Abstract
One of the major problems facing clinical nephrology currently throughout the world is an exponential increase in patients with end-stage renal disease (ESRD), which is largely related to a high incidence of diabetic nephropathy. The latter is characterized by a multitude of metabolic and signaling events following excessive channeling of glucose, which leads to an increased synthesis of extracellular matrix (ECM) glycoproteins resulting in glomerulosclerosis, interstitial fibrosis and ultimately ESRD. With the incidence of nephropathy at pandemic levels and a high rate of ESRD, physicians around the world must treat a disproportionately large number of diabetic patients with up-to-date innovative measures. In this regard, identification of genes that are crucially involved in the progression of diabetic nephropathy would enhance the discovery of new biomarkers and could also promote the development of novel therapeutic strategies. Over the last decade, we focused on the recent methodologies of high-throughput and genome-wide screening for identification of relevant genes in various animal models, which included the following: (1) single nucleotide polymorphism-based genome-wide screening; (2) the transcriptome approach, such as differential display reverse transcription polymerase chain reaction (DDRT-PCR), representational difference analysis of cDNA (cDNA-RDA)/suppressive subtractive hybridization, SAGE (serial analysis of gene expression) and DNA Microarray; and (3) the proteomic approach and 2-dimensional polyacrylamide gel electrophoresis (2D-PAGE) coupled with mass spectroscopic analysis. Several genes, such as Tim44 (translocase of inner mitochondrial membrane-44), RSOR/MIOX (renal specific oxidoreductase/myo-inositol oxygenase), UbA52, Rap1b (Ras-related GTPase), gremlin, osteopontin, hydroxysteroid dehydrogenase-3β isotype 4 and those of the Wnt signaling pathway, were identified as differentially expressed genes in kidneys of diabetic rodents. Functional analysis of these genes and the subsequent translational research in the clinical settings would be very valuable in the prevention and treatment of diabetic nephropathy. Future trends for identification of the biomarkers and therapeutic target genes should also include genome scale DNA/histone-methylation profiling, metabolomic approaches (e.g. metabolic phenotyping by 1H spectroscopy) and lectin microarray for glycan profiling along with the development of robust data-mining strategies.
Background of Diabetic Nephropathy
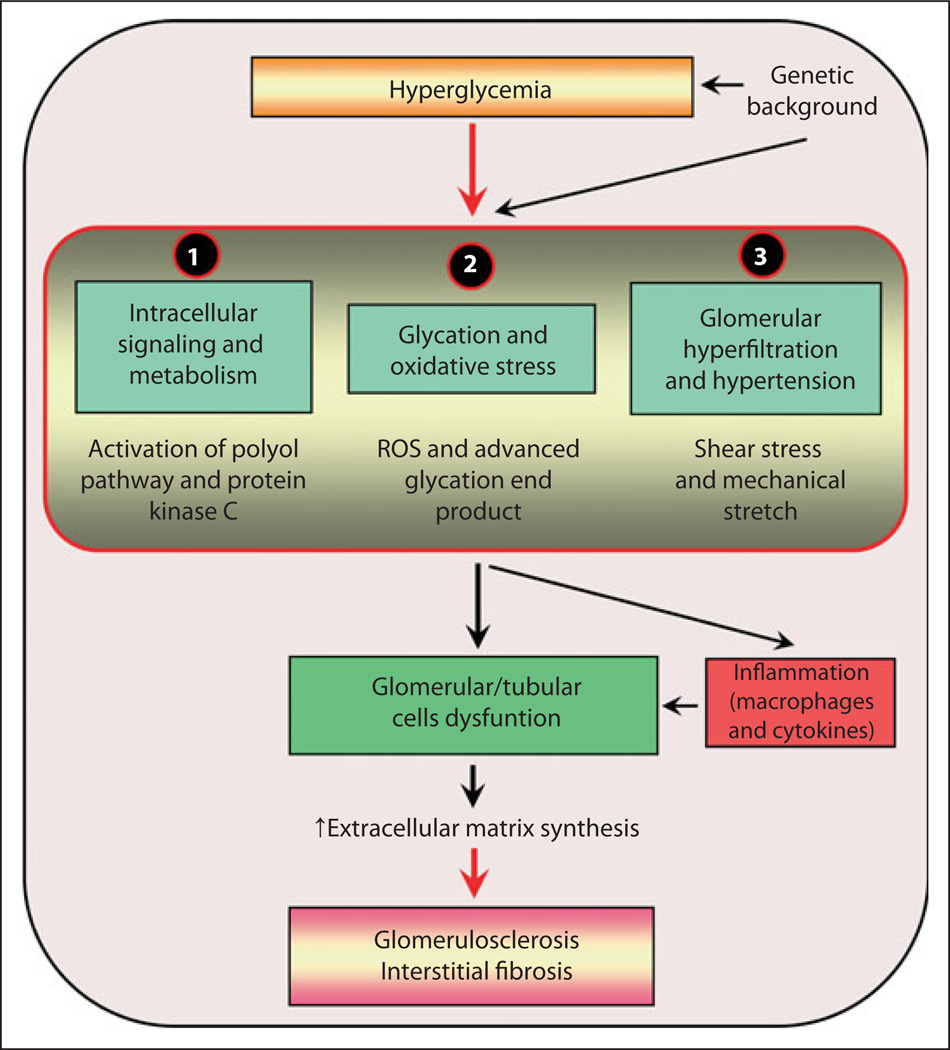
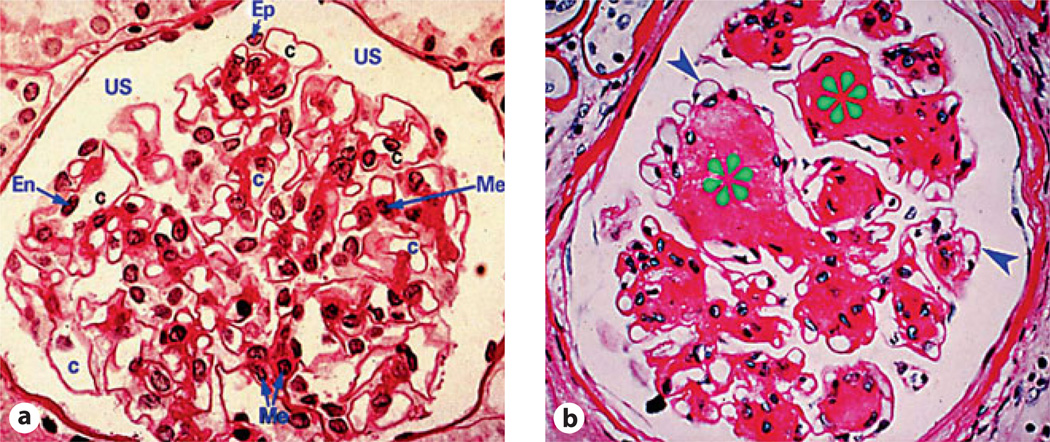
The most problematic issue in clinical nephrology is the progressive worldwide increase in patients with end-stage renal disease (ESRD) that is commonly associated with diabetic nephropathy. Over the last 30 years, extensive investigatory efforts have been made to delineate the mechanisms involved in the pathogenesis of diabetic nephropathy, resulting in the identification of three major signaling pathways (fig. 1) [1]. Upstream of these pathways, hyperglycemia seems to be the major driving force for initiating various signaling events. The down-stream events include setting in of microinflammation and increased synthesis of extracellular matrix (ECM) glycoproteins by the renal glomerular, tubular and interstitial cells. This leads to the amassing of the ECM in the glomerulus resulting in thickening of the glomerular basement membranes, mesangial expansion and nodule formation, a characteristic of Kimmelstiel-Wilson lesion and a hallmark of diabetic nephropathy [2] (fig. 2). At the same time, the tubulointerstitial compartment is also affected. One can observe a thickening of the tubular basement membranes and increased interstitial fibrosis, adding further to the glomerular injury and, consequentially, chronic irreversible progressive renal failure. Conceivably, it is likely that the lesions in both compartments are the result of common signaling pathways initiated by excessive intracellular channeling of glucose.
