleotide polymorphism in the TRIM5α linker 2 region (rs11038628), which substituted aspartic acid (D) for glycine (G) at position 249, with susceptibility to HIV-1 infection in Japanese and Indian subjects. rs11038628 is rare in Europeans but common in Asians and Africans. Functional analyses were performed by multiple-round replication and single-round assays, and indicated that the G249D substitution attenuated anti-HIV-1 activity of human TRIM5α. A slight attenuation of anti-HIV-2 activity was also observed in TRIM5α with 249D. The predicted secondary structure of the linker region suggested that the 249D substitution extended the α-helix in the neighboring coiled-coil domain, suggesting that human TRIM5α with 249D may lose the flexibility required for optimal recognition of retroviral capsid protein. We further analyzed the frequency of G249D in Japanese (93 HIV-1-infected subjects and 279 controls) and Indians (227 HIV-1-infected subjects and 280 controls). The frequency of 249D was significantly higher among HIV-1-infected Indian subjects than in ethnicity-matched control subjects [odds ratio (OR)=1.52, p=0.026]. A similar weak tendency was observed in Japanese subjects, but it was not statistically significant (OR=1.19, p=0.302). In conclusion, G249D, a common variant of human TRIM5α in Asians and Africans, is associated with increased susceptibility to HIV-1 infection.
Introduction
TRIM5α from rhesus monkeys restricts human immunodeficiency virus-1 (HIV-1) replication at the postentry,1 preintegration stage in the viral life cycle through rapid degradation of the HIV-1 core,2 whereas human TRIM5α restricts HIV-1 only weakly but potently restricts N-tropic murine leukemia virus.3,4 TRIM5α is a member of the tripartite motif-containing proteins and consists of RING, B-box 2, coiled-coil, and PRYSPRY (B30.2) domains. TRIM5α recognizes the multimerized capsid (CA) proteins of an incoming virus by its α-isoform-specific PRYSPRY domain. Studies of chimeric TRIM5αs have shown that the determinant of species-specific restriction against viral infection resides in the variable regions of the PRYSPRY domain.5–11
Infection by HIV-1 and progression to acquired immune deficiency syndrome (AIDS) vary among human individuals, and these phenomena are considered to be at least partially controlled by diversity in the human genome.12,13 Two common TRIM5α functional polymorphisms, H43Y and R136Q, have been studied with regard to the association with HIV-1 infection.14–21 Price et al. sequenced exon 2 of the TRIM5 gene in 1,032 women enrolled in a long-term monitored Pumwani sex worker cohort, and found that women with the R136Q polymorphism were less likely to seroconvert despite heavy exposure to HIV-1 through active sex work.15 Previous studies, including ours, showed the reduced antiviral activity of the H43Y substitution, but the associations with HIV-1 infection and disease progression were inconsistent among studies.14,16–20 Javanbakht et al. reported a paradoxical protective effect of TRIM5α with 43Y against HIV-1 transmission in African-Americans.14 Taken together, these findings indicate that anti-HIV-1 activity of human TRIM5α cannot protect humans from an HIV-1 pandemic, but may affect the rate of HIV-1 transmission.
In the present study, we investigated the association between a single nucleotide polymorphism (SNP) in the TRIM5α linker 2 region (rs11038628) between coiled-coil and PRYSPRY domains with susceptibility to HIV-1 infection. This SNP substituted aspartic acid (D) for glycine (G) at position 249. We show here that this SNP is associated with increased susceptibility to HIV-1 infection.
Materials and Methods
Cloning and expression of TRIM5α
The generation of recombinant Sendai viruses (SeVs) expressing human TRIM5α derived from MT4 cells, rhesus monkey TRIM5α derived from LLC-MK2 cells, and cynomolgus monkey TRIM5α lacking the PRYSPRY domain has been previously described.9,22 All these TRIM5αs carried a hemagglutinin (HA) tag (YPYDVPDYAA) at the C-terminus. The D-to-G substitution at the 249th position was introduced into MT4 TRIM5α by polymerase chain reaction (PCR) site-directed mutagenesis. The resultant PCR fragment was cloned into pSeV18+b(+) as a vector. Recombinant SeVs expressing human TRIM5α carrying G at position 249 were recovered according to the previously described method.23 The second passages in embryonated chicken eggs were used as stock virus for all experiments.
