Abstract
Malaria afflicts over 200 million people worldwide and its most lethal etiologic agent, Plasmodium falciparum, is evolving to resist even the latest-generation therapeutics. Efficient tools for genome-directed investigations of P. falciparum pathogenesis, including drug resistance mechanisms, are clearly required. Here we report rapid and targeted genetic engineering of this parasite, using zinc-finger nucleases (ZFNs) that produce a double-strand break in a user-defined locus and trigger homology-directed repair. Targeting an integrated egfp locus, we obtained gene deletion parasites with unprecedented speed (two weeks), both with and without direct selection. ZFNs engineered against the endogenous parasite gene pfcrt, responsible for chloroquine treatment escape, rapidly produced parasites that carried either an allelic replacement or a panel of specified point mutations. The efficiency, versatility and precision of this method will enable a diverse array of genome editing approaches to interrogate this human pathogen.
INTRODUCTION
By current estimates, 655,000 individuals die each year from severe malaria caused by the protozoan parasite Plasmodium falciparum. Recent reductions in the global disease burden, brought about by artemisinin-based combination therapies and anti-mosquito measures, are at threat of being overturned by resistance1. Defining molecular pathways of malaria pathogenesis and identifying mechanisms of treatment failure require precise manipulation of the parasite genome to rapidly and efficiently generate knockouts, targeted integrants, and allelic replacements of genes associated with disease progression. Existing methods for genomic modification of P. falciparum, while having yielded important insights, do not adequately meet these challenges2,3.
Genome editing with customized zinc-finger nucleases (ZFNs), first achieved in Drosophila4, has been successfully applied to a range of multicellular organisms5. To achieve editing, pairs of zinc-finger proteins with unique specificity for adjacent sequences on either strand of the DNA helix are linked to an endonuclease (FokI) that functions as an obligate heterodimer6,7. Engineered zinc-finger proteins typically consist of tandem arrays of up to six individual fingers that recognize 3–4 bp of DNA. Nuclease activity is dependent on the left and right zinc-finger proteins binding to each side of the chromosomal target with a precise orientation and spacing8. ZFNs induce a double-strand break (DSB) that can alter the target, either by activating the error-prone non-homologous end-joining (NHEJ) pathway or by stimulating homologous recombination when a donor template is provided. In metazoans, NHEJ can produce nuclease-mediated gene disruptions8. However, P. falciparum lacks several critical components of this pathway (Ku70/80 and DNA Ligase IV9; our unpublished observations). Thus, homologous recombination is likely the primary pathway of DSB repair in this parasite10, and gene-targeting approaches using this process have been successful, albeit highly inefficient3. We hypothesized that ZFNs could provide the needed breakthrough in efficiency by introducing DSBs in a specified target site at a much higher frequency than occurs stochastically. Here, we establish methods of genome editing in P. falciparum by defining conditions for optimal expression of ZFNs, developing strategies to edit a phenotypic marker with or without direct selection, and introducing a specific set of polymorphisms into an endogenous locus relevant to drug resistance.
RESULTS
2A peptide permits dual protein synthesis in P. falciparum
Directed genome editing requires the coordinated expression of two ZFNs that heterodimerize to cleave a unique site. Given the paucity of selectable markers for P. falciparum and the instability of propagating large plasmids containing AT-rich Plasmodium DNA in Escherichia coli3, we first tested whether a plasmid-encoded ZFN pair could be co-expressed from a single promoter using a viral 2A “ribosome skip” peptide. This short self-processing peptide permits polycistronic expression of individual polypeptides11. We generated transgenic parasites expressing a mrfp-2A–gfp fusion driven by the calmodulin promoter (Fig. 1a). Both protein products were detected in parasites by fluorescence microscopy, and immunoblotting for the downstream GFP reporter revealed its expression as the 27 kDa monomer (Fig. 1a). Deletion of a key proline residue in the 2A peptide crippled co-translational release, resulting in a mRFP-2AΔP-GFP fusion protein with a molecular mass slightly greater than the conventional fusion of these two proteins (Fig. 1a). These findings confirm efficient ribosomal skipping across the 2A site and illustrate its utility for dual protein expression from a single promoter in P. falciparum.
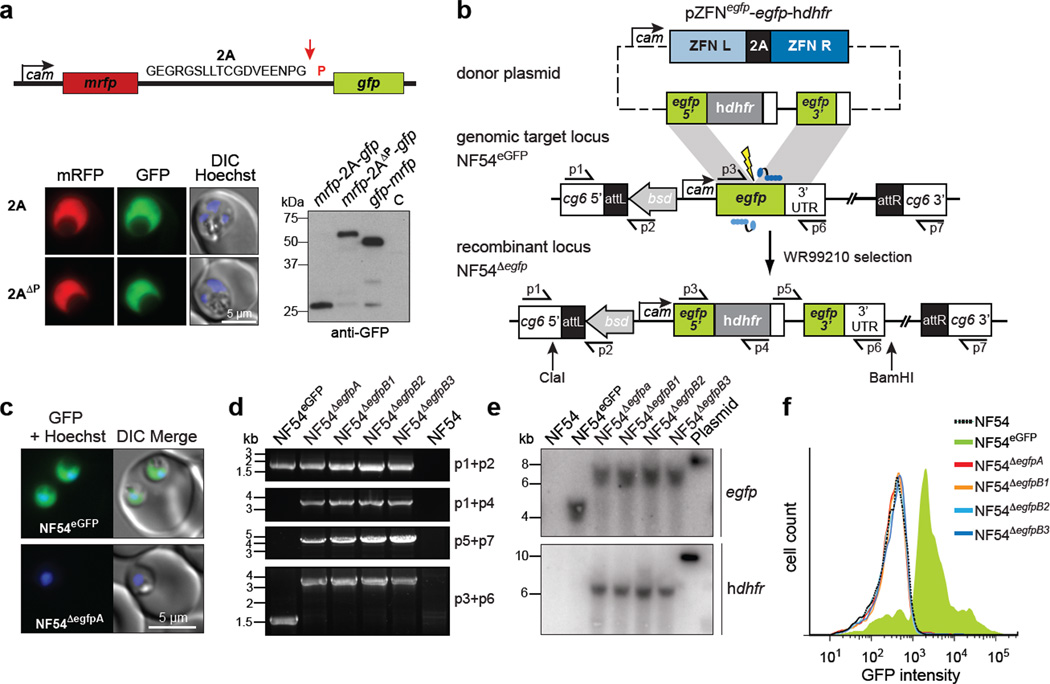
Figure 1. 2A-linked ZFNs drive efficient disruption of egfp in P. falciparum .
a) Coexpression of 2A–linked mRFP and GFP monomers from a single calmodulin (cam) promoter is demonstrated by fluorescence microscopy and immunoblotting for GFP. The red arrow indicates the ribosome skip site. C, control untransfected parasites. b) Strategy to disrupt egfp integrated at the genomic cg6 locus. The donor plasmid encodes 2A-linked left (L) and right (R) ZFNs in addition to egfp homologous regions flanking the ZFN target site (thunderbolt). Repair of the ZFN-induced DSB, via homology-directed repair using the donor as template, yielded an in-frame integration of hdhfr into the egfp locus. c) The micrographs show EGFP expression in the parental line NF54EGFP but not in the recombinant line NF54ΔegfpA. Nuclei were stained with Hoechst 33342. d) PCR analysis of the ZFN-transfected lines NF54ΔegfpA-B3 and the parental line NF54EGFP using the primers indicated in panel b. e) Southern blot hybridization of genomic DNA digested with ClaI + BamHI (panel b) demonstrates integration of hdhfr in the ZFN-transfected lines (lower panel) and the expected 2 kb size increase at the disrupted egfp locus (upper panel) f) Flow cytometry reveals complete loss of EGFP signal in the ZFN-modified parasite populations.