Fig. 1.
An overview of the signaling events induced by exposure of renal cells to high glucose ambience leading to altered expression of various genes and cellular abnormalities culminating in diabetic nephropathy. The schematic highlights three sets of major signaling events which converge to induce dysfunction of glomerular and tubular cells leading to glomerulosclerosis and interstitial fibrosis and ultimately ESRD. ROS = Reactive oxygen species.
Fig. 2.
Light photomicrographs illustrating glomerular lesions in diabetic nephropathy. a Normal glomerulus. b Thickened basement membranes (arrowheads) and nodular appearance of the mesangial regions characteristic of Kimmelstiel-Wilson lesions (asterisks). Me = Mesangium; Ep = visceral epithelial cell (podocyte); En = endothelial cell; C = capillary lumen; US = urinary space.
The first major pathway relates to metabolic abnormalities in which there is a heightened activity of the polyol pathway and activation of protein kinase C-β (PKC-β). The events related to this pathway have been described in an animal model of STZ-induced diabetic rats (model of type-1 diabetes) and db/db mice (model of type 2 diabetes) [3]. Interestingly, an ameliorative effect was observed when these animals were treated with inhibitors of PKC-β or of the polyol pathway, i.e. aldose reductase [4–6]. These agents, especially the inhibitor of PKC-β, could possibly serve as a therapeutic measure to dampen the progression of diabetic nephropathy, a subject matter that would be of attractive clinical value in future investigations.
The characteristics of the second pathway include the formation of advanced glycation end-products (AGEs) from excessive imbibing of glucose [7]. The AGEs via interaction with their receptor, RAGE, transduce a complex series of signaling events that result in cellular dysfunctions, thus generating an inflammatory response and reactive oxygen species (ROS), which in turn cause oxidative stress [7]. Both in vitro and in vivo studies support the relevance of this pathway in the pathogenesis of diabetic nephropathy [7]. The fact that several inhibitors of AGEs, such as pyridoxamine, LR-90 and KIOM-79, have been demonstrated to be beneficial in various murine models of diabetes emphasizes the role of AGE:RAGE interactions [8–10]. Although these inhibitors may be effective in murine models, their efficacy certainly needs to be evaluated in diabetic nephropathy in humans.
The oxidative stress due to ROS also induces cellular dysfunction, which compounds the glomerular and tubular injury further, thus worsening the outcome of diabetic nephropathy [11, 12]. The ROS are mainly generated via the NADPH oxidase system or at the level of the mitochondria. In this regard, the NADPH oxidase inhibitors in the amelioration of diabetic nephropathy have been reported in various animal model systems [11, 12]. During the last 10 years, glomerular hyperfiltration leading to intraglomerular hypertension has emerged as a third pathway, and this pathway adversely influences the course of diabetic nephropathy [13]. Intriguingly, reduction of glomerular hyperfiltration by renin-angiotensin system inhibitors apparently has remarkable beneficial effects in the amelioration of proteinuria and progression of nephropathy, both in animal experiments and various clinical studies in patients with diabetes mellitus [13].
These three major pathways converge to induce dysfunction of glomerular, tubular and interstitial cells. There is good evidence that cellular injury affects the endothelial cells of renal vasculature, accompanied with increased expression of adhesion molecules and chemokines, resulting in a macrophage influx into the renal parenchyma and subjecting it to low-grade inflammation [14]. There are a few reports from the literature supporting this prolonged microinflammation as a common pathway for the progression of diabetic nephropathy [14, 15]; therefore, it is likely that anti-inflammatory agents serving as supplemental agents to treat diabetic nephropathy would be another fertile area of investigation in coming years. Counterintuitive to the notion of microinflammation in diabetic nephropathy is the upregulation of a profibrogenic cytokine, transforming growth factor-β (TGF-β). TGF-β is not only a potent downregulator of an immune/inflammatory response, but it is also a well-characterized critical molecule for the increased synthesis ECM glycoproteins leading to glomerulosclerosis and interstitial fibrosis [6, 16]. As expected, the administration of neutralizing anti-TGF-β antibody has been reported to prevent the renal insufficiency in db/db mice [16]. Along these lines, the efficacy of administration of such neutralizing antibodies to other relevant growth factors, such as connective tissue growth factor and inflammatory cytokines, or the genetic deletion of adhesion molecules, such as ICAM-1 and macrophage scavenger receptor-A, on the amelioration of diabetic nephropathy and prevention of ESRD need to be aggressively evaluated since there has been a precipitous increase in the incidence of this dire renovascular complication of diabetes in recent years [14, 15].
The incidence reported for ESRD is highest in Taiwan, at 404/106 persons in 2005, followed by the United States, Jalisco (Mexico), Shanghai (China) and Japan (in 2004) with the respective rates being 351, 302, 275 and 267 for these 4 geographic regions. Most countries with a high rate of ESRD have a disproportionately large proportion of patients with diabetes mellitus. For example, in Jalisco 60% of new ESRD patients have a primary diagnosis of diabetes, while approx. 12% in Russia and Norway have such a diagnosis and only 5% in Iceland (http://www.usrds.org/adr.htm).
The impact of diabetic nephropathy on the incidence of chronic kidney disease and ESRD is enormous. To combat this problem, intensified multi-prong approaches have been performed in patients with type 2 diabetes mellitus, resulting in reduced risk of microangiopathy, cardiovascular complications and mortality in randomized studies carried out at the Steno Memorial Hospital, Denmark, by Gaede et al. [17–19]. However, the incidence of ESRD is still progressively increasing worldwide. In such a dilemma, identification of the genes that are critically involved in the progression of diabetic nephropathy is warranted, the discovery of which should eventually lead to the development of biomarkers and new treatment strategies. In view of the above considerations, we devoted our efforts to identifying genes relevant to diabetic nephropathy in various animal models using various current biological approaches, including high-throughput and genome-wide screening methods.
Single Nucleotide Polymorphism-Based Genome-Wide Screening
Attempts have been made to identify the susceptibility gene variants for diabetic nephropathy by single nucleotide polymorphism (SNP)-based case-control association screening in patients with type 1 and 2 diabetes mellitus. The advent of high-throughput and low-cost genotyping technologies, discovery of sufficiently dense SNP markers, and vast improvements in collection methods for DNA analysis has made genome-wide screening readily feasible. For example, a combination of multiplex PCR and Invader assay has enabled the genotyping of one SNP with as low as 0.4 ng of genomic DNA, and theoretically more than 200,000 SNPs can be genotyped in one cycle with100 µg [20, 21]. In addition, microarray systems, such as Genechip Arrays (Affymetrix) and Infinium beads array (Illumina) can genotype hundreds of thousands of SNPs using 250–750 ng of genomic DNA.
