Abstract
Here we show potent inhibition of HIV-1 replication in a human T cell line and primary human CD4+ cells by expressing a single antiviral protein. Nullbasic is a mutant form of the HIV-1 Tat protein that was previously shown to strongly inhibit HIV-1 replication in nonhematopoietic cell lines by targeting three steps of HIV-1 replication: reverse transcription, transport of viral mRNA, and trans-activation of HIV-1 gene expression. Here we investigated gene delivery of Nullbasic, using lentiviral and retroviral vectors. Although Nullbasic could be delivered by lentiviral vectors to target cells, transduction efficiencies were sharply reduced primarily because of negative effects on reverse transcription mediated by Nullbasic. However, Nullbasic did not inhibit transduction of HEK293T cells by a murine leukemia virus (MLV)-based retroviral vector. Therefore, MLV-based virus-like particles were used to transduce and express Nullbasic-EGFP or EGFP in Jurkat cells, a human leukemia T cell line, and Nullbasic-ZsGreen1 or ZsGreen1 in primary human CD4+ cells. HIV-1 replication kinetics were similar in parental Jurkat and Jurkat-EGFP cells, but were strongly attenuated in Jurkat-Nullbasic-EGFP cells. Similarly, virus replication in primary CD4+ cells expressing a Nullbasic-ZsGreen1 fusion protein was inhibited by approximately 8- to 10-fold. These experiments demonstrate the potential of Nullbasic, which has unique inhibitory activity, as an antiviral agent against HIV-1 infection.
Apolloni and colleagues show that retroviral delivery of a mutant Tat protein (Nullbasic) mediates potent inhibition of HIV-1 replication in a human T cell line and in primary CD4+ cells.
Introduction
Since the beginning of the epidemic, HIV-1 has infected more than 60 million people and killed more than 25 million (UNAIDS, 2009). The primary therapy used to treat HIV-1 infection and AIDS is combinatorial antiviral chemotherapy, using drugs that act by inhibiting at least two steps of virus replication. Although this therapy has been incredibly effective at limiting virus replication and disease progression, challenges remain when treating chronic infections, including the inability to eliminate virus infection. In the absence of a potent vaccine to prevent HIV-1 infection, strategies to eliminate infection are under investigation. Alternatively, the cure of HIV-1 infection in the “Berlin patient” has raised interest in alternative genetic therapies capable of introducing an HIV-1-resistant immune system to people living with HIV-1 and AIDS (Hutter et al., 2009).
Among the approaches tested to render human immune cells resistant to HIV-1, protein-based antiviral inhibitors that act by targeting viral proteins as dominant (or trans-dominant) negative proteins or engineered innate cellular inhibitors have been described. A dominant negative protein is typically an altered form of a protein that can inhibit the normal function of its wild-type counterpart. Dominant negatives for many viral proteins have been described including Tat (Green et al., 1989; Pearson et al., 1990; Modesti et al., 1991), Rev (Bevec et al., 1992; Malim et al., 1992), Vif (Walker et al., 2010), Nef (Fackler et al., 2001), and Gag (Trono et al., 1989; Furuta et al., 1997; Lee et al., 2009; Checkley et al., 2010). For example, engineered Tat proteins with altered basic domains possess trans-dominant negative phenotypes against wild-type Tat function in transcription (Pearson et al., 1990), and a trans-dominant-negative Rev mutant called RevM10 has been described that was shown to inhibit wild-type Rev function (Malim et al., 1991). The RevM10 mutant protein is dominant negative because it retains the ability to bind the HIV-1 Rev response element (RRE) but is unable to promote CRM1 (chromosome region maintenance 1)-mediated export of the viral mRNA from the nucleus, thereby inhibiting HIV-1 replication (Malim et al., 1991; Stauber et al., 1998). RevM10 mutant, the first trans-dominant negative protein to undergo a pilot clinical trial in HIV-1-infected patients (Woffendin et al., 1996; Ranga et al., 1998), was beneficial to cell survival in the context of HIV infection (Podsakoff et al., 2005). A human engineered TRIM5 (tripartite motif 5)–cyclophilin protein introduced into primary CD4+ T cells and macrophages provided extraordinary protection from HIV-1 infection in vitro and in immune-compromised mice (Neagu et al., 2009). TRIM5–cyclophilin inhibits HIV-1 infection in the newly infected cell by binding to capsid protein (CA), leading to downregulated viral reverse transcription. In summary, various antiviral proteins have been described, each of which targets a single step of the virus life cycle, often by targeting a single viral or cellular protein.
We described a mutant of the two-exon HIV-1 Tat protein, termed Nullbasic, that inhibited multiple steps of the HIV-1 replication cycle (Meredith et al., 2009), a unique feature compared with other antiviral proteins. Nullbasic was created by replacing the entire arginine-rich basic domain of wild-type Tat with glycine/alanine residues. Like similarly mutated one-exon Tat mutants, Nullbasic exhibited dominant negative effects on Tat-dependent HIV-1 gene expression. However, unlike previously reported mutants (Green et al., 1989; Pearson et al., 1990; Modesti et al., 1991; Ulich et al., 1996), Nullbasic inhibited reverse transcription and also effectively suppressed the steady state levels of unspliced and singly spliced viral mRNA, an activity caused by inhibition of HIV-1 Rev activity (Meredith et al., 2009). HeLa CD4+ cells constitutively expressing Nullbasic were strongly protected from a spreading infection by HIV-1 (Meredith et al., 2009). This study investigated whether Nullbasic could be used to effectively protect human T cells from HIV-1 infection. We show that Nullbasic can provide strong and sustained protection from HIV-1 infection.
Materials and Methods
Cells and HIV-1 viruses
HEK293T, Phoenix-A (also known as ΦNX-A) (Swift et al., 2001), and PG13-myc-αerb-CD28ζ (von Kalle et al., 1994; Teng et al., 2004) cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% (v/v) fetal bovine serum (FBS) (referred to as DF10 medium). HeLa and Jurkat cells were grown as described previously but using RPMI 1640 medium supplemented as described previously (RF10 medium). All cells were grown at 37°C in humidified incubators, using 5% CO2.
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coat supplied by the Australian Red Cross Blood Service. CD4-positive cell selection was performed with human CD4 MicroBeads and magnetic separation columns according to the instructions of the manufacturer (Miltenyi Biotec, Bergisch Gladbach, Germany). Isolated cells were determined to be >98% CD4+ by flow cytometric analysis. CD4+ cells were incubated in RPMI 1640 containing 20% FBS (RF20) and human interleukin-2 (IL-2, 20 units/ml; Roche, Indianapolis, IN) at 1 million cells per milliliter in flasks precoated with purified anti-human CD3 (clone OKT3) and purified anti-human CD28 (clone CD28.2) antibodies (BioLegend, San Diego, CA). After 3 days, cells were transferred to an uncoated flask and grown in RF20 and IL-2 as previously described.
HIV-1pNL4-3 and HIV-189.6 viral stocks were made by transfection of proviral DNA plasmids in HEK293T cells (Adachi et al., 1986; Doranz et al., 1996). All viral stocks were stored in small aliquots at −80°C.
Oligonucleotides and plasmids
All oligonucleotide primers used are shown in Supplementary Table S1 (supplementary data are available online at www.liebertonline.com/hum). pLOX-CW-Nullbasic-EGFP, pLOX-CW-EGFP, pCMVΔR8.91, and pCMV-VSV-G plasmids used for production of lentiviral VLPs (virus-like particles) have been described elsewhere (Zufferey et al., 1997; Salmon et al., 2000). The pCDN3.1-Nullbasic-FLAG plasmid was previously described (Meredith et al., 2009). The plasmid pGCsamEN has been described (Treisman et al., 1995). The pGCsamEN-Nullbasic-EGFP construct was made by PCR of Nullbasic-EGFP from pcDNA3.1-Nullbasic-EGFP (Meredith et al., 2009), using primers P1 and P2, which introduce unique NotI and SnaBI restriction sites compatible with the multiple cloning site of pGCsamEN (Supplementary Table S1). Similarly, pGCsamEN-EGFP was made by PCR of an EGFP cassette from pCDN3.1-EGFP, using primers P2 and P3, and pGCsamEN-ZsGreen1 was made by PCR of ZsGreen1 from pLVX-IRES-ZsGreen1 (Clontech, Mountain View, CA), using primers P4 and P5. A precursor plasmid, pcDNA3.1-Nullbasic-ZsGreen1 was constructed by PCR of pLVX-ZsGreen, using primers P6 and P7, which introduce unique XhoI and XbaI restriction to replace EGFP in pCDN3.1-NB-EGFP with ZsGreen1. Next, primers P1 and P5 were used to generate the Nullbasic-ZsGreen1 DNA, which was then ligated into the NotI/SnaBI sites of pGCsamEN to make pGCsamEN-Nullbasic-ZsGreen1.