ZFNs mediate gene ablation in P. falciparum parasites
We set out to achieve ZFN-mediated gene disruption in P. falciparum by targeting a chromosomal egfp locus, yielding a readily quantifiable phenotype. First, we integrated the target egfp gene into the cg6 locus of NF54 parasites using attB×attP integrase-mediated recombination12, generating the NF54EGFP line that was uniformly EGFP-positive (Fig. 1b – d). We then designed a composite pZFNegfp-egfp-hdhfr donor plasmid, comprising our 2A–linked egfp-specific ZFN expression cassette as well as two homology regions (egfp 5′ and 3′) that flanked the ZFN cleavage site. To select for egfp-disrupted parasites, the 5′ homology region was fused in frame with human dihydrofolate reductase (hdhfr)13, such that resistance to the antifolate drug WR99210 would be contingent on the egfp-hdhfr fusion integrating downstream of the genomic calmodulin promoter (Fig. 1b). Importantly, targeted integration of hdhfr into the egfp locus should produce a GFP-negative parasite.
Following electroporation of the ZFN donor plasmid into the NF54EGFP line, parasites were selected directly with WR99210 (yielding the NF54ΔegfpA line) or first supplemented with red blood cells preloaded with plasmid DNA (reported to increase transformation efficiency14 and yielding the three lines NF54Δegfp-B1/B2/B3). With all four lines, WR99210-resistant parasites were detected on day 15 post-electroporation. We confirmed ZFN-driven disruption of the egfp gene by fluorescence microscopy, PCR and Southern blotting (Fig. 1c–e). Flow cytometry of the bulk cultures revealed the complete loss of fluorescence in all NF54Δegfp lines (Fig. 1f). Three independent transfections with a ZFN-deficient control pegfp-hdhfr plasmid failed to yield parasites after 60 days.
To assess potential off-target activity of the ZFNs, we sequenced the genomes of two recombinant lines (NF54ΔegfpA and NF54ΔegpfB1) as well as the parent (NF54EGFP). Sequence analysis revealed a depth of coverage of hdhfr (56 × and 42 × for NF54ΔegfpA and NF54ΔegfpB1 respectively) that mirrored the average coverage across the entire genome (54 × and 69 ×), consistent with the presence of a single genomic copy of hdhfr (Supplementary Table 1). Furthermore, flanking sequence reads that partially overlapped hdhfr could only be mapped to the egfp-hdhfr locus, consistent with the specific disruption of egfp (Supplementary Fig. 1a, b).
Gene replacement in the absence of a selectable phenotype
Gene disruption by in-frame integration of a selectable marker is limited to targets that are expressed during the asexual blood stage. We sought to develop a broader strategy for gene manipulation, irrespective of expression pattern during the parasite life cycle and independent of a selection event. We first aimed to replace the egfp reporter with monomeric rfp (mrfp) fused to the cytosolic ATPase pfvps4. This fusion was placed on a donor plasmid (pmrfp-vps4) flanked by egfp untranslated regions (UTRs) and plasmid backbone sequences (3.5 kb and 2.8 kb on the 5′ and 3′ ends respectively) that provided templates for homologous recombination (Fig. 2a). ZFNs were expressed from a separate plasmid (pZFNegfp-hdhfr) containing the hdhfr selectable marker. The plasmids were co-electroporated, and WR99210 pressure applied for 6 days to transiently enrich for parasites that expressed the ZFNs. Parasite proliferation was detected microscopically 12 days post-electroporation.
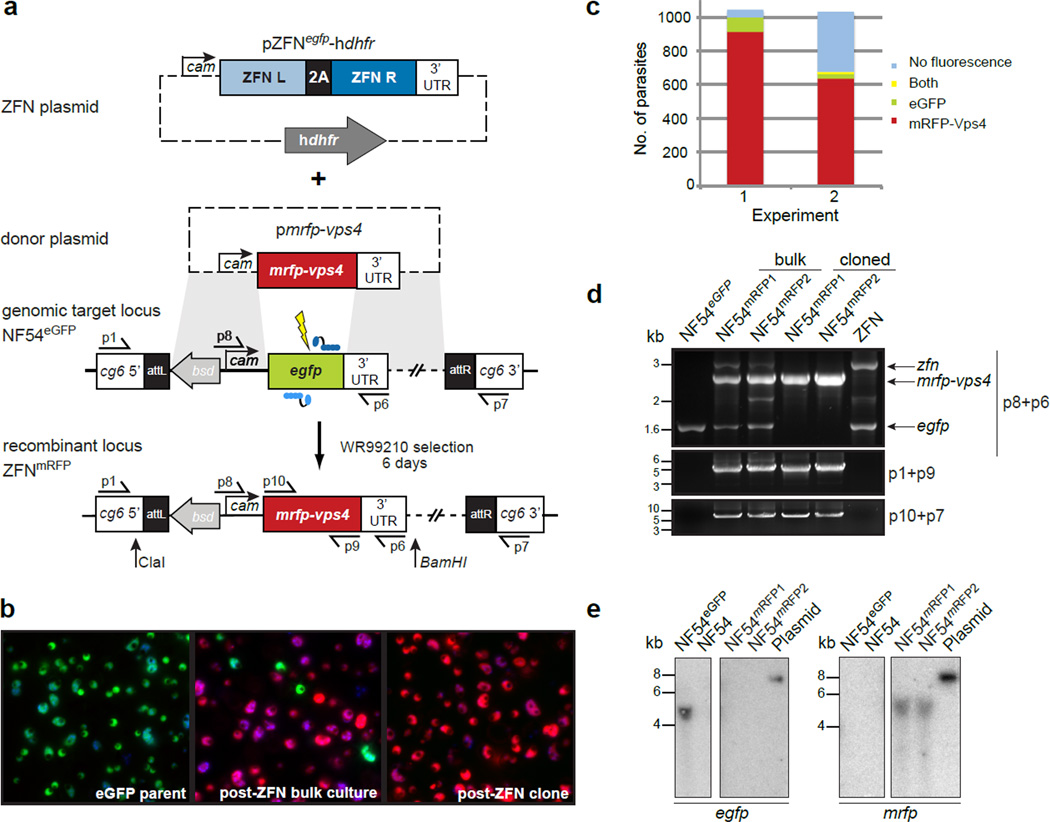
Figure 2. ZFNs mediate efficient gene replacement of egfp.
a) Schematic of the egfp replacement strategy. ZFNs were expressed from the calmodulin promoter on the pZFNegfp-hdhfr plasmid and cotransfected with the mrfp-vps4 donor sequence. Homology-directed repair of the ZFN-induced DSB, using the flanking regions on the donor as template, resulted in replacement of egfp with the mrfp-vps4 fusion construct. b) The parental line NF54EGFP, a post-ZFN bulk culture and a clonal line were imaged for EGFP and mRFP expression. Nuclei were stained with Hoechst 33342. c) Quantification of parasite fluorescence in the bulk culture in two independent experiments (n = 1042 and n = 1032). d) PCR analysis of parental NF54EGFP and ZFN-transfected parasites bulk culture and individual parasite clones. Primer positions are shown in panel a. e) Southern blot hybridization of genomic DNA from the indicated parasite lines digested with ClaI + BamHI (panel a), using an egfp probe (left panel) and a mrfp probe (right panel). Linearized transfection plasmids served as positive controls.