In line with the development of genotyping technology, the International HapMap Project (IHMP) has further promoted the identification of SNPs [22]. The goal of IHMP is to compare the gene sequences and to identify chromosomal regions where the genetic variants are shared among different individuals. By making this information freely accessible, the IHMP will enable investigators to search for genes involved in various diseases and will also aid in delineating the responsiveness to differential therapies.
Utilizing genotyping technologies and SNP methods in the Genetics of Kidneys in Diabetes (GoKinD) study, a large number of individuals with type 1 diabetes were screened to identify biomarkers in two subsets of patients: one with well-documented kidney disease and the other with normal kidney functions despite long-term diabetes [23]. A single marker, rs11886047 (located upstream of PLEKHH2 promoter), was found to be associated with diabetic nephropathy by transmission disequilibrium testing in the case trios (p = 0.0307). Also, there was a increase of heterozygous genotypes in cases, relative to controls, from the 601 case and 577 control GoKinD singleton case/control population (p = 0.00256) [24]. The results of the GoKinD study suggest that the PLEKHH2, which is exclusively expressed in the kidney glomerulus, may be a genetic risk factor for susceptibility to diabetic nephropathy. In another study by Maeda et al. [25], patients with type 2 diabetes were divided into two groups: one having retinopathy with overt nephropathy, and the other control group having diabetic retinopathy with no renal dysfunctions. They genotyped over 80,000 gene-based SNPs and found that substitution of Arg913 to Gln in the gene, belonging to solute carrier family 12 member 3 (SLC12A3), seems to reduce the risk in the development of diabetic nephropathy [26]. They also noted that SNP is also prevalent in the engulfment and cell motility 1 gene (ELMO1), and thus concluded that it is a likely candidate in conferring susceptibility for diabetic nephropathy [27].
Transcriptome Approach Using Cultured Cells, Animal Models and Human Samples
Gene expression profiling has been used to identify genes that are up- or down-regulated in cultured cells subjected to various forms of pathophysiological stresses. The differentially expressed genes that are induced under such stresses may serve as good candidates to develop potential novel biomarkers and targets for new therapies. Such gene profiling information has also been forthcoming from various animal models, such as streptozotocin (STZ)-induced diabetes in C57B6/J and ICR mice [28, 29] and Wistar rats [30], as well as rodents developing diabetes spontaneously, i.e. non-obese diabetic, db/db and KK/TA mice [28, 31–34], and OLETF rats [35].
The STZ-induced diabetic animals are a model of type 1 diabetes which have rapid onset of hyperglycemia within a couple of days. Since these mice develop renal changes several weeks or months later, the differential gene expression can be readily assessed during the early phases of diabetic nephropathy. However, one cannot absolutely rule out the possibility that the altered gene expression may also be related to the ROS that are induced upon STZ administration during the early phases of diabetic nephropathy. On the other hand, in spontaneous type 1 and 2 rodent models, one needs to take into account the time course-dependent changes in the gene expression that would be correlative of the histological lesions seen during various phases of renal hypertrophy versus sclerosis or fibrosis.
The insurmountable problem in rodent animal studies is that there is really no good model which mimics lesions in human diabetic nephropathy, such as glomerular hypertrophy, basement membrane thickening, increase in mesangial matrix and nodular sclerosis. In this regard, the triple transgenic mice over-expressing megsin, RAGE (receptor for advanced glycation end-products) and iNOS (inducible nitric oxide synthase) exhibit severe albuminuria and renal changes comparable to human diabetic nephropathy in addition to inflammatory cell infiltration and interstitial fibrosis [36]. Conceivably, these mice may be the most suitable model for identifying genes that may be critical in the pathogenesis of diabetic nephropathy.
Another critical problem in gene expression profiling studies is that there are many cell types in the kidney, each contributing to varying degrees in the pathogenesis of diabetic nephropathy; thus, it seems that analyses of each of the cells under high glucose ambient cultures would be the most appropriate measure for gene profiling relevant to diabetic nephropathy. Along this notion, many investigators have indeed utilized mesangial cells [37], conditionally immortalized mouse podocytes [38], human proximal tubular cells (HK-2 cells) [39, 40] and rat-immortalized renal proximal tubular cells to study the pathogenesis of diabetic nephropathy [30]. For human samples, peripheral blood mononuclear cells of diabetic patients [41], renal biopsy tissues [42], laser capture microdissection and analysis of the glomeruli in these biopsy specimens [32], and examination of glomerular podocytes in the urinary sediment [43] may serve as the next group of potential targets of investigation for gene profiling studies.
DDRT-PCR and RDA-cDNA/SSH
The elegant method of DDRT (differential display reverse transcription)-PCR to study the differential gene regulation between two populations of RNA was originally described by Liang and Pardee [44] in the early 90s. In this technique, two sets of oligonucleotide primers are used: one anchored to the polyadenylate tail of a subset of mRNAs, and the other being short and arbitrary in sequence so that it can anneal at different positions relative to the first primer. The mRNA subpopulations are then amplified following reverse transcription. The cDNAs are then resolved by sequencing gel and the bands from two sets of populations are compared side by side. With the application of DDRT-PCR for cDNAs isolated from mesangial cells grown in 5.5 mm (low) and 25 mm (high) d-glucose, Song et al. [45] discovered that the munc13s gene was differentially regulated.
Although DDRT-PCR was considered a powerful method to identify the differentially regulated genes, it had certain technical problems such as false-positivity for certain irrelevant genes. To overcome this problem, Hubank and Schatz [47] devised the PCR-coupled subtractive process of representational difference analysis (RDA) and adapted it for the use with mammalian cDNA [46]. Along the principles of cDNA-RDA, the suppressive subtractive hybridization (SSH) technique was developed later [47]. By applying RDA/SSH for the cDNAs prepared from STZ-induced diabetic mice kidneys, we identified a number of genes, such as Tim44 (translocase of inner mitochondrial membrane-44) [48–50], RSOR/MIOX (renal specific oxido-reductase/myo-inositol oxygenase) [51, 52], UbA52 [53] and Ras-like GTPase, Rap1b [54, 55]. All of these were differentially regulated in hyperglycemic state. Similarly, McMahon et al. [56] and Murphy et al. [57] reported gremlin to be upregulated in mesangial cells in culture subjected to high glucose ambience. The above approaches are certainly very useful in identifying the differentially expressed genes; however, they lack the capacity for gene profiling of global mRNA expressions. Therefore, microarray methods were developed over the past decade.
Serial Analysis of Gene Expression and DNA Microarray
Serial analysis of gene expression (SAGE) allows the quantitative and simultaneous analysis of a large number of transcripts. The method is based on the isolation of unique sequence tags (9- to 13-bp in length) from individual mRNAs and concatenation of tags serially into long DNA molecules for lump-sum sequencing [58]. So far, the usage of SAGE has been restricted to the expression profiling of kidney transcriptomes [59], in particular the renal collecting duct cells of mice subjected to potassium depletion [60], and comparative gene analyses between various cell types, such as glomerular versus aortic endothelia [61]. In this regard, the advent of high-throughput next generation sequencers, such as the 1G Genome Analyzer (Illumina/Saleza) and SOLID System (Applied Biosystems) have greatly facilitated gene expression profiling by SAGE tag with higher efficiency and lower costs. The application of these sequencers should enhance the capabilities of profiling mRNA expressions as well as discovery of miRNAs relevant to a given disease process.