VLP production
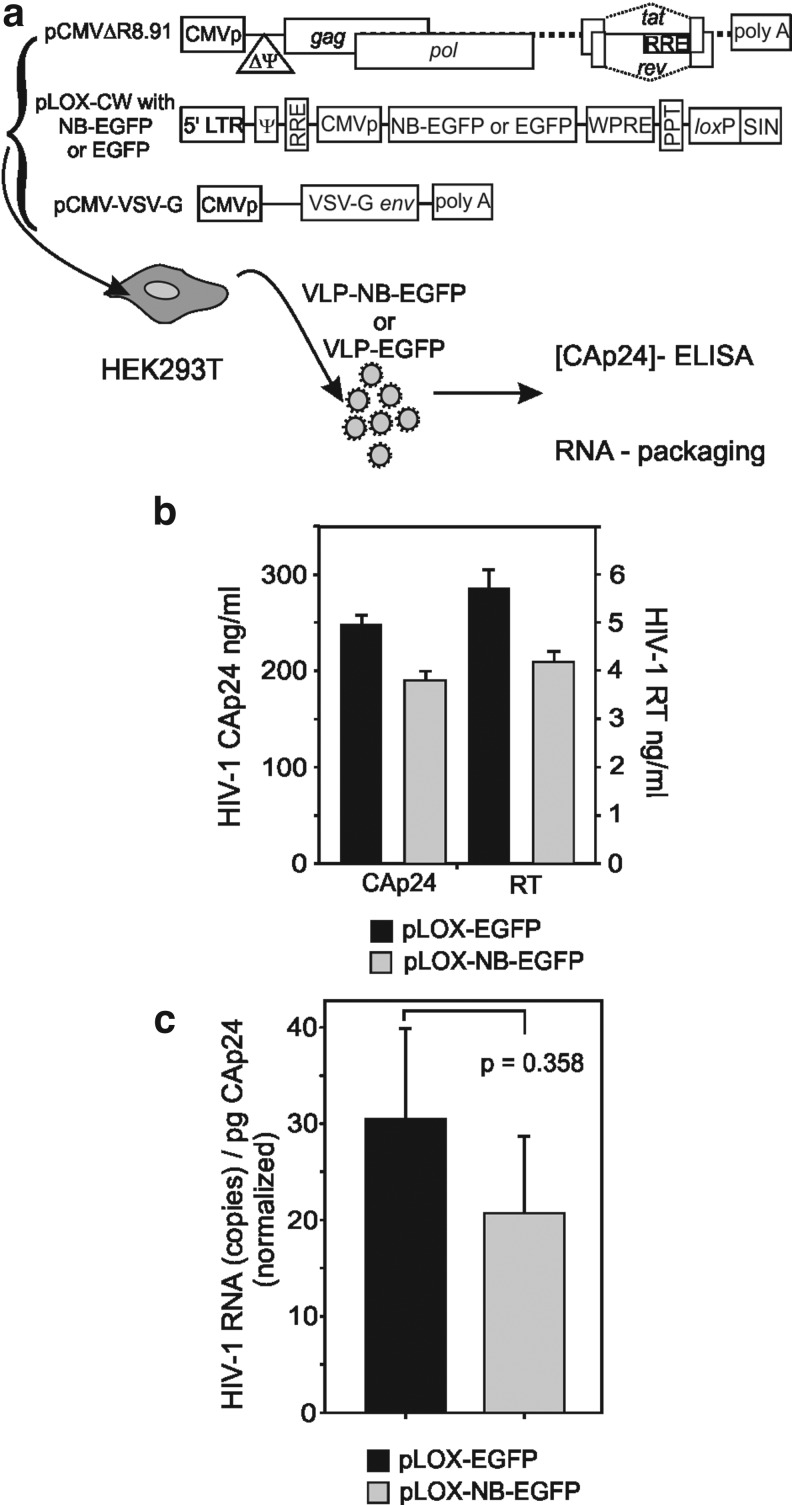
HEK293T cells grown in 10-cm dishes were cotransfected with 15 μg of pCMVΔR8.91 plasmid, 5 μg of pCMV-VSV-G, and 5 μg of pLOX-CW-Nullbasic-EGFP or pLOX-CW-EGFP, using a calcium phosphate method as previously described (Fish and Kruithof, 2006). After 16 hr, the transfected cells were incubated in 10 ml of DF10 medium and 48 hr later lentiviral VLP stocks were harvested and stored in aliquots at −80°C. The HIV-1 CAp24 concentration in lentiviral VLP supernatants was measured with an HIV-1 p24 ELISA kit (Zeptometrix, Buffalo, NY).
PG13-myc-αerb-CD28ζ cells produce retroviral VLPs as previously described (von Kalle et al., 1994; Teng et al., 2004). This cell line was transduced secondarily with CAp24 (100 or 500 ng) containing lentiviral VLPs conveying EGFP or Nullbasic-EGFP, respectively, in hexadimethrine bromide-coated 24-well plates. Transduced cells expressing high levels of EGFP were isolated with a FACSAria (BD Biosciences, San Jose, CA), and the purified cells were expanded. Flow cytometry was used to monitor EGFP or Nullbasic EGFP expression. To make ΦNX-A VLPs, cells were transfected with 10 μg of pGCsamEN-EGFP, pGCsamEN-Nullbasic-EGFP, GCsamEN-ZsGreen1, or GCsamEN-Nallbasic-ZsGreen1. Where indicated, 5 μg of pCDNA3.1-Nullbasic-FLAG or the parental plasmid was cotransfected. VLPs were collected after 48 hr and stored in aliquots at −80°C. VLP titers were determined according to a previously described method (Fassati, 2009), and reverse transcriptase (RT) levels for all VLP and HIV-1 stocks were determined by RT colorimetric assay (Roche) according to the manufacturer's instructions.
Lentiviral VLP RNA packaging
Lentiviral VLP stock containing 1 μg of CAp24 was treated with DNase I (Worthington Biochemicals, Lakewood, NUJ) and pelleted through 1×phosphate-buffered saline (PBS) containing 20% (v/v) sucrose at 100,000×g for 2 hr, using an SW 41 Ti rotor (Beckman Coulter, Brea, CA). The VLP pellet was washed with 1×PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4; pH 7.4) and resuspended in 500 μl of 1×PBS overnight at 0°C. The concentrated VLP stock was used for RNA packaging and endogenous reverse transcription (ERT) assays (described below). To measure VLP RNA, VLPs containing 200 ng of CAp24 were solubilized with TRIzol (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions and a synthetic kanamycin RNA (10 ng; Promega, Madison, WI) was added to the mixture before isolation of RNA. RT-PCR was performed with SuperScript III RT and random hexamer oligonucleotides. The synthesized DNA was analyzed by qPCR in triplicate, using primers P8 and P9. A serially diluted pLOX-CW plasmid was used to determine VLP cDNA levels, using primers P8 and P9. Primers P10 and P11 were used to determine kanamycin cassette RNA recovery and RT-PCR by the comparative CT method (Schmittgen and Livak, 2008).
ERT assays
The ERT assay is described in detail elsewhere (Warrilow et al., 2008). ERT reactions were performed in triplicate with concentrated VLP stocks that were normalized to RT. A no-dNTP ERT reaction was always included. Purified viral cDNA levels were measured by real-time qPCR using primers P8 and P9.
Western blot analysis
Expression of EGFP and Nullbasic-EGFP was confirmed by Western blot using lysates from cells prepared and electrophoresed as previously described (Meredith et al., 2009). Lysates were equalized for total protein concentration. Nullbasic was detected with a rabbit anti-Tat antibody (Diatheva, Fano, Italy). EGFP was detected with a rabbit anti-GFP antibody (Cell Signaling Technology, Danvers, MA). Endogenous β-tubulin was detected with a mouse anti-β-tubulin antibody (Sigma-Aldrich, St. Louis, MO). Primary antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit antibodies (Life Technologies).
Transduction assays and cell sorting
Lentiviral and retroviral VLP stocks (1 ml) normalized to RT activity were added to hexadimethrine bromide (Sigma-Aldrich)-coated 24-well plates and centrifuged at 1000×g. For ΦNX-A VLP transductions, VLP stocks (2 ml) normalized to RT activity were applied to plates coated with RetroNectin (TaKaRa Bio, Otsu, Japan) according to the manufacturer's instructions. Cells were added (1.5×105 HEK293T, HeLa, or Jurkat cells; 3.5×105 activated primary CD4+ cells) in 2×medium as indicated. ZsGreen1 and EGFP expression was measured 48 hr posttransduction with a FACSCalibur flow cytometer (BD Biosciences). Expression of myc-αerb-CD28ζ was determined by staining with a mouse anti-Myc–Alexa Fluor 488 monoclonal antibody (Life Technologies) and flow cytometry. Where indicated, transduced cells were collected after 72 hr with a FACSAria (BD Biosciences). EGFP and ZsGreen1 expression in cells was monitored by flow cytometry with a FACSCalibur (BD Biosciences). Cells collected by fluorescence-activated cell sorting (FACS) were incubated for 48 hr before infection with HIV-1.