Imaging and quantification of parasite fluorescence from the bulk cultures was consistent with a gene replacement efficiency of 88 % and 62 % in two independent experiments (Fig. 2b,c). This level of efficiency was confirmed by analysis of clonal lines, which expressed mRFP and not EGFP in 19/27 (70 %) and 21/39 (54 %) of cloned parasites from the two experiments. This recombination event involves DNA end resection of greater than 260 bp from at least one side of the DSB, leading to invasion of the mrfp flanking sequences common to both the donor plasmid and the chromosomal egfp locus (Fig. 2a). These flanking sequences were shared with the ZFN expression vector, which could compete with the pmrfp-vps4 plasmid as a template for homology-directed repair and could account for the minority of non-fluorescent parasites observed in the bulk cultures (Fig. 2c). PCR and Southern blot analyses nonetheless confirmed replacement of egfp with the mrfp fusion in the majority of parasites, shown in two representative clones (Fig. 2d,e).
Allelic replacement of an endogenous parasite gene
We next sought to employ ZFNs for allelic replacement at an endogenous parasite locus. An important drug resistance determinant in P. falciparum is the chloroquine (CQ) resistance transporter PfCRT15, which localizes to the digestive vacuole where hemoglobin degradation and formation of toxic CQ-heme adducts occurs16. The extensive worldwide use of CQ in malaria treatment has led to the selection of multiple mutations in pfcrt, generating geographically distinct alleles17. Genetic engineering of isogenic parasites expressing various pfcrt alleles is required to fully analyze their phenotypic impact on drug response, but this has proven exceptionally time- and labor-intensive18,19.
We designed ZFNs against pfcrt using an archive of pre-validated modules, each consisting of two zinc fingers8,20. Based on results from yeast proxy cleavage assays21 (data not shown), we selected the two most active nuclease pairs (13/15 and 14/15; Supplementary Table 2). These pairs, which target the boundary of intron 1 and exon 2, were cloned into a plasmid expressing a blasticidin S-deaminase (bsd) selectable marker, yielding pZFNcrt-bsd (Fig. 3a). The pfcrt donor sequence was inserted on a second plasmid (pcrtDd2-hdhfr), consisting of the pfcrt cDNA from the CQ-resistant (CQR) strain Dd2 and the 3′ UTR from the P. berghei crt ortholog, followed by a hdhfr expression cassette that served as an independent selectable marker. Upstream and downstream regions of homology, derived from the pfcrt promoter and terminator sequences, flanked these elements to promote ZFN-mediated replacement of the entire 3.1 kb gene with the donor-provided pfcrt 1.2 kb cDNA and the downstream hdhfr selectable marker (Fig. 3a).
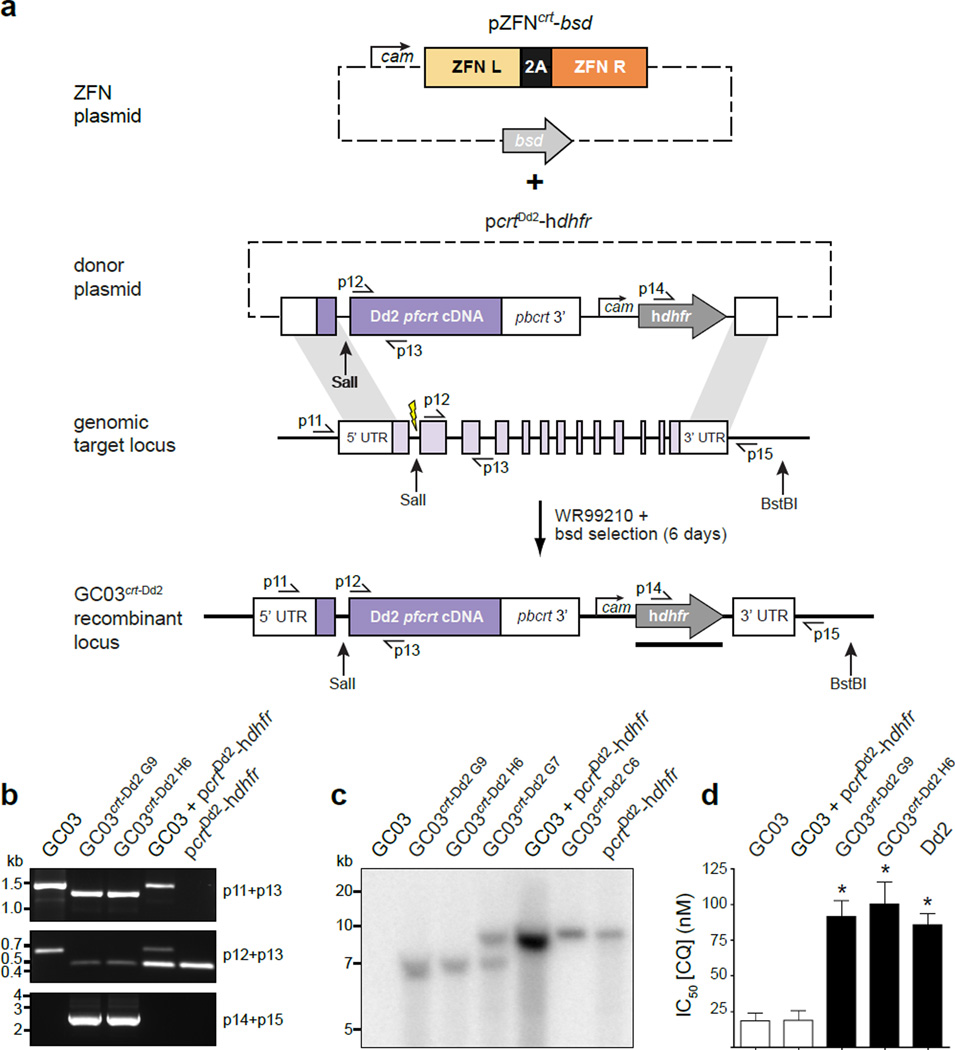
Figure 3. ZFN-driven allelic replacement of pfcrt.
a) Schematic of pfcrt allelic replacement strategy. The pZFNcrtbsd plasmid encodes pfcrt-specific ZFNs, driven by the calmodulin promoter. The pcrtDd2-hdhfr donor plasmid contains the 1.2 kb coding sequence of the Dd2 pfcrt allele, followed by 0.7 kb of the pbcrt 3′ UTR, and the hdhfr selectable marker. These cassettes are flanked by two homology regions: 0.4 kb upstream of the DSB and 1 kb of the pfcrt 3′ UTR. ZFN-driven homology-directed repair yielded the pfcrt-modified GC03crt-Dd2 locus. b) PCR analysis of two independent clones. Primer positions are shown in panel a. c) Southern blotting of genomic DNA from the indicated parasite lines digested with SalI + BstBI and probed for hdhfr (black bar in panel a). The band size (6.7 kb) observed with clones G9 and H6 is consistent with pfcrt replacement. The pcrtDd2-hdhfr plasmid was linearized with SpeI (8.1 kb) and is present as episomes in clones G7 and C6, as well as the ZFN-minus control line, GC03 + pcrtDd2-hdhfrd) The plot shows half-maximal inhibitory concentration (IC50 ± SEM) values for the indicated parasite lines. Asterisks indicate significant differences between the two representative pfcrt allelic replacement clones GC03crt-Dd2G9 and GC03crt-Dd2H6 and the GC03 parental line (*P = 0.0286, Mann-Whitney U test, two-tailed, n = 4).