In recent years, cDNA microarray has been the most commonly used method for profiling mRNA expression compared to SAGE [62]. The microarray molecular profiling of human kidney [42] and rodents [29] has resulted in the generation of a large amount of data, and in such a scenario it may be difficult to pinpoint the gene(s) that are relevant to the pathogenesis of diabetic nephropathy. These difficulties can be overcome when microarray is combined with reliable analytical procedures that can correlate with a given phenotype.
Susztak et al. [28] employed weighted vote-based analytical methods to delineate the genes whose expression can be correlated with the presence or absence of mesangial matrix expansion. They identified hydroxysteroid dehydrogenase-3β isotype 4 and osteopontin as the lead classifier genes that are correlative of mesangial matrix expansion phenotype. Along these lines Cohen et al. [63] performed Affymetrix oligonucleotide microarrays (HG-U133A) by standard robust multi-array analysis (RMA) on microdissected human renal tissues and compared the gene profiles of patients with and without diabetic nephropathy.
These studies indicated limited detection of certain biological processes that are also relevant to the pathogenesis of diabetic nephropathy. These included genes pertinent to inflammation and angiogenesis. The limited detection was thought to be attributed to the apparent lack of sensitivity that was associated with the gene-oriented averaging probe signals. This shortcoming was rectified by the use of ChipInspector, which is based on single probe analysis and de novo gene annotation that bypasses the probe set definition based on the out-of-date genomic data. In doing so, the single probe-based analysis yielded reduced background noise with enhanced sensitivity and fewer false positives. It also successfully identified the Wnt signaling pathway activated in diabetic nephropathy [63].
Another approach to identify the biologically relevant genes is to investigate pathway analysis of the gene expression. Naito et al. [64] compared gene expression of glomerular cells harvested from kidneys of db/db and db/m mice by laser capture microdissection. They then performed pathway analysis of 550 upregulated probes using the Ingenuity signal analysis. The results pointed out that the mitochondrial oxidative phosphorylation pathway is the most significantly affected canonical pathway in diabetic nephropathy [64]. The above studies emphasize that the efficient use of microarray data for identification of the relevant genes would require further improvements for the specificity and sensitivity of the mRNA detection and specifically the development of new software for the analysis of the vast amount of the available current data sets.
Proteomic Approaches
Proteomic analysis using two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) coupled with Mass spectroscopic analysis has been used to survey the changes in protein expression in diabetic nephropathy. Thongboonkerd et al. [65] compared the renal proteome of 120 day-old OVE26 transgenic mice manifesting hypoinsulinemia, hyperglycemia, hyperlipidemia and proteinuria with background FVB non-diabetic mice. Proteins derived from whole-kidney lysate were separated by 2D-PAGE and identified by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. They demonstrated marked accumulation of elastin in the macula densa, collecting ducts and pelvicalyceal epithelia of diabetic kidneys [65]. Tilton et al. [66] analyzed the renal cortical proteome of db/db mice using 2D-PAGE combined with MALDI-TOF, MALDI-TOF/TOF and LC-MS/MS. The top-ranked network described by Ingenuity Pathway Analysis indicated that PPAR-α may be the common nodal point of interactions for several enzymes whose expression levels are influenced by diabetic state [66].
Besides renal tissues, urine proteome analysis in diabetic patients may also allow early detection of diabetic nephropathy and could thus provide certain prognostic information. Along these lines, an increase in α1-anti-trypsin, UbA52 and type I collagen expression has been reported in patients with diabetic nephropathy [67–69]. In view of the above observations, it is conceivable that the panel of biomarkers identified in patients with diabetic nephropathy with their signatures in the serum and urinary proteins/peptides along with their unique pattern of expression and profiling could serve as a novel clinical diagnostic approach for the detection of early diabetic nephropathy [69, 70].
Newly Identified Genes Relevant in the Progression of Diabetic Nephropathy
The cellular events such as increased flux of polyols and hexosamines; generation of AGEs; increased activity of PKC, transforming growth factor-β-Smad-MAPK (mitogen-activated protein kinase) pathway and GTP-binding proteins; G1 cell cycle arrest associated with altered expression of cyclin kinases and their inhibitors; and generation of ROS are responsible for a final outcome of increased synthesis and deposition of ECM. The ROS, whether mitochondrial or cell membrane-derived, are also responsible for the activation of the reninangiotensin system that eventually contributes to glomerular hyperfiltration and subsequent renal fibrosis (fig. 1) [71]. In addition to these macromolecules, newly identified genes, such as RSOR/MIOX, Tim44 and Rap1b, may also be an integral part of the hyperglycemia-induced cytosolic and mitochondrial processes that culminate in the development of diabetic nephropathy [48–55].
Under high glucose ambience, there is an increased activity of accessory cytosolic polyol pathway; as a result, glucose is reduced to sorbitol by a NADPH-dependent enzyme, aldose reductase. The sorbitol is oxidized to fructose by sorbitol dehydrogenease utilizing NAD+ as a cofactor. Although aldose reductase inhibitors have been used for diabetic neuropathy in Japan and other countries, the relevance of the polyol pathway is somewhat unclear in diabetic nephropathy. Another enzyme that has come to light in the context of glucose metabolism is RSOR/MIOX [52, 72]. It is expressed in the renal proximal tubules, and is upregulated in diabetic mouse kidneys. It is responsible for the oxidation of myo-inositol generated from glucose-6-phosphate. Phosphatidylinositol, a metabolic product of myo-inositol is involved in cellular signaling and osmoregulatory functions of the kidney. MIOX expression is upregulated in the experimental diabetes, while renal concentration of myo-inositol decreases and its supplementation normalizes glucose-induced proliferation and collagen synthesis in tubular cells, thus implicating the role of the cytosolic MIOX in diabetic nephropathy [73].
