Abstract
Biomarkers reflective of the molecular and genetic heterogeneity in colorectal cancers now guide certain aspects of clinical management and offer great potential for enrichment, stratification, and identification of novel therapeutic targets in drug development. Using case-based examples, this article reviews biomarkers that have an established role in the clinical management of colorectal cancer: mismatch repair protein testing and KRAS and BRAF mutational analysis. A selection of biomarkers undergoing validation for future clinical application is presented, and the dynamic and challenging interface between biomarkers in research and clinical practice is discussed.
Keywords: Colon cancer, colorectal cancer, biomarker, KRAS, BRAF, microsatellite instability, mismatch repair
Despite the development of multiple new therapeutics over the past decade, colorectal cancer continues to be the second leading cause of cancer mortality in the United States and worldwide, with median survival less than 2 years for patients with advanced disease.1,2 The evolving understanding of the genetic heterogeneity of colorectal tumors suggests that a purely anatomic classification may be obsolete.3 Recent identification of molecular tumor subtypes with distinctive treatment responsiveness and prognosis in some cancers has led to an emerging paradigm of genetically tailored clinical management, and promises to guide future drug development in oncology. A prominent example in colorectal cancer is the finding that an activating mutation in the KRAS oncogene is a biomarker for tumor resistance to monoclonal antibodies targeting the epidermal growth factor receptor (EGFR); this has led to the widespread adoption of tumor KRAS mutational analysis before treatment with this class of drugs.4–7
Biomarkers are measures, such as the presence of a mutation in the KRAS gene found through polymerase chain reaction (PCR) testing, associated with a clinically distinct prognosis, diagnosis, or response to a specific treatment. Conventional analyses such as tumor immunohistochemistry for cytokeratin profile or serum carcinoembryonic antigen (CEA) measurement are also types of biomarkers according to this broad definition. So-called predictive markers are associated with response (or lack thereof) to a particular therapy, whereas prognostic markers are baseline measurements associated with future disease trajectory.8,9 This article focuses on newer, genetically based tests of tumor, host, or both used to evaluate prognosis and/or to predict the likelihood of treatment response.
Using case-based examples, this article reviews biomarkers with an established role in clinical colorectal cancer management, presents a selection of biomarkers undergoing validation for future clinical application, and concludes with a discussion of the dynamic interface between biomarkers in research and clinical practice.
Overview of Established Biomarkers in Colorectal Cancer Clinical Management
The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) for Colon Cancer10 and Rectal Cancer11 have incorporated the following tumor genetic tests as biomarkers to guide specific clinical decisions for patients with colon and rectal cancers:
In patients with stage II colon cancer, mismatch repair (MMR) protein testing or microsatellite instability (MSI) testing should be considered to guide decisions on whether to administer adjuvant single-agent fluoropyrimidine chemotherapy.11 Patients with MMR deficiency have a favorable prognosis and do not seem to benefit from adjuvant single-agent fluoropyrimidine therapy.12–17
KRAS mutational analysis for patients with metastatic colorectal cancers is recommended as a predictive marker for nonresponse to EGFR-targeted therapy with cetuximab or panitumumab.11 Patients whose tumors harbor certain mutations in the KRAS gene do not respond to cetuximab or panitumumab and should not be treated with either of these agents.4–7,18
BRAF mutational analysis is also included as an option for patients with KRAS wild-type metastatic colorectal cancers as a strong negative prognostic factor and for possibly predicting a lower likelihood of benefit from EGFR-targeted therapy after progression on first-line therapy.10,18–23
These tests are reviewed below in the context of 3 constructed cases, along with selected examples of other commercially available biomarkers that have not been incorporated into the NCCN Guidelines because of a lack of consistent evidence for clinical validity or efficacy.
Case 1: MMR Analysis in Stage II Colon Cancer
A healthy 73-year-old man presents for medical oncology consultation 6 weeks after undergoing an uncomplicated right hemicolectomy with the finding of pathologic T3, N0 (out of 18 lymph nodes retrieved), M0 colon adenocarcinoma. He is considered to have average-risk AJCC stage II disease based on the clinical and pathologic features of his tumor.11,24 Although his family history is negative for malignancy, his oncologist requests mismatch repair protein testing of his tumor specimen to inform decision-making regarding adjuvant chemotherapy. His tumor is found to have deficiency in MLH1 through immunohistochemistry along with high-level MSI (MSIH) according to PCR. What are the implications of these findings on his recurrence risk and risk reduction from adjuvant chemotherapy, if administered?