We chose to modify the CQ-sensitive (CQS) strains 106/1 and GC03, which harbor distinct alleles and exhibit characteristic drug response phenotypes15. Instead of conventional co-transfection, we first electroporated the donor plasmid pcrtDd2-hdhfr and applied WR99210 to select for episomally transformed parasites (Fig. 3a). These parasites were then electroporated with pZFNcrt-bsd, and blasticidin was applied for 6 days to enable transient ZFN expression and consequent homology-directed repair. Prolonged selection for the ZFN plasmid (12 days) caused a delay in parasite re-emergence post-electroporation (data not shown), potentially due to repeated chromosome cleavage. After removal of blasticidin, but not WR99210, parasite proliferation was detected microscopically after 13–16 days. To quantify the efficiency of pfcrt allelic replacement, clones were generated by limiting dilution and analyzed by PCR. We observed replacement events in 13/82 (15.9 %) 106/1 clones and 4/83 (4.8 %) GC03 clones (Fig. 3b). Southern blotting of two representative clones (GC03crt-Dd2 G9 and GC03crt-Dd2 H6) demonstrated acquisition of the donor-provided CQR pfcrt allele (Fig. 3c). We confirmed the CQ resistance phenotype of these two clones, which both displayed a 4– to 5– fold shift in CQ IC50 values compared to the GC03 parent (Fig. 3d). Notably, in three independent transfections, 106/1 and GC03 parasites that only received the pfcrt donor plasmid but not the ZFN plasmid failed to yield allelic replacement parasites after more than 6 months.
Site-specific editing of a parasite drug resistance locus
We next assessed whether our engineered pfcrt-targeted ZFNs could drive a subtle gene-editing event that delivers a single point mutation to the targeted site from an episomal donor template. In contrast, conventional allelic exchange strategies for P. falciparum typically result in significant modification of the endogenous locus by crossover-mediated incorporation of the entire plasmid (often as a concatamer), including a selectable marker and other sequence elements3.
To achieve gene editing in P. falciparum, we exploited the CQ resistance-conferring properties of mutant pfcrt. PfCRT mediates resistance by effluxing CQ from the digestive vacuole, dependent on mutation of residue K76 to T (in the case of field isolates) or I (observed in CQ-pressured 106/1 parasites15,22,23). pfcrt alleles from CQR parasite strains also possess at least 3 additional, potentially compensatory mutations24. For this experiment, we chose the CQS 106/1 strain whose pfcrt allele encodes six out of seven CQR mutations observed in Asian and African strains while retaining the CQS K76 codon (Fig. 4a). Based on prior studies15,22, mutation of pfcrt codon 76 from K to I in this isolate was predicted to confer resistance.
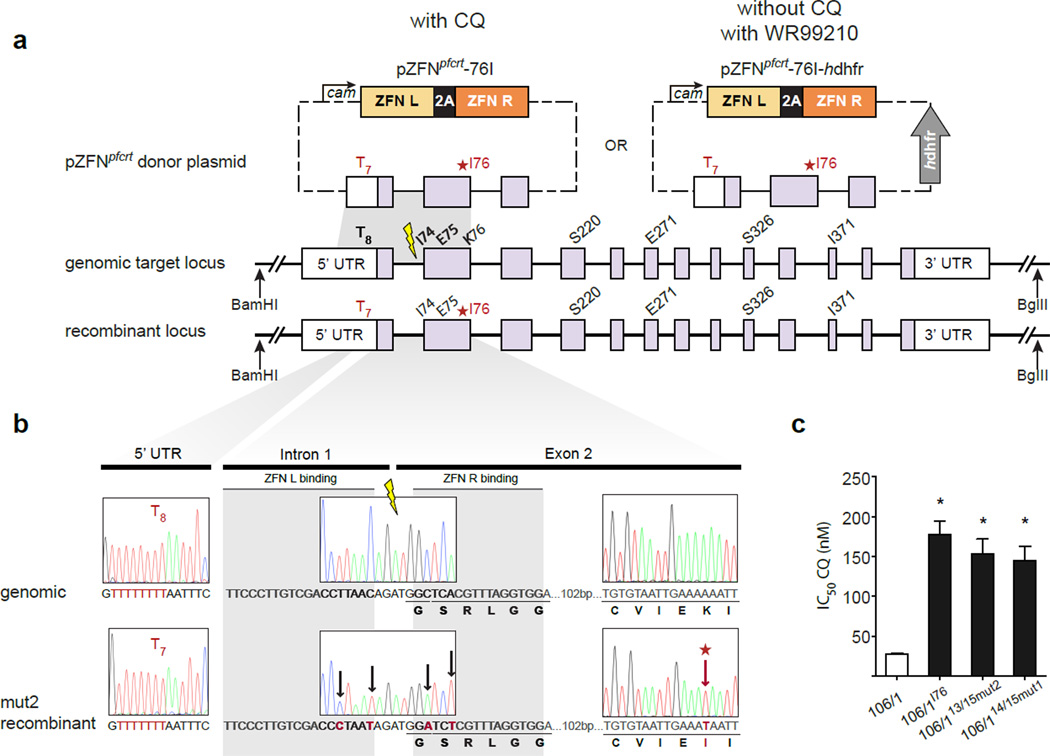
Figure 4. ZFN editing of pfcrt with and without chloroquine selection.
a) Schematic of pfcrt editing strategy. The calmodulin promoter drives expression of the pfcrt-specific ZFN pairs from plasmids with (pZFNcrt-76I–hdhfr) or without (pZFNcrt-76I) the selectable marker hdhfr. The homologous donor sequence for DSB repair comprises a fragment of pfcrt stretching 0.4 kb upstream and 0.6 kb downstream of the ZFN target site (thunderbolt). One version of the donor (termed mut1) is identical to the genomic locus but contains the mutant I76 codon (starred) conferring CQ resistance, and a single nucleotide deletion, T7 versus T8, in the endogenous 5′ UTR. An alternate donor construct (mut2, not shown) is mutated at the ZFN binding site. Homology-dependent repair of a ZFN-induced DSB leads to incorporation of donor-provided SNPs. The chromatograms show sequence analysis of genomic and mut2 recombinant DNA. The 5′ UTR deletion as well as the mutations at the ZFN binding site and the CQ resistance-conferring I76 codon are indicated. c) The plot shows half-maximal inhibitory concentration (IC50 ± SEM) values for the indicated parasite lines. The asterisk indicates that the 106/1 parental line is significantly different (P < 0.0286, Mann-Whitney U test, n = 4, two-tailed) from the gene-edited parasites as well as the previously established CQR line 106/1I76
We generated the ZFN expression plasmid pZFNcrt-bsd with a 1 kb pfcrt donor sequence that spans the ZFN cleavage site and encodes the K76I mutation (yielding the plasmid pZFNcrt-76I) (Fig. 4a). We tested two versions of the donor sequence: one with an intact ZFN binding site (“mut1”), and another with four silent mutations (“mut2”). The latter was designed to prevent ZFN binding and cleavage of a successfully modified chromosomal target, thereby potentially enhancing editing efficiency. One day post-electroporation, 106/1 parasites were placed under 33 nM CQ to eliminate unmodified CQS parasites. Parasite proliferation was detected microscopically 16 to 33 days post-electroporation. In contrast, similar CQ exposure of six independent non-transfected 106/1 cultures (beginning with parasite numbers equivalent to those used for ZFN-mediated gene editing) yielded no parasites after 90 days.