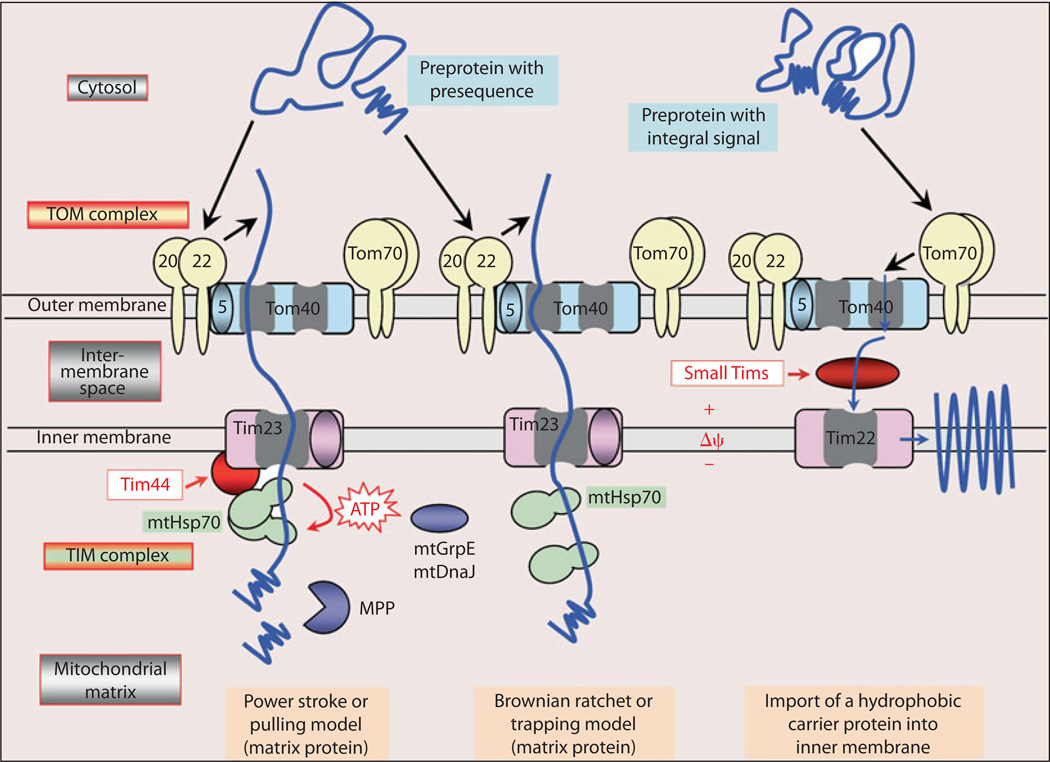
In the mitochondria, during oxidative phosphorylation, the electron donors generate a high membrane potential by pumping protons across the mitochondrial inner membrane. As a consequence, the electron transport is inhibited and the half-life of free-radical intermediates of ubiquinone increases, which reduces O2 to with ensuing oxidant stress in high glucose ambience. Rotenone, an inhibitor of electron transport, blocks the DCF-sensitive ROS generation and the fact that a similar effect is observed by overexpression of mitochondrial manganese superoxide dismutase (Mn-SOD) or uncoupling protein-1 (UCP-1) supports the notion that mitochondria are the major source of ROS following increased glucose flux into the cell. The ROS produced can thus modulate various cellular processes and expression of proteins, including those involved in the mitochondrial transport. One of the transport proteins, known as Tim44, is upregulated in hyperglycemic state [50]. It functions as a membrane anchor of mtHsp70 to the TIM23 complex and is involved in the import of preproteins with mitochondrial-targeted pre-sequences into the mitochondrial matrix (fig. 3). Its importance in the pathogenesis of diabetic nephropathy is underscored by the fact that gene delivery of Tim44 by the HVJ-envelope vector inhibits proteinuria, renal hypertrophy, cell proliferation and apoptosis, and superoxide production in STZ-induced diabetes in CD-1 mice [48]. Also, gene delivery of Tim44 in human proximal tubule cell line HK-2 facilitates the import of antioxidative enzymes such as SOD and glutathione peroxidase along with the inhibition of the ROS production, reduction of ATP contents and normalization of alterations in inner mitochondrial membrane potential [48].
Fig. 3.
Main pathways of protein import into mitochondrial matrix and inner membrane. Mitochondria matrix preproteins with an amino-terminal presequence as well as proteins with integral target sequence translocate through the translocase outer membrane (TOM) complex, made up of 5-, 20-, 22-, 40-and 70-kDa proteins. Preproteins with presequence are translocated across the inner membrane by the Tim23 complex. Two models are proposed: power stroke and Brownian ratchet. The process requires the inner membrane potential (Δψ) and ATP-dependent action of mtHsp70. In the power stroke model, Tim44 seems to function as a membrane anchor of mtHsp70 to the TIM23 complex. The proproteins with integral signals are guided by small Tims proteins across the intermembrane space to the Tim22 complex of the inner membrane and are then inserted into the membrane in a Δψ-dependent step. Tim44 is upregulated in hyperglycemia and its overexpression reduces proteinuria, renal hypertrophy, cell proliferation and apoptosis, and superoxide production in STZ-induced diabetes in CD-1 mice [48–50].
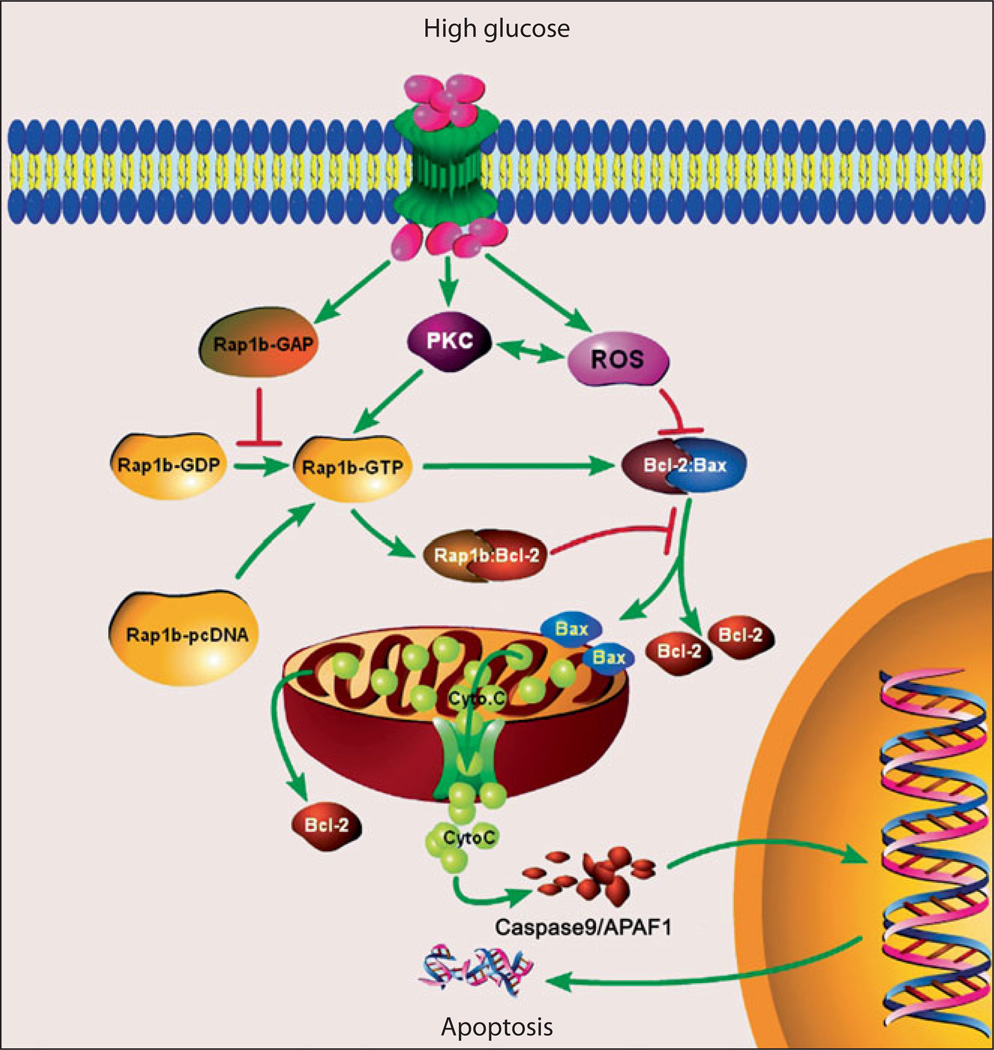
In recent years, the role of small GTP-binding proteins in the pathobiology of hyperglycemia-induced renovascular injury has attracted much attention of many investigators. Major small GTP-binding proteins activated in hyperglycemic state belong to the Ras and Rho family of small GTPases. They exert pleiotropic effects and regulate growth, morphogenesis, cell motility, axonal guidance, cytokinesis and intracelullar trafficking by cycling between an inactive (GDP-bound) and active (GTP-bound) state. Rap1b is a homolog of Ras-related family members that are endowed with anti-apoptotic properties mediated via their effectors (e.g. Raf) [54]. Rap1b is known to stimulate B-Raf activity, thus serving as a modulator of cell proliferation and apoptosis [74]. In the kidney of diabetic mice, apoptosis of tubular cells and dysmorphic mitochondria are observed to be associated with reduction of Bcl-2 and increased Bax expression [54]. Interestingly, the GTPase activity seems to be reduced significantly, although Rap1b expression is increased. Overexpression of Rap1b in HK-2 cells partially reverses these high glucose-induced abnormalities, such as increased DNA fragmentation, altered mitochondrial morphology and disrupted Bcl-2-Bax and Bcl-2-Rap1b interactions (fig. 4) [54]. Overall, it seems that these newly identified genes, RSOR/MIOX, Tim44 and Rap1b, are indeed functionally linked to various intracellular events in kidney tissues subjected to high glucose ambience. The discovery of these new genes relevant to diabetic nephropathy in various animal models should give some impetus to future translational research and to the development of pharmaceutical interventions where these can serve as molecular targets.