Approximately 15% to 20% of colorectal cancers have sporadic or inherited deficiency of a mismatch repair protein, most commonly MLH1, MSH2, MSH6, or PMS2.25–28 Sporadic MMR deficiency is from inactivation of MLH1 in approximately 95% of cases, most often by promoter hypermethylation.29,30 Inherited MMR deficiency (Lynch syndrome) is from germline mutation in MLH1 or MSH2 in approximately 90% of cases, and in MSH6 in almost 10%.31
MMR deficiency is significantly more prevalent in stage II tumors compared with stage III, with a recent pooled analysis from the CALGB 9581 and 89803 studies showing a prevalence of 21.3% versus 14.4%, respectively (P < .001).15 MMR deficiency results in an inability to correct DNA replication errors, leading to variability in the length of naturally occurring repetitive DNA sequences (microsatellites) located throughout the genome; this is known as MSI.25 MMR-deficient tumors are characteristically located proximally and have a mucinous histology with tumor-infiltrating lymphocytes present.32,33 Testing for loss of a MMR protein using immunohistochemistry and MSI testing with PCR are concordant, with MSI-H being associated with deficiency in a MMR protein or proteins, whereas microsatellite stable (MSS) and tumors with lowlevel MSI (MSI-L) show MMR proficiency.28,34,35 By convention and for clarity, this article uses the terms MMR-deficient and MMR-proficient to describe both MMR testing results using immunochemistry and MSI results using PCR.
Historically, MMR testing has been reserved for patients meeting the Revised Bethesda Guidelines for Lynch syndrome screening.26,28 Independent of familial cancer risk, however, subset analyses of multiple large, randomized trials consistently show that MMR-deficient tumors have a favorable prognosis compared with stage-matched MMR-proficient tumors.12,13,15,27,36,37 Although the prognostic value of MMR deficiency is apparent in stage II and III disease, its impact on clinical decision making is greater for patients with stage II colon cancer, given the modest value of adjuvant chemotherapy overall, the limitations of conventional clinicopathologic risk assessment, and the increased prevalence of MMR deficiency in this group.12,14 In the randomized phase III QUASAR study, the overall recurrence risk for patients with stage II colon cancer with MMR deficiency was 11% (25 of 218) compared with 26% (438 of 1695) for those without MMR deficiency (P < .001).12 Meta-analyses and pooled data sets suggest that patients with stage II colon cancer with MMR deficiency do not derive benefit from adjuvant single-agent 5-FU or capecitabine therapy.13,14,27,38,39 In the largest study of pooled data from randomized trials of adjuvant 5-FU compared with a control arm without adjuvant treatment (N = 1027), Sargent et al.14 showed that the overall survival of patients with stage II colon cancer with MMR deficiency treated with adjuvant 5-FU (n = 47) was actually reduced compared with those treated with surgery alone (n = 55; hazard ratio [HR], 2.95; P = .04). Although diseasefree survival was also lower in the treated group, it did not reach statistical significance (P = .09). The biologic mechanism underlying this finding is unknown, however, and other datasets have not shown a negative interaction between MMR deficiency and outcomes with the use of adjuvant 5-FU.12,37 In QUASAR, the subset of patients with stage II colon cancer with MMR deficiency treated with adjuvant 5-FU showed a nonsignificant trend toward decreased recurrence compared with the no-treatment arm, but no difference was seen in overall survival between the arms, and interpretation is limited by very small numbers of patients and recurrences.12
Overall, these data strongly support the conclusion that MMR deficiency is a prognostic biomarker associated with a lower recurrence risk in patients with stage II colon cancer, and a predictive biomarker for lack of significant benefit from adjuvant fluoropyrimidine chemotherapy in this population. In this context, and acknowledging the findings of Sargent et al.14 suggesting a possible detriment in patients with MMR-deficient tumors, testing for this biomarker should be considered for all patients with stage II colon cancer being considered for adjuvant chemotherapy. The NCCN Guidelines concur with the cumulative data that patients with MMR-deficient tumors do not benefit from adjuvant single-agent fluoropyrimidine therapy.10 Notably, these recommendations for MMR testing do not apply to patients with stage II rectal cancer, who require a different treatment algorithm, including chemoradiation because of higher local and distant recurrence risk.