To confirm acquisition of the K76I mutation in transfected parasites, we PCR-amplified the pfcrt locus from the bulk culture and subcloned these products for sequence analysis. In five independent parasite transfections, we observed 100 % K76I conversion rates (Fig. 4a; Table 1). No alternate mutations were detected at this position. Editing of the K76 codon was equally successful using either ZFN pair (13/15 or 14/15, Supplementary Table 2), irrespective of whether or not the ZFN binding site was mutated in the donor construct (Table 1). Of note, the additional 4 silent mutations in the mut2 template were always incorporated at the pfcrt locus. In addition, both the mut1 and mut2 donor templates contained a pfcrt 5′ UTR sequence harboring a single base pair deletion (a string of seven Ts (T7), compared to T8 in the endogenous locus). This deletion, located ∼300 bp upstream of the ZFN cut site, was transferred into the edited gene sequence with a mean efficiency of 51 % (Table 1). By comparison, mutations located an equivalent distance from the ZFN cleavage site have been captured with considerably lower frequency in mammalian cells (e.g. 5 % in mouse embryonic stems cells25). Importantly, the T7 deletion was captured despite its presence on the side opposite the DSB relative to the selected K76I mutation (located 140 bp downstream of the ZFN cleavage site). Incorporation into the chromosomal target of all mutations on the donor plasmid could be explained by gene editing proceeding via synthesis-dependent strand annealing or other non-crossover events (Fig. 4a; Supplementary Fig. 2)10.
Table 1.
Efficiency of ZFN-mediated gene editing of pfcrt
| Successful editing event |
|||||||
|---|---|---|---|---|---|---|---|
| Strain | ZFN pair | Donor | Sequences analyzed |
Binding Site mutations |
K76I | T (deletion) |
|
| CQ | 106/1 | 13/15 | pcrt-76I–mut1 | 29 pGEM-T | N/A | 29/29 | 0/29 |
| 106/1 | 13/15 | pcrt-76I–mut2 | 25 pGEM-T | 25/25 | 25/25 | 9/25 | |
| 106/1 | 14/15 | pcrt-76I–mut1 | 38 pGEM-T | N/A | 38/38 | 38/38 | |
| 106/1 | 14/15 | pcrt-76I–mut1 | 28 pGEM-T | N/A | 28/28 | 28/28 | |
| 106/1 | 14/15 | pcrt-76I–mut2 | 31 pGEM-T | 31/31 | 31/31 | 6/31 | |
| Targeting efficiency with CQ selection: | 100 % | 100 % | 51 % | ||||
| no CQ | Dd2 | 13/15 | pcrt-76I–mut2 | 36 parasite lones | 4/36 | 2/36 | 4/4* |
| Dd2 | 14/15 | pcrt-76I–mut2 | 40 parasite clones | 10/40 | 10/40 | 10/10* | |
| Targeting efficiency without CQ selection: | 18.4 % | 15.8 % | (18.4 %) | ||||
| Distance from ZFN cut site: | 3–6 bp | 140 bp | 296 bp | ||||
The table shows ZFN mediated gene-targeting efficiency in the presence and absence of chloroquine. The site of cleavage of the ZFN pairs is described in Fig. 4. The pfcrtI76-mut1 donor contains the K76I CQ resistance mutation, in addition to a single nucleotide (T) deletion in the 5’ UTR of pfcrt, located 0.3 kb upstream of the ZFN cut site. pfcrtI76-mut2 differs from pfcrtI76-mut1 by the presence of four additional mutations in the ZFN binding site to prevent recleavage of the integrated donor sequence and the donor itself (see Fig. 4). To analyze gene-targeting efficiency, the pfcrt locus was PCR amplified from either a bulk culture and the products cloned into pGEM-T or from parasites cloned by limiting dilution and sequenced.
only lines identified as edited at the binding site were analyzed with a second sequencing reaction for the T (indel).
We confirmed the CQ resistance phenotype of two gene-edited lines, 106/113/15mut2 and 106/114/15mut1 (Fig. 4b). Both lines displayed a 5- to 6-fold shift in CQ IC50 values relative to the 106/1 parental strain. This shift in drug response was comparable to a CQR mutant of 106/1 (termed 106/1I76) bearing the K76I mutation, derived by CQ pressure15,22.
Whole-genome sequencing revealed no detectable off-target activity of the pfcrt-targeting ZFN pairs in two representative recombinant lines (106/113/15mut1 and 106/114/15mut1). Illumina next-generation sequencing yielded a 15 × coverage for > 97 % of all three genomes (Supplementary Table 1). We found no evidence of any rearrangement of the pfcrt locus in these edited lines, and confirmed 100% incorporation of the K76I mutation (Supplementary Fig. 1c).
To demonstrate the applicability of ZFNs to generate SNPs that may not confer a selectable phenotype, we repeated the ZFN-mediated pfcrt K76I editing event described above without applying CQ pressure. To select for transfected parasites and ensure ZFN expression, we added the hdhfr selection cassette to the mut2 version of the pZFNcrt-76I plasmid (yielding pZFNcrt-76I–hdhfr) (Fig. 4a). Transfected Dd2 parasites were selected with WR99210 for 6 days, and parasite proliferation was observed 11 days after removal of drug. From two independent experiments, we generated a total of 76 clones and used these to PCR-amplify the pfcrt genomic locus (Table 1). This analysis identified the ZFN binding site mutations in 18.4 % and the K76I mutation in 15.8 % of clones. The upstream T7 deletion was also found in all edited clones. These data suggest that non-selected gene editing events can be generated with sufficient efficiency to readily permit the isolation of modified parasite clones by limiting dilution, thus expanding the range of potential targets beyond those related to drug resistance.
DISCUSSION
Here, we have established diverse ZFN-driven strategies with broad utility for interrogating the P. falciparum genome. These include gene disruption and replacement approaches that can be used to verify the essentiality of parasite genes, and site-specific editing approaches to probe gene function. ZFN-mediated genome editing of P. falciparum differs fundamentally from prior methods of genetic manipulation, which have typically relied on stochastic integration of plasmids designed for gene disruption or allelic exchange via homologous recombination (Supplementary Fig. 2)26. In these cases, integration likely takes place after a rare and random DSB occurs in the gene of interest. Plasmid integration is typically observed after 2–4 months, and any fitness costs resulting from genome manipulation jeopardize the acquisition of the recombinant line. Extreme cases include engineering the pfcrt CQ resistance locus, which required two rounds of genetic manipulation and over 18 months of continuous culture to obtain the desired clones18. Furthermore, the non-directed nature of this earlier method of allelic exchange meant that integration could take place at any genomic site with homology to the transfection plasmid, often yielding undesired outcomes. Even if integration occurs into the desired locus, single-site crossovers could result in a recombinant locus that harbors the plasmid-borne mutations in a non-functional duplicated fragment and not the functional gene27.
Using the approaches we describe, parasites carrying the desired specific editing event can be obtained rapidly and with an efficiency that approaches 100 % in cases where the outcome yields a selectable phenotype. Importantly, we were able to readily obtain gene deletion, allelic exchange and single nucleotide editing events even in the absence of a selectable phenotype. The efficiencies of these editing events ranged from relatively high in the case of the egfp-mrfp replacement, when ∼3 kb homology arms were provided either side of the target locus, to more modest levels (∼18 % efficiency) for the pfcrt K76I editing, when a 1 kb sequence was utilized as the donor template. These editing efficiencies were sufficient to readily isolate parasite clones bearing the desired modifications. Furthermore, the capture of mutations several hundred base pairs from the DSB site provides some flexibility when selecting ZFN pairs. The shortest region of homology was 0.5 kb (for the egfp disruption strategy), and the potential increase in editing efficiency by using longer regions must be balanced by constraints on the size and stability in E. coli of AT-rich Plasmodium transfection plasmids.