HIV-1 infections
All infections were performed in triplicate, using 4×105 Jurkat cells or 1×106 primary CD4+ cells. Cells were infected with HIV-1pNL4-3 or HIV-189.6 containing 20 ng of CAp24 in 24-well plates in a volume of 500 μl of Opti-MEM I with 5% FBS at 37°C for 2 hr. After infection, the medium was replaced with RF10 or RF20 with IL-2, respectively. Culture supernatant and cells were collected as indicated and assayed with an HIV-1 CAp24 ELISA kit (Zeptometrix); EGFP and ZsGreen1 expression was measured by flow cytometry.
MTS assay
The CellTiter 96 AQueous One Solution cell proliferation assay was used following the manufacturer's instructions. CellTiter 96 AQueous One Solution reagent was directly to a sample of culture supernatant incubating for 2 hr and then recording absorbance at 490 nm with a 96-well plate reader.
Transduction and reverse-transcription efficiencies of NB-GFP and GFP VLPs by PCR and RT-PCR
VLPs equivalent to 0.6 μg of CAp24 were treated with DNase I at 37°C for 1 hr before addition to a 60-mm dish of 239T cells. Where indicated, VLPs were inactivated by heating at 70°C for 10 min. If the HIV RT inhibitor nevirapine was used, cells were incubated with a final concentration of 200 μM nevirapine for 2 hr at 37°C before transduction. Transduction was carried out at 4°C for 1 hr and then at 37°C for 3 hr. The cells were washed three times with PBS containing 0.5 mM EDTA and lysed in 0.5 ml of hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, and 1 mM dithiothreitol [DTT]) for DNA PCR, or lysed in TRIzol reagent for RNA extraction. To isolate DNA from cytoplasmic lysate, the washed cells were incubated in hypotonic buffer for 10 min at 4°C and then lysed with a 2-ml homogenizer (20 strokes). The lysate was centrifuged at 7500×g for 20 min at 4°C to remove nuclei and the supernatant was assayed by qPCR using primers targeting the EGFP region. Nucleic acid concentrations of lysates were normalized by PCR amplification of the mitochondrial DNA-encoded cytochrome c oxidase subunit II gene, using oligonucleotide primers P12 and P13. To extract RNA, the washed cell pellet was lysed in 0.5 ml of TRIzol reagent and the RNA was obtained by passage through a Direct-zol RNA MiniPrep column (ZYMO Research, Irvine, CA). Kanamycin RNA (100 ng) was added to each lysate to monitor the purification process and analysis. In-column DNase I treatment was carried out before a final wash to remove DNA contaminations. The RNA was eluted in 35 μl of water and used to make cDNA, using random primers, followed by qPCR. A cDNA reaction lacking reverse transcriptase was always included to monitor for DNA contamination.
Results
Nullbasic downregulates infectivity of lentiviral vectors by inhibiting reverse transcription
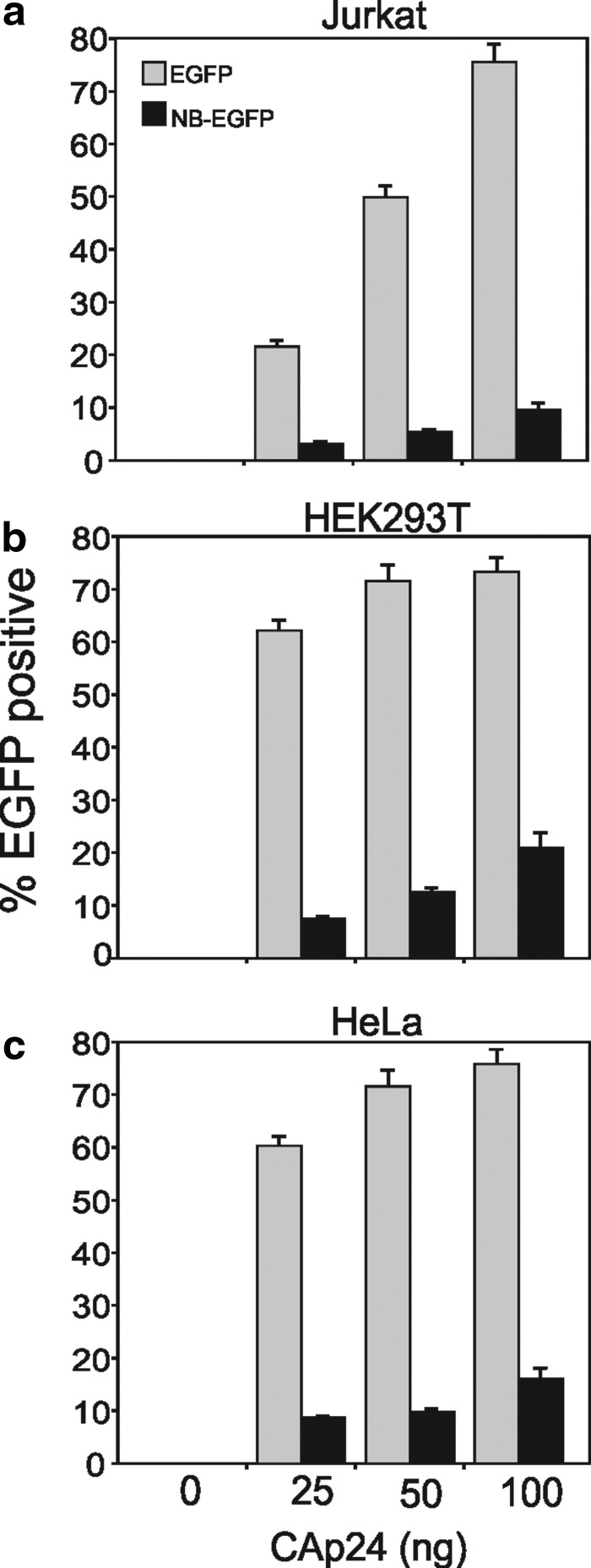
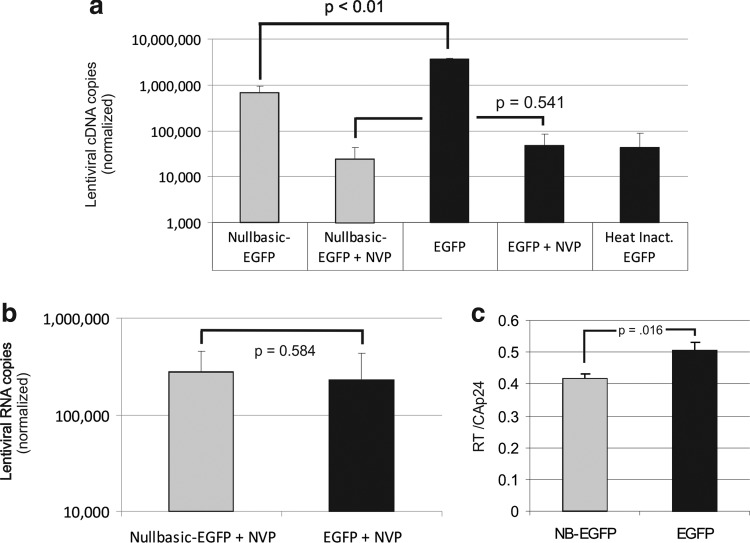
Lentiviral vectors are commonly used to deliver genes to human cells. As many lentiviral vectors are derived from HIV-1, we sought to determine whether Nullbasic negatively affected lentiviral virus-like particle (VLP) production or infectivity as with HIV-1 (Meredith et al., 2009). A Nullbasic-EGFP or EGFP gene cassette was inserted in pLOX-CW and cotransfected with pCMVΔ8.91 and pCMV-VSV-G in HEK293T cells to make infectious lentiviral VLPs (Fig. 1a). Supernatant were recovered after 48 hr and assayed for capsid protein, reverse transcriptase (RT), and packaged mRNA made by pLOX-CW vectors (Fig. 1b and c). We observed no significant difference in the level of VLP production, indicating that Nullbasic-EGFP expression did not interfere with viral protein expression or particle assembly. An equivalent amount of Nullbasic-EGFP or EGFP viral supernatant, normalized to total capsid protein, was used to infect three human cell lines: Jurkat, HEK293T, and HeLa cells (Fig. 2). Sharp decreases in the levels of EGFP-positive cells were observed for all cell lines transduced with Nullbasic-EGFP compared with EGFP VLPs irrespective of the target cell line. This outcome was consistent with our observation that Nullbasic reduced HIV-1 infectivity when viral particles were made by cells expressing Nullbasic or Nullbasic-EGFP, primarily by having a pronounced negative effect on HIV-1 reverse transcription (Meredith et al., 2009). To determine whether Nullbasic had a similar effect on lentiviral particles, endogenous reverse transcription (ERT) reactions were performed with equivalent amounts of viral supernatant normalized to RT levels as measured in a standard homopolymeric primer:template RT assay. As shown in Fig. 3a, pLOX-NB-EGFP VLPs produced approximately 4-fold less negative-strand strong stop DNA than pLOX-EGFP VLPs, suggesting that Nullbasic decreased the infectivity of VLPs largely by inhibiting reverse transcription. Considerably less viral DNA was detected if nucleotides were excluded from the ERT reactions, indicating DNA synthesis was de novo. To confirm that the reduced amount of ERT DNA was not somehow influenced by the Nullbasic-EGFP gene cassette, lentiviral VLPs were made using pLOX-CW-EGFP that was cotransfected with an expression vector containing Nullbasic-FLAG or the empty vector pCDNA3.1 as a control (Fig. 3b). ERT assays were performed with viral supernatant collected 48 hr posttransfection and normalized to RT concentration. As shown in Fig. 3b, lentiviral VLPs conveying EGFP that were made in cells expressing Nullbasic-FLAG also had a significantly reduced ability to undergo reverse transcription, supporting the observation that Nullbasic-EGFP could inhibit reverse transcription by VLPs. As shown previously, much less viral DNA was detected if nucleotides were excluded from the reactions, indicating DNA synthesis was de novo, and Western blot analysis confirmed that Nullbasic-FLAG was expressed by the transfected cells (Fig. 3c). We also tested the comparative transduction efficiency and reverse transcription capacity of VLPs conveying Nullbasic-EGFP or EGFP in HEK293T cells that were transduced in parallel so that reverse transcription products and total lentiviral RNA in cells could be isolated and quantified. As shown in Fig. 4a, a 4- to 5-fold defect in reverse transcription was observed after transduction with HIV-1-based lentiviral VLPs conveying Nullbasic-EGFP compared with EGFP. Reverse transcription was strongly reduced if the cells were treated with nevirapine or if VLPs were heat-treated, indicating that cDNA synthesis was de novo. However, we observed no significant difference in lentiviral RNA levels in cells, indicating that transduction efficiencies were similar (Fig. 4b). Interestingly, total RT activity was inhibited by a minor but statistically significant amount as measured with a homopolymer primer:template [a poly(A) and oligo(dT)-based] system. However, a much larger reverse transcription defect was observed in ERT or cell transduction experiments, suggesting that Nullbasic affects the viral reverse transcription complex rather than RT per se. We conclude that Nullbasic can inhibit reverse transcription of lentiviral particles, resulting in reduced infectivity by lentiviral VLPs derived from HIV-1. Therefore standard lentiviral vectors conveying Nullbasic are not suitable for gene delivery to human cells.