In Case 1, the finding of a MSI-H tumor identifies this patient as having a favorable prognosis, with an approximately 11% recurrence risk at 5 years according to the results from the QUASAR subset analysis. The patient is unlikely to benefit from adjuvant chemotherapy. Genetic evaluation should be offered, although sporadic MMR deficiency is more likely based on his age and negative family history. Tumor BRAF V600E mutation testing can be performed; if present, this mutation confirms sporadic MMR deficiency and excludes Lynch syndrome.40 The prognostic significance of BRAF mutation together with MMR deficiency is unclear, however, and is likely to be mediated by a complex interplay of other factors, including methylation status.12,36,41,42
Case 2: KRAS, BRAF, and Other Tests Available for Patients With Metastatic Disease
A 62-year-old woman is found to have unresectable, metastatic disease involving the liver and peritoneum 6 months after completing adjuvant FOLFOX chemotherapy for stage III colon adenocarcinoma. Before initiating chemotherapy for metastatic disease, her oncologist sends a paraffin-embedded specimen of archival tissue from her prior primary tumor resection specimen for KRAS mutational analysis. This is performed as part of a commercially available panel of tests, including PCR testing for KRAS and BRAF mutations along with immunohistochemistry for tumor EGFR, excision repair cross-complementation group 1 (ERCC1), thymidylate synthase (TS), and vascular endothelial growth factor receptor 2 (VEGFR2) expression levels. Her tumor is found to be wild-type for KRAS but harbors a mutation in BRAF at V600E. EGFR, ERCC1, and TS expression are reported as high, whereas VEGFR2 expression is low. Which (if any) of these results should influence choice of chemotherapy based on currently available evidence?
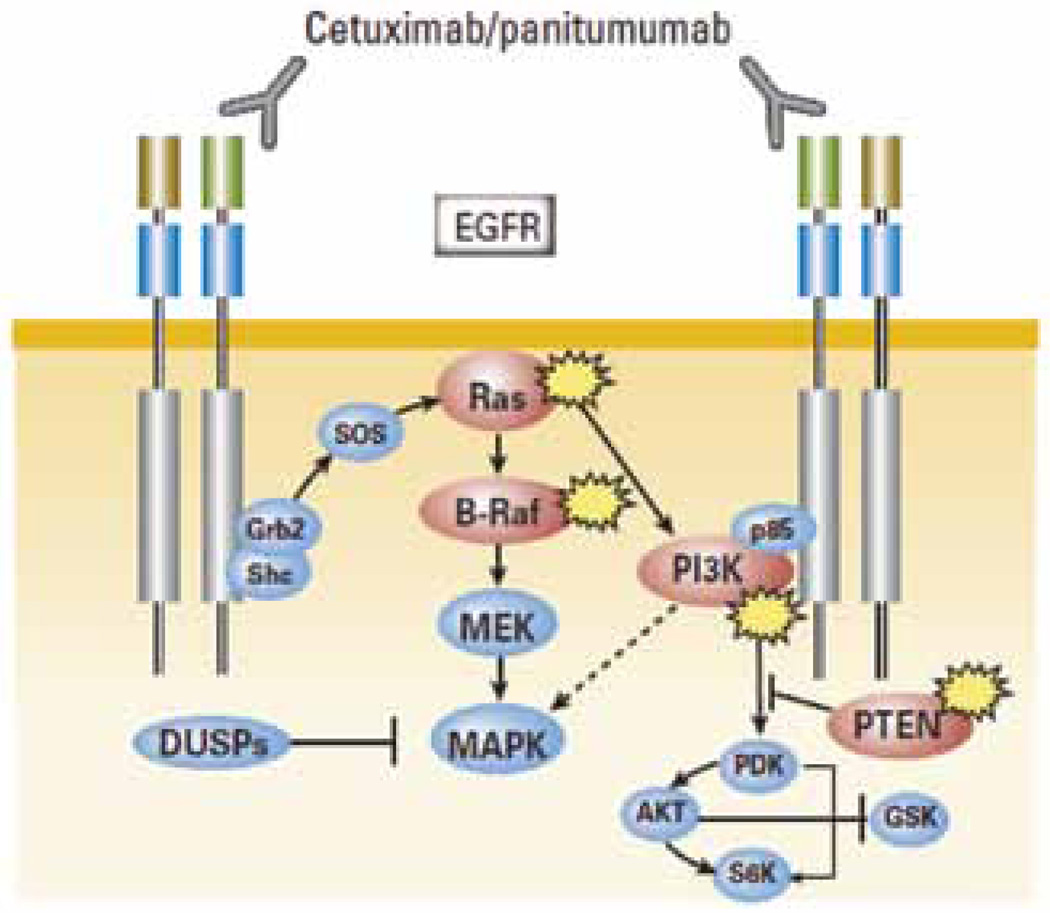
An activating mutation in codon 12, 13, or 61 of the KRAS oncogene is present in approximately 40% of colorectal adenocarcinomas and has recently been shown to be highly associated with resistance to the anti-EGFR antibodies, cetuximab and panitu-mumab.4–7 Figure 1 depicts the potential mechanism underlying this effect.