ZFN-mediated editing can now be leveraged to generate knockouts or more subtle modifications (including fusions with fluorescent reporters20 or affinity tags) that probe parasite gene function across a broad range of biological contexts. This includes the ability to inactivate genes without integration of a selectable marker, fulfilling an important requirement for genetically attenuated vaccines. ZFNs can also be deployed for sequence-specific editing28, for example to assess the involvement of candidate loci and identify causal polymorphisms to survey the spread of antimalarial drug resistance in endemic areas. ZFN-mediated genome editing now adds a powerful reverse-genomic methodology for dissecting the etiologic agent of a major human infectious disease.
METHODS
Zinc-finger nuclease engineering
ZFNs specific for egfp have been described previously29. ZFNs targeting pfcrt (MAL7P1.27) were designed and assembled using an archive of validated modules8, and screened for activity using a yeast proxy system21.
Plasmid construction
To test whether the Thosea asigna virus 2A peptide11 could mediate co-translational polypeptide separation in P. falciparum, we generated the pDC2-cam-mrfp-2A–gfp-attP plasmid (whose expression cassette is illustrated in Fig. 1a). The 2A and mrfp fragments were first amplified separately and linked by “splicing by overlap extension” (SOE) PCR. The 2A sequence was amplified from the plasmid pVAX-ZFN-L-2A–ZFN-R21 using primers p18 + p19 (Supplementary Table 3), and mrfp was amplified from pML230 using primers p21 + p22. The linked mrfp-2A sequence was amplified using primer p21 and either p19 (full 2A sequence) or p20 (2AΔP21). Finally, the mrfp-2A fusions were digested with AvrII and BglII and inserted into a pDC2-based expression vector30 following excision of the 5′ gfp full-length sequence in the tandem gfp-gfp plasmid pDC2-cam-gfp(N-C)-attP. To generate the construct used to produce the NF54EGFP line, the egfp coding sequence was amplified from the His-EGFP-FokI plasmid30 using primers p23 + p24 and cloned into the AvrII and XhoI restriction sites of pDC2-cam-gfp(N-C)-attP. This cloning yielded the egfp integration plasmid pDC2-cam-egfp-attP used to generate the NF54EGFP line. Expression of ZFNs in P. falciparum was achieved by cloning these downstream of a calmodulin (Pf14_0323) promoter and upstream of an hsp86 (PF07_0030) 3′ UTR in a pDC2 vector with or without the hdhfr or bsd selectable marker. ZFNs linked with the 2A peptide were digested with NheI and XhoI and cloned into the compatible restriction sites AvrII and XhoI, yielding the intermediate plasmids pZFNegfp and pZFNcrt and the final transfection plasmids pZFNegfp-hdhfr and pZFNcrt-bsd. The donor templates provided for the ZFN-induced DSB repair were either placed on the same plasmid as the ZFN expression cassette or on a separate plasmid, as indicated below.
To generate the egfp-hdhfr donor template (Fig. 1b), we first amplified 5′ and 3′ regions of homology flanking the ZFN cleavage site in the integrated egfp locus. The 5′ region was PCR amplified from pDC2-cam-egfp-attP using primers p3 + p25 (yielding a 543 bp product), while the 3′ region (denoted egfp 3′) was amplified using primers p27 + p28 (yielding a 795 bp product). A promoter-less hdhfr selection cassette was then amplified with primers p26 + p4 and fused in frame to egfp 5′ using primers p3 + p4 in a SOE PCR reaction. The egfp-hdhfr template was then cloned into pZFNegfp using ApaI and SacII restriction sites. The second homologous region egfp 3′ was then inserted using the restriction sites BstAPI and ZraI, yielding the transfection plasmid pZFNegfp-egfp-hdhfr (Fig. 1b). To produce the pegfp-hdhfr control plasmid we excised the egfp-specific ZFN segment following HindIII digestion of pZFNegfp-egfp-hdhfr and agarose gel electrophoresis and then religated the plasmid.
To assemble the mrfp-vps4 donor template (Fig. 2a), we amplified mrfp and vps4 separately and then linked the two products by SOE PCR. The mrfp sequence was amplified from pML230 using primers p29 + p30, and vps4 was amplified from NF54 genomic DNA using primers p31 + p32. The linked mrfp-vps4 sequence was amplified with p29 + p32 and cloned into the AvrII and XhoI restriction sites of pDC2, yielding the replacement plasmid pmrfp-vps4.
To generate the pcrtDd2-hdhfr donor plasmid for pfcrt allelic replacement (Fig. 3a), we first amplified intron 1 through exon 13 of pfcrt Dd2 cDNA and pbcrt 3′ UTR from an expression plasmid31 using p33 + p34. The PCR product was cloned into the restriction sites NotI and SpeI of the pCC1 plasmid (containing a hdhfr cassette). The left homology region, comprising the pfcrt 5′ UTR to intron 1 sequence (431 bp), was subcloned from the pZFNcrt-76I plasmid with ApaI and SalI (see below). The pfcrt 3′ UTR served as right region of homology and was amplified from 106/1 gDNA using primers p35 and p36 (yielding a 1050 bp product) and then inserted into EcoRI and ClaI to generate the allelic replacement plasmid pcrtDd2-hdhfr (Fig. 3a).
We generated two donor constructs to introduce a single point mutation (K76I) in pfcrt, located 140 bp downstream of the ZFN cut site. A 1 kb donor fragment without additional mutations at the ZFN target site (mut1) was PCR amplified from genomic DNA isolated from 106/1I76 parasites15 using primers p37 + p38 and cloned into the ApaI and SacI restriction sites in pZFNcrt. We also generated a second donor template with four silent mutations at the ZFN binding site (mut2) via SOE PCR using primers p37 + p39 and p38 + p40 for the first round of PCR and p37 + p38 for the second round (Supplementary Table 3). The same restrictions sites ApaI and SacI were used to clone donor fragment mut2. Both donor constructs contained a single nucleotide (T) deletion in the 5’ UTR of pfcrt located 0.3 kb upstream of the ZFN cut site. The two ZFN pairs (13/15 and 14/15; Supplementary Table 2) were expressed from a plasmid (pZFNcrt-76I) containing either the mut1 or mut2 donor.
Parasite cultures and transfections
Asexual blood stage parasites were propagated in human red blood cells in RPMI-1640 malaria culture medium13 with 0.5% (w/v) Albumax II (Invitrogen) under 5% O2 / 5% CO2 / 90% N2. Parasites were electroporated with purified circular plasmid DNA as described13. The 2A test line expressing mRFP-2A–GFP (Fig. 1a) was generated by attB×attP crossover-mediated integration32 of the pDC2-cam-mrfp-2A–gfp-attP plasmid into the cg6 locus of the recipient line Dd2attB. The NF54EGFP target line (Fig. 1b) was similarly generated by attB×attP-mediated integration of the EGFP expression plasmid (pDC2-cam-egfp-attP) into the NF54attB line12. attB×attP crossover events were mediated by Bxb serine integrase, which was supplied by cotransfection with the plasmid pINT32.
To test for ZFN-mediated cleavage of our integrated egfp sequence, we electroporated NF54EGFP parasites with the plasmid pZFNegfpegfp-hdhfr. This plasmid was designed to integrate a functional egfp-hdhfr fusion (Fig. 1b), expressed from the upstream calmodulin promoter and conferring resistance to WR99210 (Jacobus Pharmaceuticals). Parasites were selected with 2.5 nM WR99210 for 24 (line A), 48 (line B1), 96 (line B2) or 120 hours (line B3) post-electroporation. To potentially increase the efficiency of plasmid delivery and gene disruption, lines B1–3 were supplemented (1:1) with red blood cells preloaded with additional plasmid (50 µg) 24 hours post-electroporation14. In a control experiment we electroporated the donor plasmid pegfp-hdhfr lacking the egfp-specific ZFNs and started selection with 2.5 nM WR99210 the following day. All phenotypic and genotypic analyses of this set of parasite lines were performed on bulk cultures.