Western blotting analysis
MT4 cells (1×106) infected with recombinant SeVs expressing HA-tagged TRIM5α proteins were lysed in lysis buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P40, 0.5% sodium deoxycholate). TRIM5α proteins in the lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins in the gel were then electronically transferred onto a membrane (Immobilon; Millipore, Billerica, MA). Blots were blocked and probed with anti-HA high-affinity rat monoclonal antibody (Roche, Indianapolis, IN) overnight at 4°C. Blots were then incubated with peroxidase-conjugated anti-rat IgG (American Qualex, San Clemente, CA), and bound antibodies were visualized with a Chemilumi-One chemiluminescent kit (Nacalai Tesque, Kyoto, Japan).
Viral infection
MT4 cells (1×106) were infected with SeVs expressing MT4-derived human TRIM5α (249D), human TRIM5α (249G), rhesus monkey TRIM5α, or cynomolgus monkey TRIM5α lacking the PRYSPRY domain [CM-TRIM5α-SPRY(−)] at a multiplicity of infection (MOI) of 10 plaque-forming units (PFU) per cell and incubated at 37°C for 9 h. Aliquots of 1×105 cells were then superinfected with HIV-1 NL43 or HIV-2 GH123. Each superinfection used a titer of virus corresponding to 7 ng of p24 of NL43 or 20 ng of p25 of GH123. Experiments were performed with triplicate samples. The culture supernatants were collected periodically and the level of p24 or p25 was measured using a RETROtek antigen ELISA kit (ZeptoMetrix, Buffalo, NY). For the single-round infection assay, hamster TK-ts13 cells were infected with SeV expressing TRIM5α as described above, and superinfected with a vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped HIV-1 vector expressing green fluorescence protein (GFP) under the control of the cytomegalovirus (CMV) promoter. The original HIV-1 vector was based on the BH10 strain.24,25 To construct the lentivector possessing CA of NL4-3, we replaced the EcoRI–ApaI fragment corresponding to MA and CA of the pMDLg/p.RRE packaging vector with that of NL4-3.26 In case of HIV-2, we used a canine cell line Cf2Th and VSV-G pseudotyped HIV-2 vector expressing GFP under the control of the LTR promoter.27 Two days after infection, the cells were fixed with formaldehyde, and GFP-expressing cells were counted by a flow cytometer.
Human DNA subjects
The protocol for the present study was approved by the Ethics Review Board of the Medical Research Institute of Tokyo Medical and Dental University and that of the All India Institute of Medical Science. At setup of the cohort of HIV-1-infected Japanese subjects with hemophilia in 1995, all patients had been infected for longer than 10 years but were asymptomatic without any antiviral measures. Blood samples were collected from 93 well-characterized patients who were selected from the cohort after obtaining written informed consent.28,29 Control DNA samples were prepared from Epstein–Barr virus-transformed human B cell lines established from randomly selected healthy donors (n=279) and obtained from the Japan Health Sciences Foundation. DNA samples from HIV-1-infected individuals were prepared from the blood samples using a QuickGene DNA whole blood kit S (Fujifilm, Tokyo, Japan). In addition, blood DNA samples were obtained from 227 HIV-1-infected Indian subjects and 226 healthy Indian volunteers with informed consent in related hospitals with the All India Institute of Medical Sciences, New Delhi.
Identification and genotyping of nucleotide variations in TRIM5α exon 5
Primer sets were designed to amplify the genomic segments covering the entire TRIM5α exon 5 as follows: sense primer (5′-GATGCGGTCATGCTATGTTG-3′) and antisense primer (5′-CGAATGCTGATTTATGACCATA-3′). Genomic DNA was subjected to PCR amplification followed by sequencing using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Polymorphisms were identified using the Sequencher program (Gene Code Co., Ann Arbor, MI).
Statistical analysis
All statistical analyses in this study were performed using GraphPad InStat version 3.06 for Windows (GraphPad Software, San Diego, CA). Pairwise linkage disequilibrium (LD) (r2) was estimated using SNPAlyze version 6.0 standard (Dynacom Co., Ltd., Chiba, Japan).
Prediction of the peptide secondary structure
The Chou–Fasman methods were used to predict the secondary structure of TRIM5α using GENETYX-MAC version 15 software (Genetyx Corporation, Tokyo, Japan).