Fig. 4.
Schematic drawing of various events following exposure to high glucose ambience and overexpression of Rap1b-GTPase in kidney cells. High glucose ambience leads to activation of PKC and generation of ROS. The latter perturb Bcl2:Bax interactions that leads to the release of mitochondrial cytochrome C, activation of caspases, fragmentation of DNA and consequential apoptosis. Transfection of GTPase Rap1b conceivably stabilizes Bcl2:Bax and Rap1b:Bcl2 interactions and inhibit the release of cytochrome C, oxidant stress, DNA fragmentation and apoptosis [54, 74].
Conclusions and Future Perspectives
Current approaches such as transcriptome and proteome profiling, as well as molecular genetics, using various cell lines, animal models and human samples have greatly facilitated the understanding of the mechanism(s) relevant to the progression of diabetic nephropathy. Based on the data generated by using these techniques, the newly discovered biomarkers could serve as therapeutic targets for the amelioration of diabetic nephropathy, which certainly contribute to the reduction in mortality and morbidity in chronic kidney disease patients that progress to ESRD. In addition to transcriptome and proteome approaches, the future trends for the identification of the biomarkers and therapeutic target genes could include genome-scale DNA methylation profiling [75]. The emerging role of epigenome control of the cancer cells, germ cells and pluripotent stem cells has been emphasized in the transcriptional regulation of various genes that receive sustained long-term injury for years and decades. Intensive long-term versus conventional short-interval symptomatic therapy seems to have remarkable beneficial effects on the risk of cardiovascular disease in patients with type 1 diabetes and this suggests that there may be alterations in the genomic DNA-or histone-methylation pattern which may be linked to the long-term ‘metabolic memory’ for the progression of vascular complications of diabetes [76]. Such a methylation-related profiling would certainly advance the field, especially with respect to development of new biomarkers and various therapeutic strategies. In addition to the delineation of epigenome control of the genes, metabolic phenotyping using 1H spectroscopy [77] and lectin microarray [78] for the glycan profiling would also promote the identification of the new biomarkers of diabetic nephropathy. Finally, integration of the information from different sources using system biology approaches would be an important step in data-mining for the identification of relevant genes that are pertinent to the diagnosis and therapy for diabetic nephropathy.
Acknowledgements
Supported by NIH grants DK28492 and DK60635.
References
- 1.Balakumar P, Arora MK, Reddy J, Anand-Srivastava MB. Pathophysiology of diabetic nephropathy: involvement of multifaceted signaling mechanism. J Cardiovasc Pharmacol. 2009;54:129–138. doi: 10.1097/FJC.0b013e3181ad2190. [DOI] [PubMed] [Google Scholar]
- 2.Reddy AS. Diabetic Nephropathy: Theory and Practice. East Hanover: College Book Publishers; 2004. pp. 1–563. 2004. [Google Scholar]
- 3.Kikkawa R, Koya D, Haneda M. Progression of diabetic nephropathy. Am J Kidney Dis. 2003;41(3) Suppl 1:S19–S21. doi: 10.1053/ajkd.2003.50077. [DOI] [PubMed] [Google Scholar]
- 4.Sugimoto H, Shikata K, Hirata K, Akiyama K, Matsuda M, Kushiro M, Shikata Y, Miyatake N, Miyasaka M, Makino H. Increased expression of intercellular adhesion molecule-1 (ICAM-1) in diabetic rat glomeruli: glomerular hyperfiltration is a potential mechanism of ICAM-1 upregulation. Diabetes. 1997;46:2075–2081. doi: 10.2337/diab.46.12.2075. [DOI] [PubMed] [Google Scholar]
- 5.Koya D, Haneda M, Nakagawa H, Isshiki K, Sato H, Maeda S, Sugimoto T, Yasuda H, Kashiwagi A, Ways DK, King GL, Kikkawa R. Amelioration of accelerated diabetic mesangial expansion by treatment with a PKC beta inhibitor in diabetic db/db mice, a rodent model for type 2 diabetes. FASEB J. 2000;14:439–447. doi: 10.1096/fasebj.14.3.439. [DOI] [PubMed] [Google Scholar]
- 6.Sharma K, McCue P, Dunn SR. Diabetic kidney disease in the db/db mouse. Am J Physiol Renal Physiol. 2003;284:F1138–F1144. doi: 10.1152/ajprenal.00315.2002. [DOI] [PubMed] [Google Scholar]
- 7.Chung AC, Zhang H, Kong YZ, Tan JJ, Huang XR, Kopp JB, Lan HY. Advanced glycation end-products induce tubular CTGF via TGF-beta-Independent Smad3 Signaling. J Am Soc Nephrol. 2010;21:249–260. doi: 10.1681/ASN.2009010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alderson NL, Chachich ME, Youssef NN, Beattie RJ, Nachtigal M, Thorpe SR, Baynes JW. The AGE inhibitor pyridoxamine inhibits lipemia and development of renal and vascular disease in Zucker obese rats. Kidney Int. 2003;63:2123–2133. doi: 10.1046/j.1523-1755.2003.00027.x. [DOI] [PubMed] [Google Scholar]
- 9.Figarola JL, Loera S, Weng Y, Shanmugam N, Natarajan R, Rahbar S. LR-90 prevents dyslipidaemia and diabetic nephropathy in the Zucker diabetic fatty rat. Diabetologia. 2008;51:882–891. doi: 10.1007/s00125-008-0935-x. [DOI] [PubMed] [Google Scholar]
- 10.Kim YS, Kim J, Kim CS, Sohn EJ, Lee YM, Jeong IH, Kim H, Jang DS, Kim JS. KIOM-79, an inhibitor of AGEs-protein cross-linking, prevents progression of nephropathy in Zucker diabetic fatty rats. Evid Based Complement Alternat Med. 2009 doi: 10.1093/ecam/nep078. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satoh M, Fujimoto S, Haruna Y, Arakawa S, Horike H, Komai N, Sasaki T, Tsujioka K, Makino H, Kashihara N. NAD(P)H oxidase and uncoupled nitric oxide synthase are major sources of glomerular superoxide in rats with experimental diabetic nephropathy. Am J Physiol Renal Physiol. 2005;288:F1144–F1152. doi: 10.1152/ajprenal.00221.2004. [DOI] [PubMed] [Google Scholar]
- 12.Asaba K, Tojo A, Onozato ML, Goto A, Quinn MT, Fujita T, Wilcox CS. Effects of NADPH oxidase inhibitor in diabetic nephropathy. Kidney Int. 2005;67:1890–1898. doi: 10.1111/j.1523-1755.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 13.Wolf G. New insights into the pathophysiology of diabetic nephropathy: From hemo-dynamics to molecular pathology. Eur J Clin Invest. 2004;34:785–796. doi: 10.1111/j.1365-2362.2004.01429.x. [DOI] [PubMed] [Google Scholar]