Figure 1.
Potential Mechanisms of Resistance to EGFR-Targeted Therapy. Schematic representation of the monoclonal antibodies cetuximab/panitumumab and of the epidermal growth factor receptor (EGFR)-mediated intracellular signaling pathways. The molecules implicated in EGFR signaling and affected by oncogenic alterations are highlighted in red.
From Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol 2010;28:1255; with permission.
Amado et al.4 compared the response rate to single-agent panitumumab in 427 patients with chemotherapy-refractory, metastatic colorectal cancers according to KRAS mutational status in a randomized study of panitumumab versus best supportive care. In the panitumumab treatment arm, patients whose tumors were wild-type for the KRAS gene showed a response rate of 17% compared with 0% for patients whose tumors harbored a mutation. In the first-line setting, randomized phase III studies have shown that addition of cetuximab or panitumumab to the FOLFIRI or FOLFOX regimens, respectively, significantly improves progression-free survival in patients with KRAS wild-type tumors, whereas those with mutant tumors do not benefit.5,7 Although KRAS mutation at c.38G>A (p.G13D) is included as a predictor of resistance with the other mutations in these analyses, it may not confer resistance to EGFR-targeted therapy, although data are limited.43 The FDA labels for cetuximab and panitumumab now restrict these agents to patients with tumors wild-type for the KRAS gene.44 Additional data are required to determine the significance of the G13D mutation.
The BRAF gene encodes a protein located downstream of KRAS (see Figure 1). Activating mutations in BRAF (most of which are c.1799T>A, leading to p.V600E) are present in 5% to 10% of patients with colorectal cancers and are almost never present in tumors with activating KRAS mutations.20,36,45,46 Table 1 displays incidence data for both mutations. Although single-arm studies suggested that the BRAF V600E mutation confers similar resistance to anti-EGFR therapies as an activating KRAS mutation, data from randomized clinical trials now show that this mutation is instead a very strong negative prognostic factor in patients with advanced disease.18,21,22,47–49 BRAF V600E mutation is also associated with significantly decreased overall survival in patients with stage II and III disease based on a subset analysis of the PETACC-3 study, particularly in patients without MMR deficiency (HR, 2.19; P = .00034 for patients with BRAF V600E mutation and without MSI-H).36 Responses to anti-EGFR therapy in the first-line setting have been described despite the presence of this mutation, however, albeit with a significantly lower rate than in patients with wild-type tumors.18–20,49 In a retrospective consortium analysis of 649 samples from patients with metastatic, refractory colorectal cancer treated with cetuximab plus chemotherapy, response rate in the BRAF mutant cohort was 8.3% compared with 38% for patients with BRAF and KRAS wild-type tumors.20 The CRYSTAL and CAIRO2 studies showed nonsignificant increases in response rate and survival in patients with BRAF mutant tumors treated with first-line cetuximab plus chemotherapy compared with patients treated with the same chemotherapy without cetuximab.18,49 Pooled analysis of the CRYSTAL and OPUS studies showed a similar nonsignificant trend toward benefit.19 Currently, the clinical role of BRAF testing remains inconclusive. BRAF mutational analysis is not required for treatment decision making, although it may be informative as a prognostic marker.10,11
Table 1.
KRAS and BRAF Mutation Incidence in Selected Randomized Studies
| Study | N | Stage |
KRAS Mutation Rate |
BRAF Mutation Rate |
|---|---|---|---|---|
| European consortium dataset20 | 773* | IV | 40% | 4.7% |
| COIN47 | 1316† | IV | 43% | 8% |
| CAIRO218 | 559 | IV | 39.4% | 8.7% |
| PETACC-336 | 1564 | II and III | 37% | 7.9% |
649 samples were eligible for outcome analyses.
Evaluable for mutation analyses
In Case 2, the results from the panel tested suggest that the patient is an appropriate candidate to receive cetuximab or panitumumab in combination with first-line chemotherapy because of the wild-type KRAS result. Unfortunately, the presence of a BRAF V600E mutation indicates that, although response to first-line therapy including an EGFR-targeted monoclonal antibody is possible, the likelihood may be significantly lower, and that the patient’s prognosis is poor. Current evidence is insufficient to support the integration of EGFR, ERCC1, thymidylate synthase (TS), or VEGFR2 expression levels into the clinical management of colorectal cancers. In the BOND study and in 2 phase II studies of panitumumab, tumor EGFR expression was not predictive of response to cetuximab or panitumumab.50,51 Evidence regarding the significance of ERCC1 expression levels in predicting response to or toxicity from oxaliplatin is conflicting; gene polymorphisms may be associated with outcomes but remain to be validated.52–55 Although data suggest an association with poor prognosis in colorectal cancers, TS expression levels have not been shown predict response or toxicity in randomized studies, and are confounded by heterogeneity in technique.55–57 Currently no validated predictive marker exists for response to bevacizumab; although high tumor VEGFR2 levels were associated with improved outcome in the BOND2 study, other studies have yielded conflicting results.58,59 Large, randomized, and controlled datasets are required to validate these and other pharmacogenomic tests as predictive biomarkers for treatment response to specific therapies. Existing data suggest that the effects are likely to be modest and modified by complex interactions with other factors, such as polymorphism. The selected examples are not currently recommended for use in clinical practice.