Results
Anti-HIV-1 activity of TRIM5α was attenuated by G249D substitution
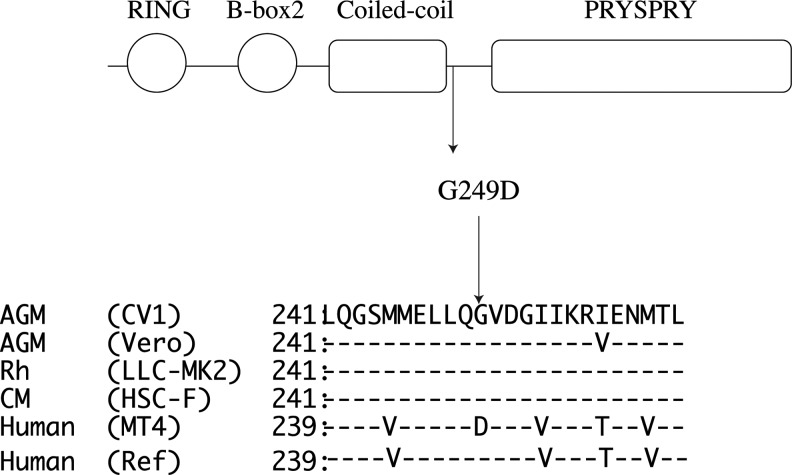
We previously cloned human TRIM5α from the CD4-positive T cell line MT4 and noted that there is a G-to-D amino acid substitution (G249D) in comparison with the reference sequence (NM_033034).9 This position is known as a polymorphic site in the human TRIM5 gene (rs11038628) located in the linker 2 region between the coiled-coil and PRYSPRY domains (Fig. 1). Initially, we speculated that this polymorphism would have no effect on antiviral activity due to its presence in the linker 2 region. Goldschmidt et al.18 reported that HeLa cells stably transduced with TRIM5α with 249D did not differ in susceptibility to HIV-1 infection. However, a tendency toward higher in vitro p24 production was observed at 7 days after infection in peripheral blood mononuclear cells from white subjects with the 249D allele, although the difference was not statistically significant mainly due to the limited number of subjects with the mutant allele.18 In addition, Old World monkey TRIM5α, including those of African green monkey, rhesus monkey, and cynomolgus monkey, also bears G at this position (Fig. 1). The HapMap project showed the 249D allele to be rare in whites (allele frequency: 0.053) but common in Japanese (allele frequency: 0.343) and African populations (allele frequency: 0.367). These findings prompted us to reevaluate the effects of this SNP on HIV-1 infection in Asians in which the frequency of G249D is higher than in whites.
FIG. 1.
Schematic presentation of TRIM5α structure. Circles and squares represent functional domains of TRIM5α. The position of the G249D polymorphism is shown by arrows. The amino acid sequences of African green monkey (AGM) TRIM5α from CV19 and Vero cells, rhesus monkey (Rh) TRIM5α from LLC-MK2,10 cynomolgus monkey (CM) TRIM5α from HSC-F,9 human TRIM5α from MT4 cells,9 and the reference sequence (NM_033034) were aligned. Dashes denote an identical amino acid to AGM TRIM5α from CV1.