FIG. 1.
Nullbasic does not inhibit the production of lentiviral VLPs. (a) Schematic diagrams of the three VLP expression plasmids and an overview of the methods used to generate lentiviral VLPs. pCMVΔR8.91 plasmid expresses structural and regulatory proteins required to form viral particles; pLOX-CW lentivector expresses the VLP-packaged RNA genome containing either the Nullbasic-EGFP (NB-EGFP) or control EGFP gene; and the pCMV-VSV-G plasmid expresses the vesicular stomatitis virus envelope glycoprotein (VSV-G env) from the cytomegalovirus promoter (pCMV) to enable pseudotyping of VLPs. Genetic elements within the pCMVΔR8.91 and pLOX-CW plasmids have been described elsewhere (Zufferey et al., 1997; Salmon et al., 2000). The plasmids were transfected into HEK293T cells in order to produce VLPs containing either the NB-EGFP lentivector genome (VLP-NB-EGFP) or the control EGFP genome (VLP-EGFP). (b) VLPs secreted into culture supernatants were collected and quantified by capsid (CAp24) ELISA and RT activity. Columns represent the means and standard deviations from three independent stocks. (c) Quantification of packaged genomic RNA, as measured by qRT-PCR, in the VLP-EGFP and VLP-NB-EGFP viral stocks. A no-dNTP control RT reaction was always included and data sets were discarded if contaminating DNA was detected. The RNA levels are normalized for CAp24 and data are expressed as copies of RNA per picogram of CAp24. Columns represent the means and standard deviations from three independent stocks. The Student t test (two-tailed distribution) was used to assess statistical significance between data sets.
FIG. 2.
Nullbasic diminishes the infectivity of lentiviral VLPs. Nullbasic-EGFP (NB-EGFP) or control EGFP VLPs, produced as per Fig. 1, were transduced into (a) Jurkat, (b) HEK293T, or (c) HeLa cells at various CAp24 inoculation amounts, as indicated. Transduced cells were subsequently quantified for EGFP expression by flow cytometry. Data indicate the proportion of total cells that were EGFP fluorescent. Columns represent the means and standard deviations from three independent transductions.
FIG. 3.
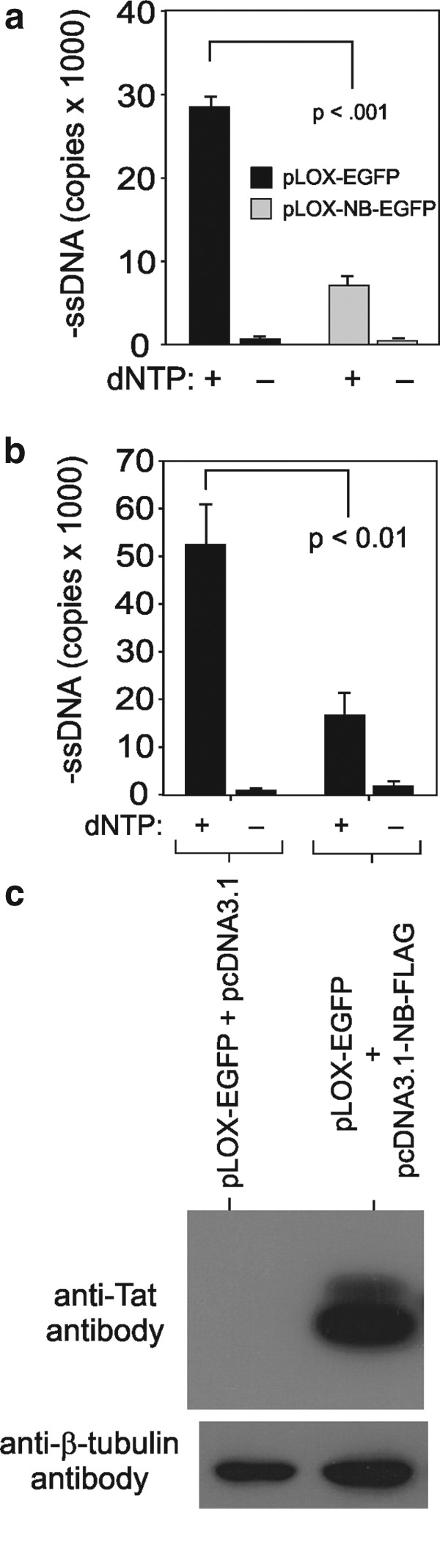
Nullbasic inhibits reverse transcription of lentiviral VLPs. (a) Using partially purified VLPs, ERT assays were performed with equivalent amounts of RT activity determined in a homopolymeric primer:template RT assay. Briefly, DNase I-treated VLP supernatant was subjected to centrifugation at 100,000×g for 1 hr. The washed viral pellets were resuspended in PBS and used in ERT reactions as described in Materials and Methods (Warrilow et al., 2008). All ERT experiments included a no-dNTP control reaction. The viral DNA was obtained and subjected to qPCRs in triplicate, using primers described in the online supplement, to measure HIV-1 negative-strand strong stop DNA. The mean value and the standard deviations are indicated. (b) Lentiviral VLPs made with pLOX-CW-EGFP were made in HEK293T cells by cotransfection with a Nullbasic expression vector, pCDNA3.1-NB-FLAG, or an empty vector. VLP supernatant was collected 48 hr posttransfection and ERT assays were performed and analyzed by qPCR as described previously. (c) Western blot analysis of protein lysates prepared from the transfected HEK293T using an anti-Tat (top) or anti-tubulin (bottom) antibody.
FIG. 4.
Nullbasic inhibits reverse transcription of lentiviral vector in cells. (a and b) HEK293T cell cultures were infected in duplicate with equal amounts of VLPs conveying either Nullbasic-EGFP or EGFP. After attachment of virus to cells at 4°C, the infections were initiated and incubated at 37°C for 3 hr. Parallel cultures transduced side by side were processed for either viral cDNA or RNA. Where indicated, cells were pretreated with 200 μM nevirapine (NVP) or VLPs were heat-inactivated. The experiments were performed three times and a representative experiment is shown. (a) The level of cytoplasmic VPL cDNA was determined by qPCR using primers specific for the EGFP region. DNA copy number was normalized to mitochondrial DNA levels measured in each sample. A representative result from three experiments with similar results is shown. (b) The level of viral RNA in transduced cells was determined. DNase I-treated RNA extracted from cells treated with NVP was used in qRT-PCRs using primers specific for the EGFP region. A synthetic kanamycin cassette RNA was included in RNA isolations to control for recovery and RT-PCR efficiency. A no-reverse transcriptase reaction was always included to control for DNA contamination. (c) VLP supernatant was measured in triplicate for CAp24 by ELISA and for RT activity, using a commercial assay. The VLP stocks used contained similar levels of CAp24, approximately 250–300 ng/ml. RT assays were performed with an equal volume of each VLP stock and the values shown represent the ratio of RT to CAp24. The experiments were performed twice with similar results and a representative experiment is shown. The Student t test (two-tailed distribution) was used to assess statistical significance between data sets.