Case 3: Biomarkers at the Interface of Clinical Research and Practice
A 57-year-old woman with unresectable metastatic sigmoid colon adenocarcinoma has rapid disease progression despite first-line treatment with FOLFIRI plus panitumumab followed by second-line treatment with FOLFOX plus bevacizumab. Her tumor is known to be wild-type for the KRAS gene but positive for a BRAF c.1799T>A p.V600E mutation. Because of her young age, excellent performance status, and desire to pursue aggressive therapy, she is referred to an academic center for possible clinical trial participation. She is enrolled on a phase II trial for patients with BRAF-mutant solid tumors studying the efficacy of a novel combination of an oral, small-molecule BRAF inhibitor combined with another new biologic agent targeting a protein downstream from BRAF.
The examples of KRAS and BRAF mutations show that tumor genotype can significantly impact a patient’s likelihood of response to therapy and overall prognosis, and that colorectal cancer should not be considered a single, homogenous entity but rather as a collection of clinically distinct molecular subsets. In addition to the findings summarized earlier pertaining to KRAS and BRAF, molecular cohort analyses from the European consortium dataset and the randomized phase III COIN and CAIRO2 trials collectively suggest different response and survival outcomes in tumors harboring PIK3CA and NRAS mutations.18,20,47
Currently, tumor genotyping beyond KRAS and BRAF mutational analyses should be considered in-vestigational and should not be performed as standard of care. In fit patients eligible for clinical trial participation, however, tumor molecular and genetic characteristics will increasingly guide clinical and translational research investigating new targeted therapies and combinations in the metastatic, refrac-tory disease setting, as exemplified by the patient in Case 3 for whom standard treatment options were exhausted.
Selected Colorectal Cancer Biomarkers in Validation Stages of Development
Table 2 displays a selection of the emerging biomarkers with evolving evidence for clinical validity, defined as a significant association with a clinical end point, such as survival or recurrence, in patients with colorectal cancer.9 The selected tests are restricted to biomarkers for patients with established disease; diagnostic and screening biomarkers are not included. These tests currently are not recommended by the NCCN Guidelines because of their unclear clinical efficacy in affecting clinical decision making or patient outcomes.9
Table 2.
Colorectal Cancer Biomarker Tests in Development
| Biomarker Name |
Type of Specimen Required |
Type of Test | Stage(s) of Disease |
Intended Use | Current Validation Status |
Commercial Availability |
|---|---|---|---|---|---|---|
| ColoPrint | Fresh, unfixed tumor tissue |
Gene expression profile by oligonucleotide array |
II | Prognostic marker for recurrence risk in stage II colon cancer |
Two non- randomized validation studies;64,65 PARSC trial underway66 |
Anticipated (Agendia) |
| CTC | Peripheral blood collected in preservative tube |
Immunomagnetic separation of epithelial cells from whole blood |
IV | Prognostic for survival in patients with metastatic colorectal cancer |
Multiple studies including meta- analysis data and randomized subset from ==CAIRO2 study67,69 |
Multiple including CellSearch System (Veridex, LLC) |
| Oncotype DX Colon Cancer Test |
FFPE tumor tissue |
Gene expression profile by RT-PCR |
II | Prognostic marker for recurrence risk in stage II colon cancer |
Independent validation in randomized subset from QUASAR study;70 second independent validation set presented at 2011 ASCO Annual Meeting71 |
Yes (Genomic Health, Inc.) |
Abbreviations: CTC, circulating tumor cells; FFPE, formalin-fixed, paraffin-embedded; PARSC, Prospective Analysis of Risk stratification by ColoPrint; RT-PCR, reverse transcriptase-polymerase chain reaction.
Conclusions
The discovery of KRAS mutation as a predictive biomarker has invigorated biomarker research in colorectal cancer, providing clinical relevance to the molecular and genetic heterogeneity of colorectal tumors and a proof of principle for targeted therapy. In the quest to discover and develop new targeted therapies, the molecular and genetic features of tumors and their hosts will be increasingly studied in efforts to identify predictive and prognostic biomarkers for stratification and enrichment in drug development and for clinical management.