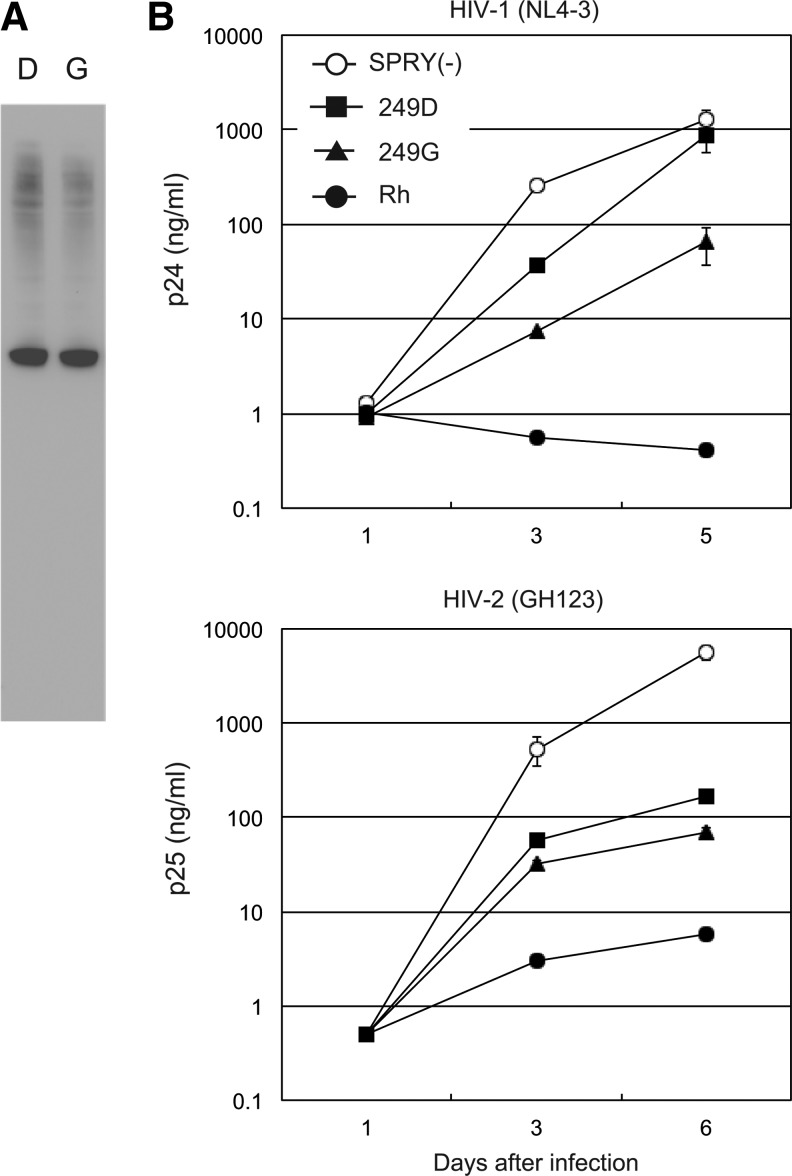
To investigate the functional significance of G249D on the anti-HIV activity of TRIM5α, we constructed SeV containing C-terminal HA-tagged human TRIM5α (249G) (Fig. 1) by site-directed mutagenesis on MT4 TRIM5α, which bears D at position 249. As shown in Fig. 2A, the expression level of TRIM5α (249G) was comparable to that of TRIM5α (249D) in recombinant SeV-infected MT4 cells.
FIG. 2.
(A) Lysates of MT4 cells infected with recombinant Sendai virus (SeV) expressing hemagglutinin (HA)-tagged human TRIM5α with 249D (lane D) and with 249G (lane G) were visualized by western blotting with an antibody against HA. Representative results of three independent experiments are shown. (B) MT4 cells were infected with SeV expressing TRIM5α lacking the PRYSPRY domain [SPRY(−); white circles], MT4-derived human TRIM5α (249D; black squares), human TRIM5α (249G; black triangles), or rhesus monkey TRIM5α (Rh; black circles). Nine hours after SeV infection, cells were inoculated with HIV-1 strain NL4-3 or HIV-2 strain GH123, and culture supernatants were periodically assayed for levels of p24 or p25, respectively. Data points are means for triplicate samples with SD. Three and six days after infection, statistically significant differences (p<0.05) of HIV-1 and HIV-2 growth were observed between human TRIM5α (249D) and human TRIM5α (249G) by unpaired t test. Representative data of at least three independent experiments are shown.
These TRIM5α constructs were tested for their ability to restrict the X4-tropic HIV-1 strain NL4-3 and HIV-2 strain GH123. MT4 cells infected with recombinant SeV expressing each of the TRIM5α constructs were superinfected with HIV-1 NL4-3 or HIV-2 GH123. We used SeV expressing cynomolgus monkey TRIM5α lacking the PRYSPRY domain as a negative control for functional TRIM5α, as overexpression of TRIM5α lacking the PRYSPRY domain was shown to exert a dominant negative effect on endogenous human TRIM5α.30 As shown in Fig. 2B, MT4-derived human TRIM5α (249D) showed only weak anti-HIV-1 activity, as we demonstrated previously.21 On the other hand, human TRIM5α (249G) showed stronger restriction activity to HIV-1 NL4-3 than human TRIM5α (249D). In the case of HIV-2, both human TRIM5α with 249G and 249D exhibited apparent anti-HIV-2 activity. The human TRIM5α (249G) showed stronger restriction activity to HIV-2 GH123 than human TRIM5α (249D), although the difference was very small (Fig. 2B, lower panel). These results indicated that the G249D variant weakened the anti-HIV-1 and anti-HIV-2 activities of human TRIM5α.