- 14.Tesch GH. Role of macrophages in complications of type 2 diabetes. Clin Exp Pharmacol Physiol. 2007;34:1016–1019. doi: 10.1111/j.1440-1681.2007.04729.x. [DOI] [PubMed] [Google Scholar]
- 15.Usui HK, Shikata K, Sasaki M, Okada S, Matsuda M, Shikata Y, Ogawa D, Kido Y, Nagase R, Yozai K, Ohga S, Tone A, Wada J, Takeya M, Horiuchi S, Kodama T, Makino H. Macrophage scavenger receptor-A-deficient mice are resistant against diabetic nephropathy through amelioration of microinflammation. Diabetes. 2007;56:363–372. doi: 10.2337/db06-0359. [DOI] [PubMed] [Google Scholar]
- 16.Ziyadeh FN, Hoffman BB, Han DC, Iglesias-De La Cruz MC, Hong SW, Isono M, Chen S, McGowan TA, Sharma K. Long-term prevention of renal insufficiency, excess matrix gene expression, and glomerular mesangial matrix expansion by treatment with monoclonal antitransforming growth factor-beta antibody in db/db diabetic mice. Proc Natl Acad Sci USA. 2000;97:8015–8020. doi: 10.1073/pnas.120055097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaede P, Vedel P, Parving HH, Pedersen O. Intensified multifactorial intervention in patients with type 2 diabetes mellitus and microalbuminuria: the Steno type 2 randomized study. Lancet. 1999;353:617–622. doi: 10.1016/S0140-6736(98)07368-1. [DOI] [PubMed] [Google Scholar]
- 18.Gaede P, Vedel P, Larsen N, Jensen GV, Parving HH, Pedersen O. Multifactorial intervention and cardiovascular disease in patients with type 2 diabetes. N Engl J Med. 2003;348:383–393. doi: 10.1056/NEJMoa021778. [DOI] [PubMed] [Google Scholar]
- 19.Gaede P, Lund-Andersen H, Parving HH, Pedersen O. Effect of a multifactorial intervention on mortality in type 2 diabetes. N Engl J Med. 2008;358:580–591. doi: 10.1056/NEJMoa0706245. [DOI] [PubMed] [Google Scholar]
- 20.Hosono N, Kubo M, Tsuchiya Y, Sato H, Kitamoto T, Saito S, Ohnishi Y, Nakamura Y. Multiplex PCR-based real-time invader assay (mPCR-RETINA): a novel SNP-based method for detecting allelic asymmetries within copy number variation regions. Hum Mutat. 2008;29:182–189. doi: 10.1002/humu.20609. [DOI] [PubMed] [Google Scholar]
- 21.Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, Nakamura Y. A high-throughput SNP typing system for genome-wide association studies. J Hum Genet. 2001;46:471–477. doi: 10.1007/s100380170047. [DOI] [PubMed] [Google Scholar]
- 22.The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 23.Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, Bucksa J, Gibson TB, Cordovado SK, Krolewski AS, Nierras CR, Warram JH. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol. 2006;17:1782–1790. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene CN, Keong LM, Cordovado SK, Mueller PW. Sequence variants in the PLEKHH2 region are associated with diabetic nephropathy in the GoKinD study population. Hum Genet. 2008;124:255–262. doi: 10.1007/s00439-008-0548-y. [DOI] [PubMed] [Google Scholar]
- 25.Maeda S. Genome-wide search for susceptibility gene to diabetic nephropathy by gene-based SNP. Diabetes Res Clin Pract. 2004;66(Suppl 1):S45–S47. doi: 10.1016/j.diabres.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka N, Babazono T, Saito S, Sekine A, Tsunoda T, Haneda M, Tanaka Y, Fujioka T, Kaku K, Kawamori R, Kikkawa R, Iwamoto Y, Nakamura Y, Maeda S. Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphisms. Diabetes. 2003;52:2848–2853. doi: 10.2337/diabetes.52.11.2848. [DOI] [PubMed] [Google Scholar]
- 27.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, Kawai K, Iiizumi T, Imanishi M, Shinosaki T, Yanagimoto T, Ikeda M, Omachi S, Kashiwagi A, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakajima M, Nakamura Y, Maeda S. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54:1171–1178. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 28.Susztak K, Bottinger E, Novetsky A, Liang D, Zhu Y, Ciccone E, Wu D, Dunn S, McCue P, Sharma K. Molecular profiling of diabetic mouse kidney reveals novel genes linked to glomerular disease. Diabetes. 2004;53:784–794. doi: 10.2337/diabetes.53.3.784. [DOI] [PubMed] [Google Scholar]
- 29.Wada J, Zhang H, Tsuchiyama Y, Hiragushi K, Hida K, Shikata K, Kanwar YS, Makino H. Gene expression profile in streptozotocin-induced diabetic mice kidneys undergoing glomerulosclerosis. Kidney Int. 2001;59:1363–1373. doi: 10.1046/j.1523-1755.2001.0590041363.x. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh TJ, Chen R, Zhang SL, Liu F, Brezniceanu ML, Whiteside CI, Fantus IG, Ingelfinger JR, Hamet P, Chan JS. Upregulation of osteopontin gene expression in diabetic rat proximal tubular cells revealed by microarray profiling. Kidney Int. 2006;69:1005–1015. doi: 10.1038/sj.ki.5000206. [DOI] [PubMed] [Google Scholar]
- 31.Wilson KH, McIndoe RA, Eckenrode S, Morel L, Agarwal A, Croker BP, She JX. Alterations of renal phenotype and gene expression profiles due to protein overload in NOD-related mouse strains. BMC Nephrol. 2005;6:17. doi: 10.1186/1471-2369-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito, Uchiyama K, Kuroda M, Mizushima K, Aoi W, Kokura S, Ichikawa H, Yoshida N, Yoshikawa T. Laser capture microdissection/GeneChip analysis of gene expression in glomerular cells in diabetic db/db mice. Redox Rep. 2004;9:307–312. doi: 10.1179/135100004225006786. [DOI] [PubMed] [Google Scholar]
- 33.Makino H, Miyamoto Y, Sawai K, Mori K, Mukoyama M, Nakao K, Yoshimasa Y, Suga S. Altered gene expression related to glomerulogenesis and podocyte structure in early diabetic nephropathy of db/db mice and its restoration by pioglitazone. Diabetes. 2006;55:2747–2756. doi: 10.2337/db05-1683. [DOI] [PubMed] [Google Scholar]
- 34.Fan Q, Shike T, Shigihara T, Tanimoto M, Gohda T, Makita Y, Wang LN, Horikoshi S, Tomino Y. Gene expression profile in diabetic KK/Ta mice. Kidney Int. 2003;64:1978–1985. doi: 10.1046/j.1523-1755.2003.00312.x. [DOI] [PubMed] [Google Scholar]
- 35.Ko GJ, Kang YS, Han SY, Lee MH, Song HK, Han KH, Kim HK, Han JY, Cha DR. Pioglitazone attenuates diabetic nephropathy through an anti-inflammatory mechanism in type 2 diabetic rats. Nephrol Dial Transplant. 2008;23:2750–2760. doi: 10.1093/ndt/gfn157. [DOI] [PubMed] [Google Scholar]