Nullbasic does not inhibit MLV-based retroviral vectors
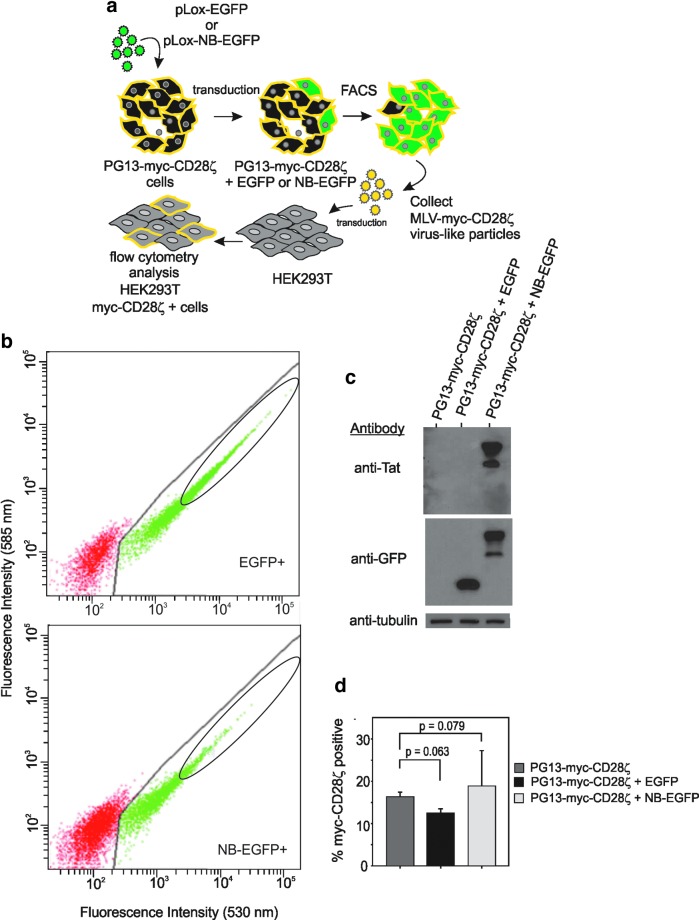
To investigate whether Nullbasic could inhibit MLV-based retroviral vectors, a PG13-myc-αerb-CD28ζ cell line was transduced with lentiviral VLPs conveying Nullbasic-EGFP or EGFP (Fig. 5a). PG13-myc-αerb-CD28ζ cells constitutively produce MLV-based VLPs that, after transduction, convey a Myc-tagged αerb-CD28ζ cell surface protein to target cells. As shown in Fig. 5b, EGFP-positive cells were collected by FACS and new PG13-myc-αerb-CD28ζ cell lines stably expressing EGFP were established. We should point out that to compensate for reduced infectivity of lentiviral VLPs with NB-EGFP, a larger volume of pLOX-CW-NB-EGFP VLPs was used to obtain the high levels of transductant cells shown in Fig. 5b (bottom). Flow cytometric analysis of the purified cells indicated that >95% of the cells were positive for EGFP or NB-EGFP (data not shown). Western blot analysis of lysates made from each cell line confirmed expression of Nullbasic-EGFP or EGFP in the cells (Fig. 5c). We noted that the two protein bands detected by an anti-Tat antibody were most likely a result of posttranslational modification known to occur on Tat such as ubiquitination, phosphorylation, and acetylation. Using supernatants containing equivalent amounts of total RT, Jurkat cells were transduced and analyzed by flow cytometry for the presence of an extracellular Myc-epitope tag. No significant difference in the number of transduced cells was observed (Fig. 5d), indicating that expression of Nullbasic in PG13-myc-αerb-CD28ζ cells did not inhibit the infectivity of MLV-based VLPs that were produced.
FIG. 5.
Nullbasic does not affect the infectivity of MLV VLPs made by PG13 cells. (a) An overview of the experimental design. PG13 cells producing MLV-myc-αerb-CD28ζ VLPs were secondarily transduced with lentiviral VLPs delivering Nullbasic-EGFP or EGFP. (b) Cells expressing Nullbasic-EGFP or EGFP were collected by FACS as indicated (designated by the oval). (c) Western blot analysis with an anti-Tat or anti-GFP antibody confirmed expression of each protein in the cell population. An anti-tubulin antibody confirmed that similar amounts of protein were used. (d) MLV-myc-αerb-CD28ζ VLP supernatant was collected from the lentivirus-transductant cell lines or the parental cell line and used to transduce Jurkat cells, which can express Myc-αerb-CD28ζ on the cell surface. Flow cytometric analysis demonstrated no significant differences in the level of transduced cells expressing myc-αerb-CD28ζ. The mean value of triplicate transduction experiments, SDs, and p values are shown. The experiment was performed twice with similar results, and a representative data set is shown.
Nullbasic-EGFP strongly inhibits HIV-1 replication in Jurkat cells
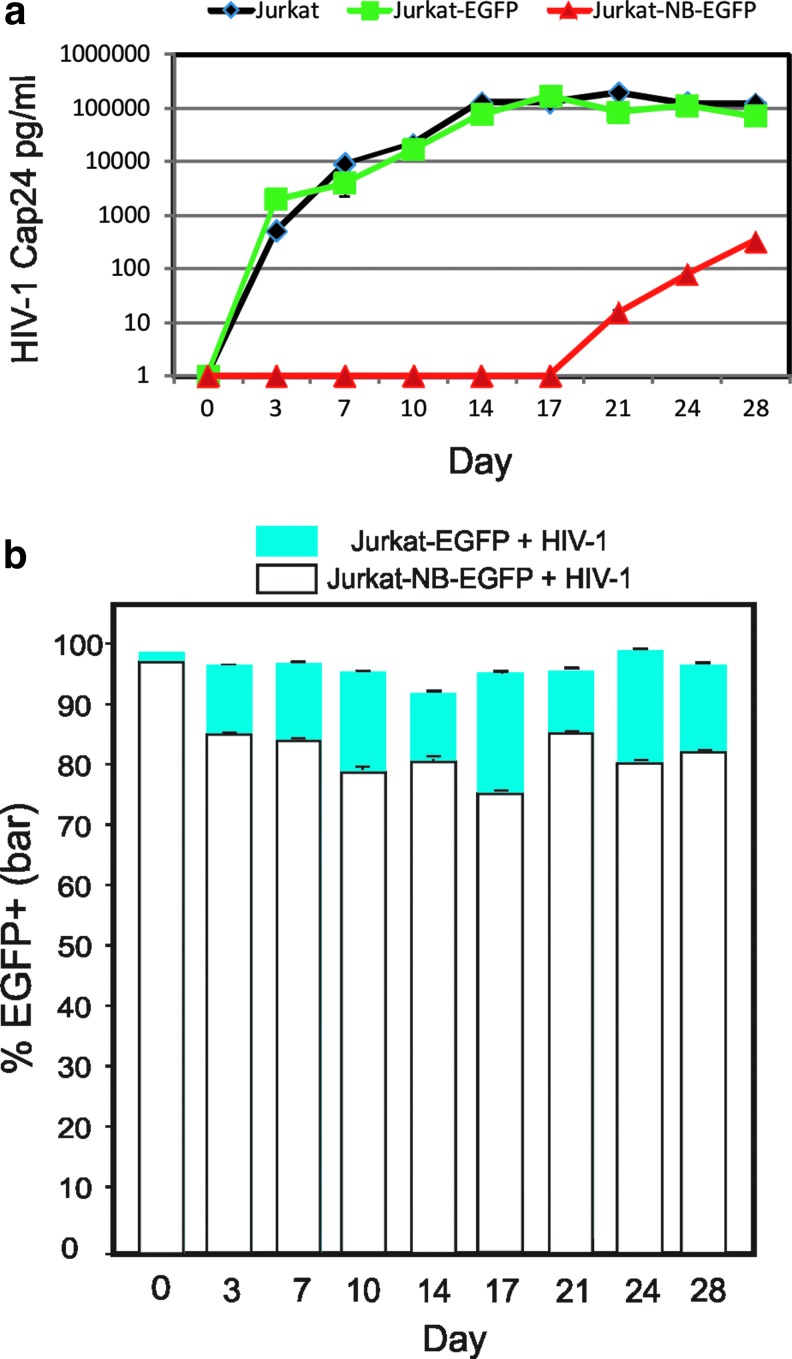
The Nullbasic-EGFP or EGFP gene cassette was inserted into the pGCsamEN (Chuah et al., 1995) retroviral vector and transfected into the MLV-A VLP-producing cell line ΦNX-amphotropic (Fig. 6a) (Swift et al., 2001). VLP-containing supernatant was obtained and relative titers were determined by serial dilution transduction of HeLa cells. We routinely obtained supernatants containing from 1×105 to 4×105 EGFP CFU/ml irrespective of the pGCsamEN construct used. The transduced Jurkat cells underwent FACS, with the EGFP-positive cell population collected as indicated (Fig. 6b). Subsequent flow cytometric analysis showed the cell populations were >97% positive for EGFP or NB-EGFP. As expected, Western blot analysis of lysates made from the cells showed that Nullbasic-EGFP or EGFP was expressed (Fig. 6c). For each infection, 106 Jurkat-Nullbasic-EGFP, Jurkat-EGFP, and parental cells were each incubated with HIV-1pNL4-3 (equivalent to 20 ng of CAp24) for 2 hr. The virus was removed and the infected cells were incubated for 4 weeks, with twice-weekly sampling of the culture medium; the samples were assayed for HIV-1 capsid protein by ELISA. As shown in Fig. 7, similar virus replication kinetics were observed in parental Jurkat and Jurkat-EGFP cells, with peaks on days 21 and 17, respectively. Virus replication was consistently inhibited in Jurkat-NB-EGFP cells, in which only low levels of CAp24 (<500 pg/ml) were detected after 3 weeks. This result was consistently observed in three independent experiments. We also noted steady expression of NB-EGFP in Jurkat cells, suggesting that the protein had little or no deleterious effects on the cells. This is in sharp contrast to HIV-1 Tat, which can accelerate HIV-1 replication and induce cytotoxic effects in human T cells (Caputo et al., 1990; Benjouad et al., 1993). We conclude that expression of NB-EGFP is well tolerated and has potent antiviral effects that can sharply delay virus replication in Jurkat cells.