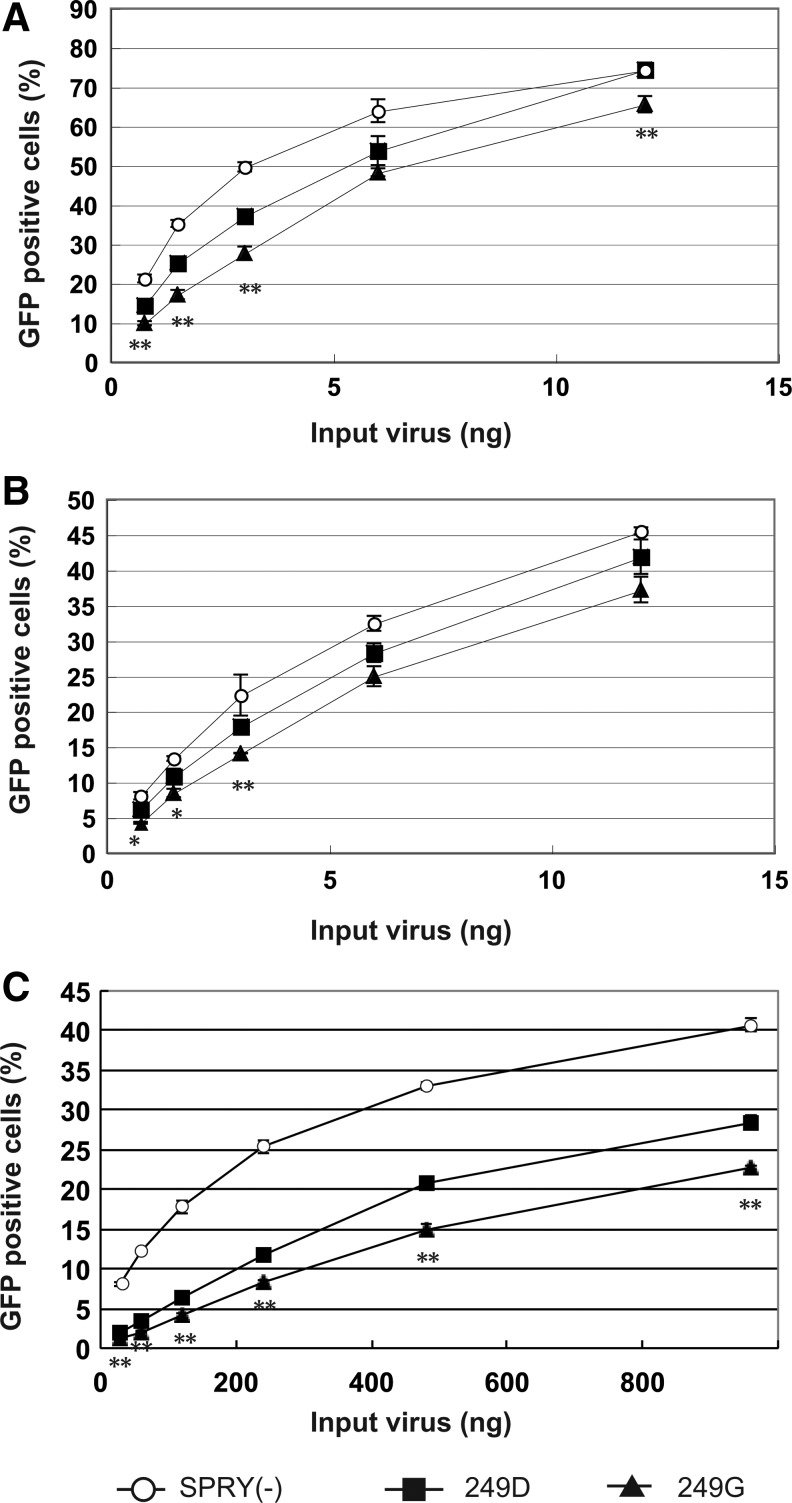
TRIM5α is known to restrict viral infection at the early steps of HIV replication. To evaluate the anti-HIV-1 activity of human TRIM5α at the early stages, we performed the single-round infection assay using a GFP expression vector (Fig. 3). The hamster cell line TK-ts13, which lacks endogenous TRIM5α expression, was infected with recombinant SeV expressing human TRIM5α. We superinfected cells with VSV-G pseudotyped lentivector expressing GFP under the control of the CMV promoter. We used HIV-1 vectors bearing CA derived from BH10 (Fig. 3A) and NL4-3 (Fig. 3B). Both HIV-1 GFP vectors were suppressed to a greater degree by human TRIM5α (249G) than by MT4-derived human TRIM5α (249D). A similar result was obtained when we used the HIV-2 GFP vector (Fig. 3C). Taken together, these observations indicated that the G249D polymorphism affected the anti-HIV-1 and anti-HIV-2 activities of human TRIM5α.