Enthusiasm for biomarkers in the research arena, however, must be tempered by a critical review of the available evidence before these biomarkers are used in clinical decision making. Appropriate adoption of new biomarkers faces the challenges of a highly dynamic evidence base and early access to commercialization.60–62 Subset analyses of randomized, controlled clinical trial datasets have confirmed the clinical validity of the biomarkers reviewed earlier through proving a statistically significant, reproducible association with a specific clinical end point, such as the likelihood of response to therapy or risk of recurrence.9 In contrast, however, the current body of clinical evidence and expert consensus do not support the clinical validity of using biomarkers such as tumor VEGFR2, EGFR, ERCC1, or TS expression levels to guide management in patients with colorectal cancers, although these tests are commercially available.
Despite robust data for their clinical validity, the examples of MMR, KRAS, and BRAF analyses also highlight the perils of early biomarker adoption in the setting of complex molecular pathways and incomplete evidence. Currently, the clinical implications of concurrent MMR deficiency and BRAF V600E mutation, which coexist in most sporadic MMR-deficient colorectal tumors, remain poorly understood and are likely to be mediated by an interplay of factors, including CpG island methylation status.41,63 Future dissection of molecular subsets within these categories will likely lead to further refinement of recommendations on risk assessment and adjuvant therapy, in patients with stage II colon cancers in particular. In the example of KRAS mutation, studies have recently shown that the c.38G>A p.G13D mutation may have a lesser degree of transforming capacity in vitro and may not influence responsiveness to EGFR-targeted therapy in the same manner as other KRAS codon 12 and 13 mutations.43 Because of small sample sizes, the significance of these findings remains limited, and currently no consensus exists regarding the use of EGFR-targeted therapy in patients with the KRAS G13D mutation.
The evolving understanding of BRAF c.1799T>A p.V600E mutation in colorectal cancers provides an example of the hazards of early interpretation of single-arm, nonrandomized data. Soon after the establishment of the KRAS mutation as a predictive biomarker for nonresponse to EGFR-targeted therapies, small single-arm studies suggested that the BRAF V600E mutation was a predictive marker for nonresponse to these agents similar to the KRAS mutation, leading to the inclusion of a statement in the NCCN Guidelines in early 2010 that patients whose tumors were known to harbor a BRAF V600E mutation seemed unlikely to benefit from anti-EG-FR monoclonal antibodies.21,22 Soon thereafter, the finding of its strong negative prognostic impact in randomized studies called into question the predictive value of the BRAF V600E mutation, leading to a modification of the statement in the NCCN Guidelines to note that data are inconsistent as to the benefit of these agents in patients with this mu-tation.13,18–20,49 In contrast to the dynamic revisions undertaken by the NCCN Guidelines in the case of BRAF mutation, however, the manufacturer information provided with commercially available panels of tests may be outdated or incomplete, leading to the potential for inappropriate clinical implementation of biomarker data. On a final cautionary note, oncologists must be aware that biomarkers with established predictive and/or prognostic value in one tumor type do not necessarily convey the same information in other tumor types.
In an era of exciting advances in molecularly targeted therapies in oncology, a robust, iterative communication between preclinical and clinical research is imperative to establish both the functional significance and the clinical validity of new biomarkers. Equally important, however, is the careful translation of this evidence into clinical practice to ensure the validity of the biomarkers used in clinical decision making. Frequently updated practice guidelines formulated by panels of experts, such as the NCCN Guidelines, are an invaluable and necessary tool to enable general oncology practitioners to interpret a rapidly evolving evidence base and identify the biomarkers that are appropriate for integration into practice.
Acknowledgments
This manuscript was supported in part by a grant from the National Cancer Institute (P01CA130818) through the Center for Translational and Policy Research on Personalized Medicine (TRANSPERS) at the University of California, San Francisco. Dr. Kelley was also supported by the Maisin Foundation. Robin K. Kelley, MD received an unrestricted educational grant from Genomic Health, Inc. for a pilot research study of the clinical utility of new molecular tests in colorectal cancer management, and an honorarium for participating in a speaker training program. Alan P. Venook, MD has held an uncompensated advisory role to and collaborated on research projects with Genomic Health, Inc.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 5.Douillard JY, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 6.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 7.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 8.McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK) Eur J Cancer. 2005;41:1690–1696. doi: 10.1016/j.ejca.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 9.Simon R. Clinical trial designs for evaluating the medical utility of prognostic and predictive biomarkers in oncology. Per Med. 2010;7:33–47. doi: 10.2217/pme.09.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NCCN Practice Guidelines in Oncology: Colon Cancer. Version 1. 2012 Available at: http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf. Accessed September 20, 2011.