To perform our ZFN-driven egfp replacement with mrfp, we co-electroporated NF54EGFP parasites with the plasmids pZFNegfp-hdhfr and pmrfp-vps4 (Fig. 2a). The pZFNegfp-hdhfr plasmid contained the 2A–linked egfp-specific ZFNs that were expressed from a calmodulin promoter. Selection for this plasmid was applied with 2.5 nM WR99210, which was applied one day post-electroporation and was maintained for six days. The pmrfp-vps4 plasmid provided the mrfp donor sequence flanked by homologous regions. No selection was applied for this plasmid. Parasite clones illustrated in Fig. 2 were generated by limiting dilution.
To replace the endogenous pfcrt allele, GC03 and 106/1 parasites were first electroporated with the donor plasmid pcrtDd2-hdhfr and selected with 2.5 nM WR99210 to enrich for episomally transformed parasites. These parasites (GCO3 + pcrtDd2-hdhfr or 106/1 + pcrtDd2-hdhfr) were then electroporated with pZFNcrt-bsd (Fig. 3a). On day one post-electroporation, parasites were selected with 2 µg/ml blasticidin (Invitrogen) and 2.5 nM WR99210 for six days, followed by selection with only WR99210. Parasite clones were generated by limiting dilution where indicated.
To edit pfcrt using CQ selection, 106/1 parasites were electroporated with pZFNcrt-76I plasmids harboring either the mut1 or mut2 sequence template (the latter harbored four ZFN binding site mutations). Transformed parasites were treated with 33 nM CQ phosphate (Sigma-Aldrich) to select for acquisition of the K76I mutation (Fig. 4a). To edit pfcrt without CQ selection, Dd2 parasites were electroporated with pZFNcrt-76I–hdhfr. One day post-electroporation, parasites were exposed to 2.5 nM WR99210 for six days to select for plasmid transformation (Fig. 4a). Analysis of parasite lines was performed on bulk cultures.
DNA analysis
P. falciparum trophozoite-infected erythrocytes were harvested and saponin-lysed. Parasite genomic DNA was extracted and purified using DNeasy Blood kits (Qiagen). Integration of the hdhfr cassette into the cg6-egfp locus of NF54EGFP parasites was confirmed by PCR amplification of the target locus using primers p1 + p2 (yielding a 1.8 kb fragment), p1 + p4 (3.9 kb), p5 + p7 (4.2 kb) and p3 + p6 (1.5 kb in NF54EGFP and 3.4 kb in ZFN-transfected parasites) (Fig. 1b, d). Replacement of the integrated egfp with mrfp at the chromosomal cg6 locus was shown by PCR amplification of the target locus using primers p10 + p7 (yielding a 5.7 kb fragment), p1 + p9 (yielding a 4.8 kb fragment) and p8 + p6 (yielding a 2.8 kb fragment for the mrfp-vps4 replacement, compared with 1.6 kb for the original egfp locus and 3.3 kb for the zfn cassette) (Fig. 2a, d).
Allelic replacement of the endogenous GC03 pfcrt locus by the pcrtDd2-hdhfr donor sequence was confirmed by PCR of the target locus with primers p11 + p13 (GC03 parent: 1.4 kb, GC03crt-Dd2 recombinant: 1.2 kb, GC03 + pcrtDd2-hdhfr: 1.4 kb, pcrtDd2-hdhfr: no product); p12 + p13 (GC03: 0.6 kb, GC03crt-Dd2: 0.4 kb, GC03 + pcrtDd2-hdhfr: 0.4 and 0.6 kb, pcrtDd2-hdhfr: 0.4 kb); and p14 + p15 (GC03: no product, GC03crt-Dd2: 2.5 kb, GC03 + pcrtDd2-hdhfr: no product, pcrtDd2-hdhfr: no product) (Fig. 3a, c).
Editing of pfcrt codon 76 (Fig. 4) was confirmed by PCR amplifying the genomic locus with primers p23 + p27 that flank the pfcrt donor sequence. These products were amplified either from bulk cultures (followed by cloning into pGEM-T), or from parasite clones (Table 1). Sequencing was performed with primers p24, p25, p26, p18 and p20 (Supplementary Table 3).
Whole-genome sequencing and data processing
Genomic DNA was prepared from the parental line NF54EGFP and the two ZFN-transfected lines NF54ΔegfpA, NF54ΔegfpB1, as well as 106/1 and 106/113/15mut1 and 106/114/15 mut1. These DNAs were sheared (10 µg per line) to obtain a fragment size of ∼200–400 bp using an E220 focused-ultrasonicator (Covaris Inc.) and the following settings: 10 % duty cycle, intensity 5, 200 cycles per burst, 180 second treatment time. Sheared gDNA was size selected on a 2 % low melting agarose gel and then purified. Barcoded libraries for Illumina TruSeq single-end sequencing were then constructed from the size-selected, sheared material using NEBNext DNA Library Preparation reagents (NEB), following the standard Illumina library preparation protocol, but omitting the final size selection step. To assess the quality and percentage of adaptor-ligated material, the final sequencing libraries were tested on an Agilent 2100 Bioanalyzer (Agilent Technologies) along with the original size-selected fragmented gDNA from the same preparation. The concentration of each library was determined using a Quant-iT dsDNA Broad-Range Assay Kit (Invitrogen). For the NF54ΔegfpB1 line the sequencing adapter ligation was determined to be less than 50 % efficient. We therefore PCR amplified the sequencing library as described33 to minimize GC bias. PCR conditions for 50 µL reactions were: 1 × Kapa HiFi Buffer, 60 nM tetramethylammonium chloride, 0.3 mM each dNTP, 0.4 µM each primer and 1 U Kapa HiFi DNA polymerase, with thermocycling conditions of 1 min at 98 °C followed by 12 cycles of [10 s 98 °C, 1 min 55 °C] and 5 min at 55 °C. The final libraries were multiplexed with 3 barcoded samples and 20 % PhiX control DNA per lane, and were sequenced using an Illumina HiSeq 2000 system.
Sequencing outputs were uploaded into Galaxy34 hosted locally at the Lewis-Sigler Institute for Integrative Genomics at Princeton University. Sequence reads were mapped to the P. falciparum 3D7 reference genome v.8.0 (http://plasmodb.org/common/downloads/release-8.0/Pfalciparum/) using the Burrows-Wheeler Alignment tool35, and files were formatted using Sequence Alignment/Maptools36. GATK tools were applied to assess depths of coverage for the overall genomes and specific regions of interest, and to perform SNP discovery and filtering using standard hard filtering parameters37. Alignments and variants were visualized using the Integrative Genomics Viewer38.
Southern blot hybridizations
Edited lines obtained from the egfp disruption and the egfp/mrfp replacement experiments and the parental control were analyzed using pairs of Southern blots, as follows. For each blot, 2 µg of genomic DNA was digested with ClaI + BamHI, electrophoresed on a 0.7% agarose gel and transferred onto a Nytran nylon membrane. Hybridization of the first blot was performed at 54°C with a 710 bp [32P]-labeled egfp probe that was PCR amplified from pDC2-cam-egfp-attP using primers p3 + p26. The second blot was either hybridized with a 560 bp [32P]-labeled hdhfr probe or a 670 bp [32P]-labeled mrfp probe amplified by PCR with primers p10 + p17 (mrfp) or p28 + p4 (hdhfr) from the respective plasmids pmrfp-vps4 or pZFNegfp-hdhfr. Linearized transfection plasmids served as positive controls (8.4 kb, 8.3 kb and 9.9kb for the egfp, mrfp and hdhfr experiment respectively; see Figs. 1e, 2e).