FIG. 3.
TK-ts13 cells infected with SeVs expressing TRIM5α lacking the PRYSPRY domain [SPRY(−); white circles], MT4-derived human TRIM5α (249D; black squares), or human TRIM5α (249G; black triangles) were exposed to green fluorescence protein (GFP)-expressing HIV-1 vector based on BH10 (A) or NL4-3 (B). (C) Cf2Th cells infected with SeVs were exposed to an HIV-2 vector based on ROD. GFP-positive cells were counted by a flow cytometer. Data points are means for triplicate samples with SD. *,** The statistically significant differences, p<0.05 and p<0.001, respectively, in unpaired t test between human TRIM5α (249D) and human TRIM5α (249G). Representative results of two independent experiments are shown.
Associations of TRIM5α G249D polymorphism with susceptibility to HIV-1 infection
We sequenced TRIM5α exon 5 and found G249D in the populations tested. The associations of G249D polymorphism with susceptibility to HIV-1 infection are summarized in Table 1. The frequency of 249D was significantly higher in the HIV-1-infected Indian subjects than in the ethnicity-matched controls [odds ratio (OR)=1.52, p=0.026]. A similar tendency was also observed in the Japanese population, but did not reach statistical significance (OR=1.19, p=0.302).
Table 1.
Association of rs10038628 (G249D) with Susceptibility to HIV-1 Infection in Japanese and Indian Populations
| |
Japanese |
Indian |
||||||
|---|---|---|---|---|---|---|---|---|
| HIV-1-infected (n=93) | Control (n=279) | Odds ratio (95% CI) | p-value | HIV-1-infected (n=227) | Control (n=280) | Odds ratio (95% CI) | p-value | |
| rs10038628 | ||||||||
| GG | 28 (30%) | 98 (35%) | 0.80 (0.48–1.32) | 0.376 | 161 (71%) | 226 (81%) | 0.58 (0.39–0.88) | 0.010 |
| DG | 47 (51%) | 137 (49%) | 63 (28%) | 49 (17%) | ||||
| DD | 18 (19%) | 44 (16%) | 1.28 (0.70–2.35) | 0.422 | 3 (1%) | 5 (2%) | 0.74 (0.17–3.12) | 0.736a |
| Allele D | 83 (45%) | 225 (40%) | 1.19 (0.85–1.67) | 0.302 | 69 (15%) | 59 (11%) | 1.52 (1.05–2.21) | 0.026 |
Fisher's exact test.
Previously, we sequenced TRIM5α exons 2 of the same subjects as above and reported the association of H43Y with susceptibility to HIV-1 infection.21 The levels of LD indicated that G249D in exon 5 and H43Y in exon 2 were not in tight linkage disequilibrium in either Japanese (r2=0.18, n=188) or Indian (r2=0.02, n=96) populations.
Discussion
The G249D polymorphism in TRIM5α is common in Asian and African populations. It was initially speculated that there was no functional effect of this SNP, as it is located outside of any functional domains of human TRIM5α. Contrary to our expectation, however, we observed attenuation of anti-HIV-1 and anti-HIV-2 activity of the G-for-D substitution with both multiround replication and single-round infection assays. Furthermore, we investigated two ethnic populations, Japanese and Indian, for the G249D polymorphism and found the association of the TRIM5α 249D allele with enhanced susceptibility to HIV-1 infection.