- 11.NCCN Practice Guidelines in Oncology: Rectal Cancer. Version 1. 2012 Available at: http://www.nccn.org/professionals/physician_gls/PDF/rectal.pdf. Accessed September 20, 2011.
- 12.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 13.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fuorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fuorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertagnolli MM, Redston M, Compton CC, et al. Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III Colon cancer-a study of CALGB 9581 and 89803. J Clin Oncol. 2011;29:3153–3162. doi: 10.1200/JCO.2010.33.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samowitz WS, Curtin K, Ma KN, et al. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev. 2001;10:917–923. [PubMed] [Google Scholar]
- 18.Tol J, Dijkstra JR, Klomp M, et al. Markers for EGFR pathway activation as predictor of outcome in metastatic colorectal cancer patients treated with or without cetuximab. Eur J Cancer. 2010;46:1997–2009. doi: 10.1016/j.ejca.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 19.Bokemeyer C, Kohne C, Rougier P, et al. Cetuximab with chemotherapy (CT) as first-line treatment for metastatic colorectal cancer (mCRC): analysis of the CRYSTAL and OPUS studies according to KRAS and BRAF mutation status [abstract] J Clin Oncol. 2010;28(Suppl 1) Abstract 3506. [Google Scholar]
- 20.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 21.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 22.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 23.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med. 2009;361:98–99. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 24.Benson ABIII, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 25.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5527. [PubMed] [Google Scholar]
- 26.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 28.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham JM, Christensen ER, Tester DJ, et al. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 30.Cunningham JM, Kim CY, Christensen ER, et al. The frequency of hereditary defective mismatch repair in a prospective series of unselected colorectal carcinomas. Am J Hum Genet. 2001;69:780–790. doi: 10.1086/323658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 32.Kakar S, Aksoy S, Burgart LJ, Smyrk TC. Mucinous carcinoma of the colon: correlation of loss of mismatch repair enzymes with clinicopathologic features and survival. Mod Pathol. 2004;17:696–700. doi: 10.1038/modpathol.3800093. [DOI] [PubMed] [Google Scholar]
- 33.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 34.Lanza G, Ferracin M, Gafa R, et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007:6–54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 36.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 37.Kim GP, Colangelo LH, Wieand HS, et al. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767–772. doi: 10.1200/JCO.2006.05.8172. [DOI] [PubMed] [Google Scholar]
- 38.Des Guetz G, Schischmanoff O, Nicolas P, et al. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45:1890–1896. doi: 10.1016/j.ejca.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46:2788–2798. doi: 10.1016/j.ejca.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Julie C, Tresallet C, Brouquet A, et al. Identification in daily practice of patients with Lynch syndrome (hereditary nonpolyposis colorectal cancer): revised Bethesda guidelines-based approach versus molecular screening. Am J Gastroenterol. 2008;103:2825–2835. doi: 10.1111/j.1572-0241.2008.02084.x. [DOI] [PubMed] [Google Scholar]
- 41.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer, in press. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Roock W, Jonker DJ, Di Nicolantonio F, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 44.Cetuximab (Erbitux) and Panitumumab (Vectibix) Administration UFaD, ed. 2009 [Google Scholar]
- 45.Tanaka N, Huttenhower C, Nosho K, et al. Novel application of structural equation modeling to correlation structure analysis of CpG island methylation in colorectal cancer. Am J Pathol. 2010;177:2731–2740. doi: 10.2353/ajpath.2010.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quasar Collaborative G, Gray R, Barnwell J, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 47.Maughan TS, Adams R, Smith CG, et al. Identification of potentially responsive subsets when cetuximab is added to oxaliplatin-fuoropyrimidine chemotherapy (CT) in first line advanced colorectal cancer (aCRC): mature results of the MRC COIN trial [abstract] J Clin Oncol. 2010;28(Suppl 1) Abstract 3502. [Google Scholar]