Parasites lines obtained following pfcrt allelic replacement were analyzed on pairs of Southern blots, for which 2 µg of genomic DNA was digested with SalI + BstBI. Digests were electrophoresed on a 0.7% agarose gel and transferred onto a Nytran nylon membrane. Hybridization was performed at 55°C with a 600 bp [32P]-labeled hdhfr probe amplified from pcrtDd2-hdhfr by PCR with primers p26 + p4.
In vitro susceptibility assays
In vitro IC50 values were determined by incubating parasites for 72 hours across a range of concentrations of CQ (2000 nM to 2.5 nM). After 72 hours of incubation, parasite growth was determined by measuring parasitemia using flow cytometry. Cells were stained with 1.6 µM Mito Tracker Deep Red and 2× SYBR Green (Invitrogen) in 1 × PBS supplemented with 5 % FBS39. In vitro IC50 values were calculated by non-linear regression analysis and Mann-Whitney U tests were employed for statistical analysis. A non-parametric statistical analysis was appropriate as experiments were independent and no normal distribution was assumed.
For the pfcrt gene replacement experiment (Fig. 3d), the half-maximal inhibitory concentration (IC50) value for the GC03 parental line (mean ± SEM 18.4 ± 5.3 nM, n = 4) was significantly different from the two pfcrt allelic replacement clones GC03crt-Dd2 G9 (91.8 ± 10.7 nM, n = 4, P = 0.028, two-tailed) and GC03crt-Dd2 H6 (100.3 ± 15.5 nM, n = 4, P = 0.028, two-tailed). No significant difference in IC50 values was observed between the parental GCO3 line and the control GC03 + pcrtDd2-hdhfr line that episomally replicated the donor plasmid (18.55 ± 6.8 nM, n = 4, P = 1.0, two-tailed).
For the pfcrt gene editing experiment (Fig. 4b), the half-maximal inhibitory concentration (IC50) value for the 106/1 parental line (mean ± SEM 27.4 ± 1.2 nM, n = 4) was significantly different (*P = 0.028, two-tailed, Mann-Whitney U test) from the gene-edited parasites 106/113/15mut2 (153.6 ± 18.3 nM) and 106/114/15mut1 (145.1 ± 17.8 nM), as well as the previously established CQR line 106/1I76 (177.9 ± 16.8 nM).
Fluorescence analysis of recombinant parasites
For live-cell imaging, resuspended cultures were washed once with RPMI-1640 media lacking Albumax II, and the cells applied to poly-L-lysine coated glass-bottom culture dishes (MatTek) and overlaid with 1 mL of RPMI media containing 2 µg/mL Hoechst 33342 (Sigma) to stain the nuclei. Images were acquired on a Nikon Ti-E inverted microscope with 100 × N.A. 1.4 PlanApo optics and a CoolSnap HQ2 camera. Images were collected with Nikon NIS Elements software and assembled using Adobe Photoshop.
To quantify the proportion of EGFP-positive parasites in the NF54EGFP and ZFN-modified lines, parasite cultures were analyzed by flow cytometry. Cells were stained for 10 min with 250 nM SYTO 61 dye (Invitrogen) in aqueous solution containing 0.2% dextrose and 0.9% sodium chloride. After a single wash 50,000 cells were counted on an Accuri C6 Flow Cytometer. Data were analyzed with FlowJo 7.6.3 gating for nuclear stain SYTO 61 (FL4) and for green fluorescence (FL1).
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Symington (Columbia University) for helpful discussions, and the sequencing core facility and staff in the Lewis-Sigler Institute for Integrative Genomics at Princeton University. M.L. is funded by a NIH Director’s New Innovators award (1DP2OD001315) and receives support from the Center for Quantitative Biology (P50 GM071508). D.A.F gratefully acknowledges support from the NIH (R01 AI50234 and AI079709). Please direct requests for ZFNs to F.D.U. (furnov@sangamo.com).
Footnotes
Note: Supplementary Information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
J.S., M.C.S.L., B.Z., A.H.L., M.L., F.D.U. and D.A.F. designed the experiments, which were performed by J.S., M.C.S.L., A.H.L. and A.E.W. B.Z., J.R.P., L.Z., E.J.R., P.D.G. and F.D.U. designed and provided the zinc-finger nucleases. J.S., M.C.S.L., A.H.L., F.D.U. and D.A.F. wrote the manuscript, with input from all authors.
COMPETING FINANCIAL INTERESTS
B.Z., J.R.P., F.D.U., L.Z., E.J.R., and P.D.G. are employees of Sangamo BioSciences.
REFERENCES
- 1.Dondorp AM, et al. The threat of artemisinin-resistant malaria. N. Engl. J. Med. 2011;365:1073–1075. doi: 10.1056/NEJMp1108322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meissner M, Breinich MS, Gilson PR, Crabb BS. Molecular genetic tools in Toxoplasma and Plasmodium: achievements and future needs. Curr. Opin. Microbiol. 2007;10:349–356. doi: 10.1016/j.mib.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Anderson T, Nkhoma S, Ecker A, Fidock D. How can we identify parasite genes that underlie antimalarial drug resistance? Pharmacogenomics. 2011;12:59–85. doi: 10.2217/pgs.10.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 7.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 8.Urnov FD, et al. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 9.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum . Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotech. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 12.Adjalley SH, et al. Quantitative assessment of Plasmodium falciparum sexual development reveals potent transmission-blocking activity by methylene blue. Proc. Natl. Acad. Sci. USA. 2011;108:E1214–E1223. doi: 10.1073/pnas.1112037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol. Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- 14.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bray PG, et al. Defining the role of PfCRT in Plasmodium falciparum chloroquine resistance. Mol. Microbiol. 2005;56:323–333. doi: 10.1111/j.1365-2958.2005.04556.x. [DOI] [PubMed] [Google Scholar]
- 17.Summers RL, Nash MN, Martin RE. Know your enemy: understanding the role of PfCRT in drug resistance could lead to new antimalarial tactics. Cell Mol. Life Sci. 2012;69:1967–1995. doi: 10.1007/s00018-011-0906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidhu ABS, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valderramos SG, et al. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum . PLoS Pathog. 2010;6:e1000887. doi: 10.1371/journal.ppat.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat. Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper RA, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum . Mol. Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 23.Martin RE, et al. Chloroquine transport via the malaria parasite’s chloroquine resistance transporter. Science. 2009;325:1680–1682. doi: 10.1126/science.1175667. [DOI] [PubMed] [Google Scholar]
- 24.Sa JM, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc. Natl. Acad. Sci. USA. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott B, et al. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carvalho TG, Menard R. Manipulating the Plasmodium genome. Curr. Issues Mol. Biol. 2005;7:39–55. [PubMed] [Google Scholar]
- 27.Sidhu ABS, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum . Mol. Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 28.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 29.Geurts AM, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MC, Moura PA, Miller EA, Fidock DA. Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Mol. Microbiol. 2008;68:1535–1546. doi: 10.1111/j.1365-2958.2008.06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ecker A, et al. Evidence that mutant PfCRT facilitates the transmission to mosquitoes of chloroquine-treated Plasmodium gametocytes. J. Infect. Dis. 2011;203:228–236. doi: 10.1093/infdis/jiq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nkrumah LJ, et al. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat. Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oyola SO, et al. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics. 2012;13:1. doi: 10.1186/1471-2164-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekland EH, Schneider J, Fidock DA. Identifying apicoplast-targeting antimalarials using high-throughput compatible approaches. FASEB J. 2011;25:3583–3593. doi: 10.1096/fj.11-187401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.