Amino acid position 249 of human TRIM5α lies within the linker region for which no three-dimensional structural data have yet been reported. Therefore, we performed secondary structure prediction by the Chou–Fasman method31 to examine the possible effect of this SNP on the protein structure. The G-to-D substitution increased the probability of α-helix formation and resulted in the extension of the α-helix from the coiled-coil region into the linker 2 region. Similar results were obtained by the PREDETOR in http://mobyle.pasteur.fr (data not shown). This suggested that TRIM5α with 249G would be more flexible than TRIM5α with 249D.
Human TRIM5α was obviously not effective in protecting against HIV-1 infection compared with the strong Old World monkey TRIM5α, as only humans are susceptible and Old World monkeys are resistant to HIV-1 infection. With experimental overexpression of human TRIM5α, the anti-HIV-1 activity of human TRIM5α was variable among previous reports.1,5,9,14,16,20,21 Our previous data showed the weakest anti-HIV-1 activity of human TRIM5α,9,20,21 even though we used the SeV system, which allowed high expression levels of inserted genes. As described in the present study, the 249D substitution would explain why our human TRIM5α derived from MT4 showed little potency against HIV-1. We examined the G249D SNP in commonly used human cell lines, CEM, HeLa, Jurkat, and 293T, and found that these were all homozygous for 249G, but MT4 was homozygous for 249D. This is not surprising because the allele frequency of 249D is high in Japan but quite rare in whites and MT4 cells were established from Japanese donor blood.32 On the other hand, MT4 is highly susceptible to HIV-1 infection,33 which is in good agreement with the present data.
Previously, Goldschmidt et al. failed to observe the attenuation of antiviral activity by the 249D mutation.18 One possible reason for the discrepancy between their results and ours is the difference in expression system used. Goldschmidt et al. used HeLa cells stably transduced with TRIM5α with various mutations.18 Transduced cell lines sometimes develop unexpected phenotypic changes during the cloning procedure. In contrast, we used the SeV system, and the conditions of cells infected with different recombinant viruses were always comparable, especially among those expressing full-length TRIM5α. It should be noted that Goldschmidt et al. also reported a tendency toward higher in vitro p24 production 7 days after infection in peripheral blood mononuclear cells from individuals with the 249D allele, which is consistent with our present results.18
We clearly showed that the 249D allele was associated with increased susceptibility to HIV-1 infection in the Indian population. However, although a similar tendency was observed in the Japanese population, the association was not significant. The precise reason why the effect of G249D was unclear in the Japanese population is not yet clear. It should be noted that our Japanese patients were infected through contaminated blood products in the early 1980s. On the other hand, the Indian patients were infected through heterosexual contact after the HIV-1 pandemic in Asia after 1990. It is possible that the difference in route of HIV-1 transmission may be responsible for this difference between Japanese and Indian patients. Further studies in well-characterized cohorts are necessary to confirm our findings regarding HIV-1 transmission and the possible effects of this SNP on AIDS progression.
Acknowledgments
We thank Ms. Setsuko Bando and Ms. Noriko Teramoto for their assistance, and Dr. Hirotaka Ode for critical suggestion. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology, and the Ministry of Health, Labour, and Welfare, Japan.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Stremlau M. Owens CM. Perron MJ. Kiessling M. Autissier P. Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Stremlau M. Perron M. Lee M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perron MJ. Stremlau M. Song B. Ulm W. Mulligan RC. Sodroski J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yap MW. Nisole S. Lynch C. Stoye JP. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Caballero D. Hatziioannou T. Yang A. Cowan S. Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawyer SL. Wu LI. Emerman M. Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stremlau M. Perron M. Welikala S. Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yap MW. Nisole S. Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama EE. Miyoshi H. Nagai Y. Shioda T. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J Virol. 2005;79:8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kono K. Song H. Shingai Y. Shioda T. Nakayama EE. Comparison of anti-viral activity of rhesus monkey and cynomolgus monkey TRIM5alphas against human immunodeficiency virus type 2 infection. Virology. 2008;373:447–456. doi: 10.1016/j.virol.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 11.Perron MJ. Stremlau M. Sodroski J. Two surface-exposed elements of the B30.2/SPRY domain as potency determinants of N-tropic murine leukemia virus restriction by human TRIM5alpha. J Virol. 2006;80:5631–5636. doi: 10.1128/JVI.00219-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Brien SJ. Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 13.Shioda T. Nakayama EE. Human genetic polymorphisms affecting HIV-1 diseases. Int J Hematol. 2006;84:12–17. doi: 10.1532/IJH97.06100. [DOI] [PubMed] [Google Scholar]