- 48.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 49.Van Cutsem E, Lang I, Folprecht G, et al. Cetuximab plus FOLFIRI in the treatment of metastatic colorectal cancer (mCRC): the influence of KRAS and BRAF biomarkers on outcome: updated data from the CRYSTAL trial. Presented at the Gastrointestinal Cancers Symposium; Orlando, Florida. 2010. Jan 22–24, [Google Scholar]
- 50.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 51.Hecht JR, Mitchell E, Neubauer MA, et al. Lack of correlation between epidermal growth factor receptor status and response to Panitumumab monotherapy in metastatic colorectal cancer. Clin Cancer Res. 2010;16:2205–2213. doi: 10.1158/1078-0432.CCR-09-2017. [DOI] [PubMed] [Google Scholar]
- 52.Yin M, Yan J, Martinez-Balibrea E, et al. ERCC1 and ERCC2 polymorphisms predict clinical outcomes of oxaliplatin-based chemotherapies in gastric and colorectal cancer: a systemic review and meta-analysis. Clin Cancer Res. 2011;17:1632–1640. doi: 10.1158/1078-0432.CCR-10-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun MS, Richman SD, Quirke P, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690–2698. doi: 10.1200/JCO.2007.15.5580. [DOI] [PubMed] [Google Scholar]
- 54.Braun MS, Richman SD, Thompson L, et al. Association of molecular markers with toxicity outcomes in a randomized trial of chemotherapy for advanced colorectal cancer: the FOCUS trial. J Clin Oncol. 2009;27:5519–5528. doi: 10.1200/JCO.2008.21.6283. [DOI] [PubMed] [Google Scholar]
- 55.Koopman M, Venderbosch S, van Tinteren H, et al. Predictive and prognostic markers for the outcome of chemotherapy in advanced colorectal cancer, a retrospective analysis of the phase III randomised CAIRO study. Eur J Cancer. 2009;45:1999–2006. doi: 10.1016/j.ejca.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 56.Popat S, Matakidou A, Houlston RS. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. J Clin Oncol. 2004;22:529–536. doi: 10.1200/JCO.2004.05.064. [DOI] [PubMed] [Google Scholar]
- 57.Salonga D, Danenberg KD, Johnson M, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 58.Asghar U, Hawkes E, Cunningham D. Predictive and prognostic biomarkers for targeted therapy in metastatic colorectal cancer. Clin Colorectal Cancer. 2010;9:274–281. doi: 10.3816/CCC.2010.n.040. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W, Azuma M, Lurje G, et al. Molecular predictors of combination targeted therapies (cetuximab, bevacizumab) in irinotecan-refractory colorectal cancer (BOND-2 study) Anticancer Res. 2010;30:4209–4217. [PubMed] [Google Scholar]
- 60.Gutman S, Kessler LG. The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer. 2006;6:565–571. doi: 10.1038/nrc1911. [DOI] [PubMed] [Google Scholar]
- 61.Phillips KA, Van Bebber SL. Regulatory perspectives on pharmacogenomics: a review of the literature on key issues faced by the United States Food and Drug Administration. Med Care Res Rev. 2006;63:301–326. doi: 10.1177/1077558706287020. [DOI] [PubMed] [Google Scholar]
- 62.Feldman MD, Petersen AJ, Karliner LS, Tice JA. Who is responsible for evaluating the safety and effectiveness of medical devices? The role of independent technology assessment. J Gen Intern Med. 2008;23(Suppl 1):57–63. doi: 10.1007/s11606-007-0275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117:1847–1854. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosenberg R, Maak M, Simon I, et al. Independent validation of a prognostic genomic profile (ColoPrint) for stage II colon cancer (CC) patients [abstract] J Clin Oncol. 2011;29(Suppl 1) Abstract 358. [Google Scholar]
- 65.Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 66.Salazar R, Marshall J, Stork-Sloots L, et al. The PARSC trial, a prospective study for the assessment of recurrence risk in stage II colon cancer (CC) patients using ColoPrint [abstract] J Clin Oncol. 2010;28(Suppl 1) Abstract TPS199. [Google Scholar]
- 67.Cohen SJ, Punt CJ, Iannotti N, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol. 2009;20:1223–1229. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- 68.Rahbari NN, Aigner M, Thorlund K, et al. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 2010;138:1714–1726. doi: 10.1053/j.gastro.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 69.Tol J, Koopman M, Miller MC, et al. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann Oncol. 2010;21:1006–1012. doi: 10.1093/annonc/mdp463. [DOI] [PubMed] [Google Scholar]
- 70.Kerr D, Gray R, Quirke P, et al. A quantitative multigene RT-PCR assay for prediction of recurrence in stage II colon cancer: selection of the genes in four large studies and results of the independent, prospectively designed QUASAR validation study [abstract] J Clin Oncol. 2009;27(Suppl 1) Abstract 4000. [Google Scholar]
- 71.Venook AP, Niedzwiecki D, Lopatin M, et al. Validation of a 12-gene colon cancer recurrence score (RS) in patients (pts) with stage II colon cancer (CC) from CALGB 9581 [abstract] J Clin Oncol. 2011;29(Suppl 1) Abstract 3518. [Google Scholar]