FIG. 6.
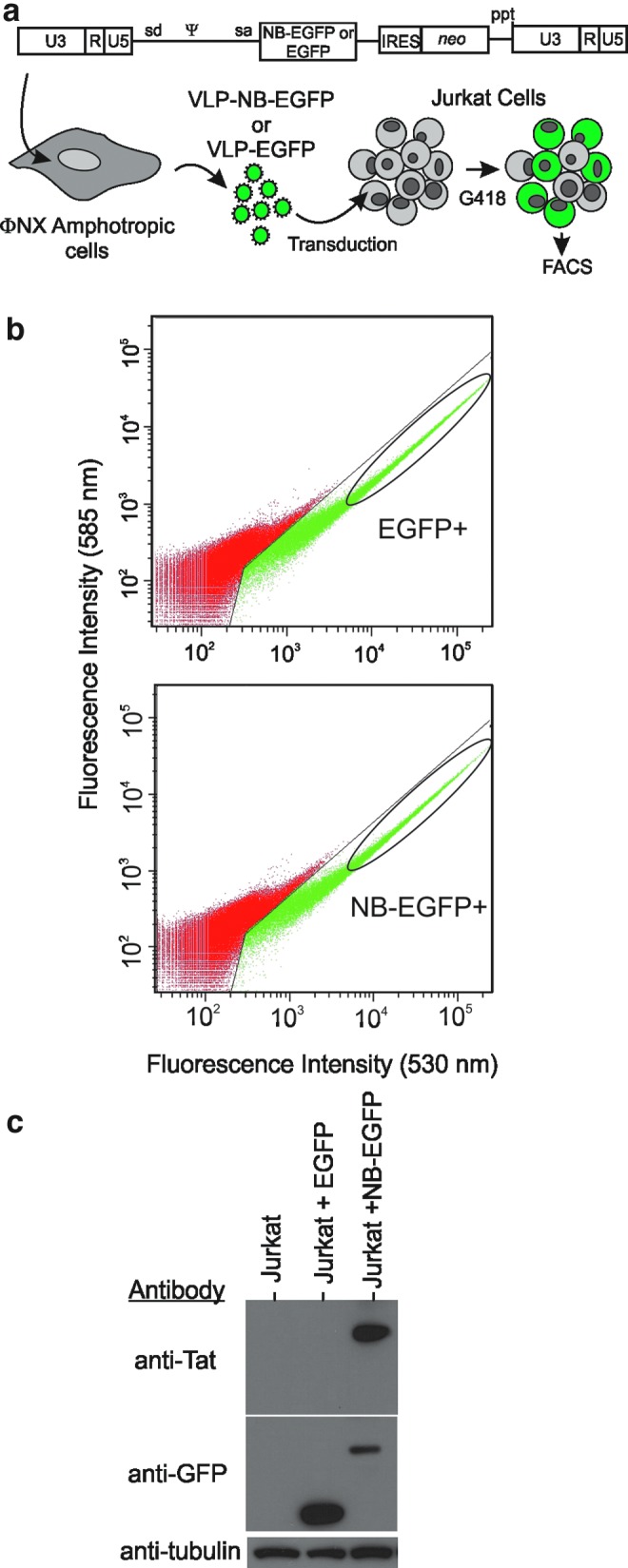
Transduction of Jurkat cells with retroviral VLPs conveying NB-EGFP or EGFP. (a) Schematic diagrams of the retroviral vector pGCsamEN (Chuah et al., 1995) carrying either the NB-EGFP or control EGFP gene cassette and an overview of the method used to generate retroviral VLPs with Phoenix (ΦNX) amphotropic packaging cells. NB-EGFP or control EGFP retroviral vectors were transfected into the ΦNX cells to produce MLV-A-based VLP-NB-EGFP or VLP-EGFP particles, respectively. VLPs were transduced into Jurkat cells, which were grown in medium containing G418 and enriched for EGFP expression by fluorescence-activated cell sorting (FACS). (b) EGFP-positive cells were collected by FACS as indicated. Representative dot plots are shown in which EGFP-positive cells (top) and NB-EGFP-positive cells (bottom) are gated (green, indicated by the oval). (c) Western blotting was performed on cell lysates prepared from postsorted transductants, detecting Nullbasic expression with an anti-Tat antibody and EGFP expression with an anti-GFP antibody. An anti-tubulin antibody was used to confirm that similar amounts of sample were used. Parental Jurkat cells were used as a negative control.
FIG. 7.
HIV replication is significantly delayed in Jurkat cell populations expressing Nullbasic-EGFP. The two Jurkat cell lines shown in Fig. 6 were infected with HIV-1pNL4-3 containing 20 ng of CAp24 and grown for 4 weeks. (a) The cell cultures were split every 3 or 4 days as indicated and a sample of cell-free supernatant was assayed for HIV-1 CAp24 by ELISA. (b) Fixed cells were assayed for Nullbasic-EGFP or EGFP expression by flow cytometry. A representative data set of three independent experiments performed in duplicate is shown. The mean value and standard deviation of the mean are indicated. Color images available online at www.liebertpub.com/hum
Nullbasic inhibits HIV-1 replication in human CD4+ primary cells
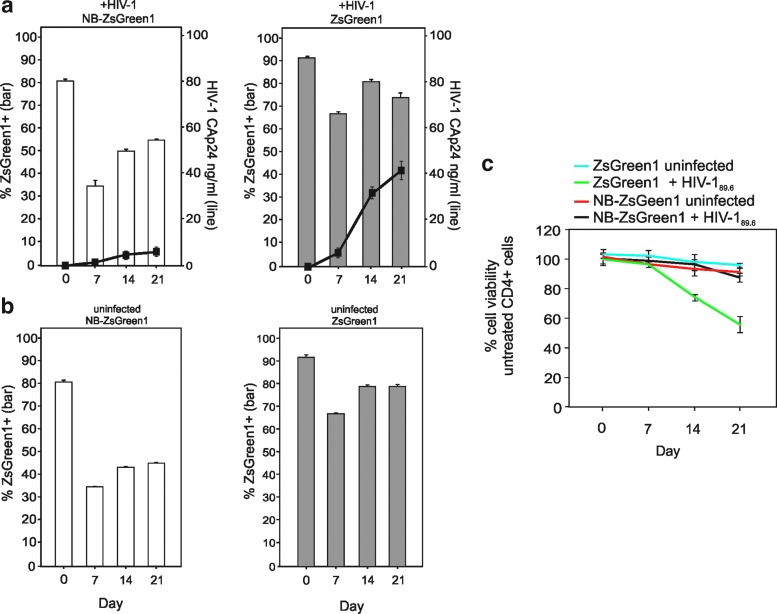
Transduction experiments were performed to determine whether Nullbasic could inhibit HIV-1 replication in primary human CD4+ cells. However, MTS assays of our initial attempts to make transductant cells with NB-EGFP or EGFP indicated a considerable level of cellular toxicity, probably caused by EGFP expression in activated CD4+ primary cells (Liu et al., 1999; and data not shown). Therefore, the EGFP fusion protein partner was replaced with the better-tolerated ZsGreen1 (Wouters et al., 2005) protein for these experiments (Figs. 8a and 9) after confirming that NB-ZsGreen1 had a similar ability to inhibit HIV-1 infectivity as previously described (data not shown; and Meredith et al., 2009). Highly enriched human CD4+ cells isolated from buffy coat cells were transduced with MLV-based retroviral vectors conveying NB-ZsGreen1 or ZsGreen1. After 72 hr, ZsGreen1-positive cells were collected by FACS as shown (Fig. 8b) and Western blot analysis confirmed expression of NB-ZsGreen1 (Fig. 8c) with an anti-Tat antibody. We noted that the overall fluorescence intensity of cells expressing ZsGreen1 or NB-ZsGreen1 was lower than that observed for comparable EGFP proteins in Jurkat cells (Fig. 6b), although ZsGreen1 has a comparable fluorescence intensity profile (Day and Davidson, 2009). We transduced 106 CD4+ cells with HIV-189.6 (a dual-tropic R5X4 laboratory HIV-1 strain) supernatant containing 20 ng of CAp24 for 2 hr. The virus was removed by washings and samples of supernatant and cells, collected on day 0 and every 7 days thereafter, were assayed for HIV-1 CAp24 by ELISA and for expression of Zs-Green1 or NB-ZsGreen1 at each time point by flow cytometry. We also compared cell viability with a commercial MTS assay (Fig. 9c). As shown in Fig. 9a (left), where cells were infected with HIV-1, the percentage of NB-ZsGreen1-positive CD4+ cells decreased from 80% on day 0 to 35% on day 7 and increased by day 21 to 54%. A similar pattern of NB-ZsGreen1 expression was observed in uninfected cells although at slightly lower levels (Fig. 9b, left). Higher percentages of CD4+ cells expressed ZsGreen1; 90% on day 0 and thereafter decreasing to 65–80% of CD4+ cells (Fig. 9a, right), irrespective of the presence of HIV-1 (Fig. 9b, right). In three identical experiments, HIV-1 levels were reduced by approximately 8- to 10-fold when NB-Zs-Green1 was expressed in CD4+ cells as compared with ZsGreen1. MTS assays of uninfected cells and HIV-1-infected NB-ZsGreen1 cells showed no significant change in cell viability compared with untreated control CD4+ cells. However, HIV-1-infected CD4+ ZsGreen1 cells demonstrated reduced viability, most likely a result of HIV-1 infection. These experiments support the conclusion that NB-ZsGreen1 and ZsGreen1 are tolerated by activated human CD4+ primary cells and that NB-ZsGreen1 strongly inhibited HIV-1 replication.