- 14.Javanbakht H. An P. Gold B, et al. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 15.Price H. Lacap P. Tuff J, et al. A TRIM5alpha exon 2 polymorphism is associated with protection from HIV-1 infection in the Pumwani sex worker cohort. AIDS. 2010;24:1813–1821. doi: 10.1097/QAD.0b013e32833b5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawyer SL. Wu LI. Akey JM. Emerman M. Malik HS. High-frequency persistence of an impaired allele of the retroviral defense gene TRIM5alpha in humans. Curr Biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 17.Speelmon EC. Livingston-Rosanoff D. Li SS, et al. Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. J Virol. 2006;80:2463–2471. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldschmidt V. Bleiber G. May M. Martinez R. Ortiz M. Telenti A. Role of common human TRIM5alpha variants in HIV-1 disease progression. Retrovirology. 2006;3:54. doi: 10.1186/1742-4690-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Manen D. Rits MA. Beugeling C. van Dort K. Schuitemaker H. Kootstra NA. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 2008;4:e18. doi: 10.1371/journal.ppat.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama EE. Carpentier W. Costagliola D, et al. Wild type and H43Y variant of human TRIM5alpha show similar anti-human immunodeficiency virus type 1 activity both in vivo and in vitro. Immunogenetics. 2007;59:511–515. doi: 10.1007/s00251-007-0217-7. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima T. Nakayama EE. Kaur G, et al. Impact of novel TRIM5alpha variants, Gly110Arg and G176del, on the anti-HIV-1 activity and the susceptibility to HIV-1 infection. AIDS. 2009;23:2091–2100. doi: 10.1097/QAD.0b013e328331567a. [DOI] [PubMed] [Google Scholar]
- 22.Song H. Nakayama EE. Yokoyama M. Sato H. Levy JA. Shioda T. A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J Virol. 2007;81:7280–7285. doi: 10.1128/JVI.00406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama EE. Tanaka Y. Nagai Y. Iwamoto A. Shioda T. A CCR2-V64I polymorphism affects stability of CCR2A isoform. AIDS. 2004;18:729–738. doi: 10.1097/00002030-200403260-00003. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi H. Takahashi M. Gage FH. Verma IM. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi H. Blomer U. Takahashi M. Gage FH. Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroishi A. Bozek K. Shioda T. Nakayama EE. A single amino acid substitution of the human immunodeficiency virus type 1 capsid protein affects viral sensitivity to TRIM5 alpha. Retrovirology. 2010;7:58. doi: 10.1186/1742-4690-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyamoto T. Nakayama EE. Yokoyama M, et al. The carboxyl-terminus of human immunodeficiency virus type 2 circulating recombinant form 01_AB capsid protein affects sensitivity to human TRIM5alpha. PLoS One. 2012;7:e47757. doi: 10.1371/journal.pone.0047757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munkanta M. Terunuma H. Takahashi M, et al. HLA-B polymorphism in Japanese HIV-1-infected long-term surviving hemophiliacs. Viral Immunol. 2005;18:500–505. doi: 10.1089/vim.2005.18.500. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T. Ohtani H. Naruse T, et al. Copy number variations of CCL3L1 and long-term prognosis of HIV-1 infection in asymptomatic HIV-infected Japanese with hemophilia. Immunogenetics. 2007;59:793–798. doi: 10.1007/s00251-007-0252-4. [DOI] [PubMed] [Google Scholar]
- 30.Maegawa H. Nakayama EE. Kuroishi A. Shioda T. Silencing of tripartite motif protein (TRIM) 5alpha mediated anti-HIV-1 activity by truncated mutant of TRIM5alpha. J Virol Methods. 2008;151:249–256. doi: 10.1016/j.jviromet.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Chou PY. Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–48. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 32.Akagi T. Ohtsuki Y. Shiraishi Y. Miyoshi I. Transformation of human fetal thymus and spleen lymphocytes by human T-cell leukemia virus type I. Acta Med Okayama. 1985;39:155–159. doi: 10.18926/AMO/31513. [DOI] [PubMed] [Google Scholar]
- 33.Harada S. Koyanagi Y. Yamamoto N. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]