FIG. 8.

Transduction of human CD4+ primary cells with MLV-A-based retroviral VLPs conveying NB-ZsGreen1 or ZsGreen1. (a) Schematic of the methods used to generate CD4+ primary cells transductants. (b) NB-ZsGreen1 or ZsGreen1 VLPs were used to transduce activated human CD4+ primary cells and ZsGreen-positive cells were collected by FACS. Representative dot plots are shown in which ZsGreen-positive cells (bottom) and NB-ZsGreen-positive cells (top) are gated (indicated by the oval). (c) Cell lysates were prepared from the purified cells and Western blot analysis was performed with anti-Tat antibody to detect Nullbasic-ZsGreen1. An anti-tubulin antibody was used to confirm that similar amounts of lysate were used.
FIG. 9.
HIV replication is inhibited in activated human CD4+ primary cells expressing NB-ZsGreen1. (a) The two CD4+ populations described in Fig. 8 were infected with HIV-189.6 containing 20 ng of CAp24 and grown for 21 days. Cell-free supernatant was collected once per week and assayed for HIV-1 CAp24 by ELISA (right y axis, lines). Fixed cells were assayed for NB-ZsGreen or ZsGreen expression by flow cytometry (left y axis, bars). (b) Uninfected cell populations as in (a) were monitored for ZsGreen1 or NB-ZsGreen1 expression by flow cytometry. (c) Cell viability of each culture compared with untreated activated human CD4+ cells was monitored weekly by MTS assay. All cultures, infected and noninfected, were performed in duplicate and the mean values and standard deviations are shown. A representative data set from three identical experiments is shown.
Discussion
Here we demonstrated that Nullbasic, a mutant Tat protein, was effectively delivered by MLV-based retroviral vectors and its expression in primary human CD4+ cells and Jurkat T cells provided strong protection from HIV-1 replication. Mutant forms of Tat that inhibit HIV-1 gene expression in T cell lines have been described but these proteins were truncated within the basic domain (amino acids 49–57), expressed only the first exon, or were truncated in the second exon (86-amino acid Tat) and had fewer substitutions in the basic domain (Pearson et al., 1990; Modesti et al., 1991). Some of these mutant Tat proteins were shown to provide modest protection from HIV-1 replication when expressed in human T cell lines (Ulich et al., 1996). Whereas this study confirmed that downregulation of virus infectivity and reverse transcription by Nullbasic was demonstrably important, previous analysis of the antiviral response induced by these shorter Tat mutant proteins was, generally, limited to effects on HIV-1 gene expression (Green et al., 1989; Pearson et al., 1990; Modesti et al., 1991), and although not demonstrated it seems possible that other dominant negative Tat proteins could negatively affect reverse transcription. In addition, the negative effect on reverse transcription by Nullbasic was limited to HIV-1 and HIV-1-derived lentiviral vectors; there was no significant effect on infectivity by MLV-based retroviral vectors made by PG13 cells, suggesting that reverse transcription was unaffected. Overall, the extended level of protection from virus replication provided by Nullbasic suggests it may be useful as an agent to inhibit HIV-1 replication.
Nullbasic was reported to inhibit HIV-1 transcription by Tat, Rev-mediated mRNA transport, and reverse transcription (Meredith et al., 2009). Meredith and colleagues showed that when Nullbasic was cotransfected with a HIV-1 proviral vector in sufficient quantity, HIV-1 virus production could be strongly reduced (Meredith et al., 2009). However, lentiviral genomic RNA incorporation and particle production were not significantly affected when Nullbasic was expressed by pLOX-CW-NB-EGFP, suggesting that levels of Nullbasic expressed by pLOX vector were insufficient to inhibit Rev. Although these analyses hinted that Nullbasic might be successfully delivered to target cells by lentiviral vectors, overall transduction ability of the lentiviral particles conveying Nullbasic were markedly reduced compared with controls, an outcome that could be largely attributed to a defect in reverse transcription. However, Nullbasic did not affect infectivity of MLV-A virus-like particles, indicating that Nullbasic most likely targets the HIV-1 reverse transcription complex (RTC), cellular components important for HIV-1 reverse transcription, or may act as a trans-dominant negative inhibitor given evidence that Tat can support improved HIV-1 reverse transcription (Harrich et al., 1997). The precise mechanism by which Nullbasic inhibits reverse transcription remains to be determined.
In all experiments, HIV-1 replication in Jurkat-NB-EGFP cells was remarkably inefficient over a 4-week period, whereas sharply reduced HIV-1 replication was observed in activated primary CD4+ cells expressing NB-ZsGreen1 compared with control cells. We suspect that one significant factor for reduced inhibition in primary cells was due to reduced expression of NB-ZsGreen1 compared with NB-EGFP in Jurkat cells. Flow cytometric analysis of the overall fluorescence intensity profiles, where the relative fluorescence intensity of ZsGreen1 is reported to be 117% of EGFP (Day and Davidson, 2009), indicated an approximate 0.5-log reduction in ZsGreen1 expression in primary CD4+ cells compared with Jurkat-NB-EGFP cells. Second, a drop in the penetrance of NB-ZsGreen1 expression was observed from day 0 to 7. Although the exact cause for the decrease in the number of ZsGreen1+ cells is unclear, we did not observe significant change is the cell viability compared with control cells. It is possible that NB-ZsGreen1 was downregulated by a gene-silencing mechanism in some CD4+ cells (Cherry et al., 2000). Nevertheless, HIV-1 replication was reduced by approximately 8- to 10-fold in CD4+-NB-ZsGreen1+ cell populations in three independent experiments. The reduced virus production, as indicated by decreased capsid protein secretion, most likely resulted from greatly suppressed virus replication after infection on day 0 when approximately 80% of cells were NB-ZsGreen1+. Expression of Nullbasic in cells can decrease virus production in infected cells, resulting in reduced virus replication kinetics (Meredith et al., 2009). To address this possibility we attempted intracellular staining of the primary cells for HIV-1 capsid protein to determine which cell population was producing viral protein. However, this analysis was unsuccessful. Although we cannot exclude HIV-1 replication in NB-ZsGreen1+ cells, delayed virus replication in unprotected cells is more likely. In future experiments, vectors capable of strong and stable expression of NB-ZsGreen1 in activated CD4+ primary cells should help address this issue. Whereas expression of HIV-1 Tat can accelerate HIV-1 replication and induce cytotoxic effects in human T cells (Caputo et al., 1990; Benjouad et al., 1993), Nullbasic-EGFP and Nullbasic-ZsGreen1 were well tolerated HIV-1 inhibitors in Jurkat and primary CD4+ cells, respectively. Further analysis of how different domains of Tat contribute to Nullbasic antiviral effects may illuminate new candidate protein inhibitors with improved expression while maintaining strong anti-HIV-1 properties.
In conclusion, here we show that expression of Nullbasic fusion proteins could provide excellent protection against HIV-1 replication in Jurkat and primary CD4+ cells. Two previous studies showed that combined expression of Tat mutant proteins and RevM10 (Ulich et al., 1996) or their fusion (Aguilar-Cordova et al., 1995) provided synergistic inhibition of HIV-1 replication, a strategy that may be relevant to Nullbasic. Therefore, the unusual properties conveyed by Nullbasic make it a candidate antiviral agent. At the very least, analysis of its mechanism of HIV-1 inhibition could advance discovery of novel antiviral strategies.
Supplementary Material
Acknowledgments
This work was funded by a grant from the Australian Centre for HIV and Hepatitis Research and by an Australian Research Council Future Fellowship to D.H.
Author Disclosure Statement
The authors declare that they have no competing interests.
References
- Adachi A. Gendelman H.E. Koenig S., et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Cordova E. Chinen J. Donehower L.A., et al. Inhibition of HIV-1 by a double transdominant fusion gene. Gene Ther. 1995;2:181–186. [PubMed] [Google Scholar]
- Benjouad A. Mabrouk K. Moulard M., et al. Cytotoxic effect on lymphocytes of Tat from human immunodeficiency virus (HIV-1) FEBS Lett. 1993;319:119–124. doi: 10.1016/0014-5793(93)80049-z. [DOI] [PubMed] [Google Scholar]
- Bevec D. Dobrovnik M. Hauber J. Bohnlein E. Inhibition of human immunodeficiency virus type 1 replication in human T cells by retroviral-mediated gene transfer of a dominant-negative Rev trans-activator. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9870–9874. doi: 10.1073/pnas.89.20.9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo A. Sodroski J.G. Haseltine W.A. Constitutive expression of HIV-1 Tat protein in human Jurkat T cells using a BK virus vector. J. Acquir. Immune Defic. Syndr. 1990;3:372–379. [PubMed] [Google Scholar]
- Checkley M.A. Luttge B.G. Soheilian F., et al. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology. 2010;400:137–144. doi: 10.1016/j.virol.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S.R. Biniszkiewicz D. Van Parijs L., et al. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol. Cell. Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuah M.K. Vandendriessche T. Morgan R.A. Development and analysis of retroviral vectors expressing human factor VIII as a potential gene therapy for hemophilia A. Hum. Gene Ther. 1995;6:1363–1377. doi: 10.1089/hum.1995.6.11-1363. [DOI] [PubMed] [Google Scholar]
- Day R.N. Davidson M.W. The fluorescent protein palette: Tools for cellular imaging. Chem. Soc. Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doranz B.J. Rucker J. Yi Y., et al. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- Fackler O.T. D'Aloja P. Baur A.S., et al. Nef from human immunodeficiency virus type 1F12 inhibits viral production and infectivity. J. Virol. 2001;75:6601–6608. doi: 10.1128/JVI.75.14.6601-6608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A. Methods of preparation and analysis of intracellular reverse transcription complexes. Methods Mol. Biol. 2009;485:107–119. doi: 10.1007/978-1-59745-170-3_8. [DOI] [PubMed] [Google Scholar]
- Fish R.J. Kruithof E.K. Evidence for serpinB2-independent protection from TNF-α-induced apoptosis. Exp. Cell Res. 2006;312:350–361. doi: 10.1016/j.yexcr.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Furuta R.A. Shimano R. Ogasawara T., et al. HIV-1 capsid mutants inhibit the replication of wild-type virus at both early and late infection phases. FEBS Lett. 1997;415:231–234. doi: 10.1016/s0014-5793(97)01132-0. [DOI] [PubMed] [Google Scholar]
- Green M. Ishino M. Loewenstein P.M. Mutational analysis of HIV-1 Tat minimal domain peptides: Identification of trans-dominant mutants that suppress HIV-LTR-driven gene expression. Cell. 1989;58:215–223. doi: 10.1016/0092-8674(89)90417-0. [DOI] [PubMed] [Google Scholar]
- Harrich D. Ulich C. Garcia-Martinez L.F. Gaynor R.B. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter G. Nowak D. Mossner M., et al. Long-term control of HIV by CCR5Δ32/Δ32 stem-cell transplantation. N. Engl. J. Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Lee S.K. Harris J. Swanstrom R. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J. Virol. 2009;83:8536–8543. doi: 10.1128/JVI.00317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.S. Jan M.S. Chou C.K., et al. Is green fluorescent protein toxic to the living cells? Biochem. Biophys. Res. Commun. 1999;260:712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- Malim M.H. McCarn D.F. Tiley L.S. Cullen B.R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M.H. Freimuth W.W. Liu J., et al. Stable expression of transdominant Rev protein in human T cells inhibits human immunodeficiency virus replication. J. Exp. Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith L.W. Sivakumaran H.R. Major L., et al. Potent inhibition of HIV-1 replication by a Tat mutant. PLoS One. 2009;4:e7769. doi: 10.1371/journal.pone.0007769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti N. Garcia J. Debouck C., et al. Trans-dominant Tat mutants with alterations in the basic domain inhibit HIV-1 gene expression. New Biol. 1991;3:759–768. [PubMed] [Google Scholar]
- Neagu M.R. Ziegler P. Pertel T., et al. Potent inhibition of HIV-1 by TRIM5–cyclophilin fusion proteins engineered from human components. J. Clin. Invest. 2009;119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson L. Garcia J. Wu F., et al. A transdominant tat mutant that inhibits Tat-induced gene expression from the human immunodeficiency virus long terminal repeat. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5079–5083. doi: 10.1073/pnas.87.13.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsakoff G.M. Engel B.C. Carbonaro D.A., et al. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34+ cells. Mol. Ther. 2005;12:77–86. doi: 10.1016/j.ymthe.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Ranga U. Woffendin C. Verma S., et al. Enhanced T cell engraftment after retroviral delivery of an antiviral gene in HIV-infected individuals. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1201–1206. doi: 10.1073/pnas.95.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon P. Oberholzer J. Occhiodoro T., et al. Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol. Ther. 2000;2:404–414. doi: 10.1006/mthe.2000.0141. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D. Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Stauber R.H. Afonina E. Gulnik S., et al. Analysis of intracellular trafficking and interactions of cytoplasmic HIV-1 Rev mutants in living cells. Virology. 1998;251:38–48. doi: 10.1006/viro.1998.9295. [DOI] [PubMed] [Google Scholar]
- Swift S. Lorens J. Achacoso P. Nolan G.P. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr. Protoc. Immunol. 2001;Chapter 10(Unit 10.17C) doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- Teng M.W. Kershaw M.H. Moeller M., et al. Immunotherapy of cancer using systemically delivered gene-modified human T lymphocytes. Hum. Gene Ther. 2004;15:699–708. doi: 10.1089/1043034041361235. [DOI] [PubMed] [Google Scholar]
- Treisman J. Hwu P. Minamoto S., et al. Interleukin-2-transduced lymphocytes grow in an autocrine fashion and remain responsive to antigen. Blood. 1995;85:139–145. [PubMed] [Google Scholar]
- Trono D. Feinberg M.B. Baltimore D. HIV-1 Gag mutants can dominantly interfere with the replication of the wild-type virus. Cell. 1989;59:113–120. doi: 10.1016/0092-8674(89)90874-x. [DOI] [PubMed] [Google Scholar]
- Ulich C. Harrich D. Estes P. Gaynor R.B. Inhibition of human immunodeficiency virus type 1 replication is enhanced by a combination of transdominant Tat and Rev proteins. J. Virol. 1996;70:4871–4876. doi: 10.1128/jvi.70.7.4871-4876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS (Joint United Nations Programme on HIV/AIDS) UNAIDS Global Facts & Figures (World Health Organization, Geneva, Switzerland) 2009. http://data.unaids.org/pub/factsheet/2009/20091124_fs_global_en.pdf http://data.unaids.org/pub/factsheet/2009/20091124_fs_global_en.pdf
- von Kalle C. Kiem H.P. Goehle S., et al. Increased gene transfer into human hematopoietic progenitor cells by extended in vitro exposure to a pseudotyped retroviral vector. Blood. 1994;84:2890–2897. [PubMed] [Google Scholar]
- Walker R.C., Jr. Khan M.A. Kao S., et al. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. J. Virol. 2010;84:5201–5211. doi: 10.1128/JVI.02318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow D. Meredith L. Davis A., et al. Cell factors stimulate human immunodeficiency virus type 1 reverse transcription in vitro. J. Virol. 2008;82:1425–1437. doi: 10.1128/JVI.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woffendin C. Ranga U. Yang Z., et al. Expression of a protective gene-prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2889–2894. doi: 10.1073/pnas.93.7.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters M. Smans K. Vanderwinden J.M. WZsGreen/+: A new green fluorescent protein knock-in mouse model for the study of KIT-expressing cells in gut and cerebellum. Physiol. Genomics. 2005;22:412–421. doi: 10.1152/physiolgenomics.00105.2005. [DOI] [PubMed] [Google Scholar]
- Zufferey R. Nagy D. Mandel R.J., et al. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.