- 36.Inagi R, Yamamoto Y, Nangaku M, Usuda N, Okamato H, Kurokawa K, van Ypersele de Strihou C, Yamamoto H, Miyata T. A severe diabetic nephropathy model with early development of nodule-like lesions induced by megsin overexpression in RAGE/iNOS transgenic mice. Diabetes. 2006;55:356–366. doi: 10.2337/diabetes.55.02.06.db05-0702. [DOI] [PubMed] [Google Scholar]
- 37.Cheng DW, Jiang Y, Shalev A, Kowluru R, Crook ED, Singh LP. An analysis of high glucose and glucosamine-induced gene expression and oxidative stress in renal mesangial cells. Arch Physiol Biochem. 2006;112:189–218. doi: 10.1080/13813450601093518. [DOI] [PubMed] [Google Scholar]
- 38.Han SH, Yang S, Jung DS, Li JJ, Kim JJ, Kwak SJ, Kim DK, Moon SJ, Lee JE, Han DS, Kang SW. Gene expression patterns in glucose-stimulated podocytes. Biochem Biophys Res Commun. 2008;370:514–518. doi: 10.1016/j.bbrc.2008.03.121. [DOI] [PubMed] [Google Scholar]
- 39.Qi W, Chen X, Zhang Y, Holian J, Mreich E, Gilbert RE, Kelly DJ, Pollock CA. High glucose induces macrophage inflammatory protein-3 alpha in renal proximal tubule cells via a transforming growth factor-beta1 dependent mechanism. Nephrol Dial Transplant. 2007;22:3147–3153. doi: 10.1093/ndt/gfm365. [DOI] [PubMed] [Google Scholar]
- 40.Qi W, Chen X, Gilbert RE, Zhang Y, Waltham M, Schache M, Kelly DJ, Pollock CA. High glucose-induced thioredoxin-interacting protein in renal proximal tubule cells is independent of transforming growth factor-beta1. Am J Pathol. 2007;171:744–754. doi: 10.2353/ajpath.2007.060813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moczulski DK, Fojcik H, Wielgorecki A, Trautsolt W, Gawlik B, Kosiorz-Gorczynska S, Oczko-Wojciechowska M, Wiench M, Strojek K, Zukowska-Szczechowska E, Grzeszczak W. Expression pattern of genes in peripheral blood mononuclear cells in diabetic nephropathy. Diabet Med. 2007;24:266–271. doi: 10.1111/j.1464-5491.2006.02067.x. [DOI] [PubMed] [Google Scholar]
- 42.Baelde HJ, Eikmans M, Doran PP, Lappin DW, de Heer E, Bruijn JA. Gene expression profiling in glomeruli from human kidneys with diabetic nephropathy. Am J Kidney Dis. 2004;43:636–650. doi: 10.1053/j.ajkd.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 43.Szeto CC, Lai KB, Chow KM, Szeto CY, Yip TW, Woo KS, Li PK, Lai FM. Messenger RNA expression of glomerular podocyte markers in the urinary sediment of acquired proteinuric diseases. Clin Chim Acta. 2005;361:182–190. doi: 10.1016/j.cccn.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 45.Song Y, Ailenberg M, Silverman M. Cloning of a novel gene in the human kidney homologous to rat munc13s: its potential role in diabetic nephropathy. Kidney Int. 1998;53:1689–1695. doi: 10.1046/j.1523-1755.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 46.Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 47.Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Wada J, Hashimoto I, Eguchi J, Yasuhara A, Kanwar YS, Shikata K, Makino H. Therapeutic approach for diabetic nephropathy using gene delivery of translocase of inner mitochondrial membrane 44 by reducing mitochondrial superoxide production. J Am Soc Nephrol. 2006;17:1090–1101. doi: 10.1681/ASN.2005111148. [DOI] [PubMed] [Google Scholar]
- 49.Matsuoka T, Wada J, Hashimoto I, Zhang Y, Eguchi J, Ogawa N, Shikata K, Kanwar YS, Makino H. Gene delivery of Tim44 reduces mitochondrial superoxide production and ameliorates neointimal proliferation of injured carotid artery in diabetic rats. Diabetes. 2005;54:2882–2890. doi: 10.2337/diabetes.54.10.2882. [DOI] [PubMed] [Google Scholar]
- 50.Wada J, Kanwar YS. Characterization of mammalian translocase of inner mitochondrial membrane (Tim44) isolated from diabetic newborn mouse kidney. Proc Natl Acad Sci USA. 1998;95:144–149. doi: 10.1073/pnas.95.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanwar YS, Akagi S, Nayak B, Sun L, Wada J, Xie P, Thakur A, Chugh SS, Danesh FR. Renal-specific oxido-reductase biphasic expression under high glucose ambience during fetal versus neonatal development. Kidney Int. 2005;68:1670–1683. doi: 10.1111/j.1523-1755.2005.00611.x. [DOI] [PubMed] [Google Scholar]
- 52.Yang Q, Dixit B, Wada J, Tian Y, Wallner EI, Srivastva SK, Kanwar YS. Identification of a renal-specific oxido-reductase in newborn diabetic mice. Proc Natl Acad Sci USA. 2000;97:9896–9901. doi: 10.1073/pnas.160266197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun L, Pan X, Wada J, Haas CS, Wuthrich RP, Danesh FR, Chugh SS, Kanwar YS. Isolation and functional analysis of mouse UbA52 gene and its relevance to diabetic nephropathy. J Biol Chem. 2002;277:29953–29962. doi: 10.1074/jbc.M204665200. [DOI] [PubMed] [Google Scholar]
- 54.Sun L, Xie P, Wada J, Kashihara N, Liu FY, Zhao Y, Kumar D, Chugh SS, Danesh FR, Kanwar YS. Rap1b GTPase ameliorates glucose-induced mitochondrial dysfunction. J Am Soc Nephrol. 2008;19:2293–2301. doi: 10.1681/ASN.2008030336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin S, Chugh S, Pan X, Wallner EI, Wada J, Kanwar YS. Identification of up-regulated Ras-like GTPase, Rap1b, by suppression subtractive hybridization. Kidney Int. 2001;60:2129–2141. doi: 10.1046/j.1523-1755.2001.00061.x. [DOI] [PubMed] [Google Scholar]
- 56.McMahon R, Murphy M, Clarkson M, Taal M, Mackenzie HS, Godson C, Martin F, Brady HR. IHG-2, a mesangial cell gene induced by high glucose, is human gremlin. Regulation by extracellular glucose concentration, cyclic mechanical strain, and transforming growth factor-beta1. J Biol Chem. 2000;275:9901–9904. doi: 10.1074/jbc.275.14.9901. [DOI] [PubMed] [Google Scholar]
- 57.Murphy M, Godson C, Cannon S, Kato S, Mackenzie HS, Martin F, Brady HR. Suppression subtractive hybridization identifies high glucose levels as a stimulus for expression of connective tissue growth factor and other genes in human mesangial cells. J Biol Chem. 1999;274:5830–5834. doi: 10.1074/jbc.274.9.5830. [DOI] [PubMed] [Google Scholar]
- 58.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 59.Schelling JR, El-Meanawy MA, Barathan S, Dodig T, Iyengar SK, Sedor JR. Generation of kidney transcriptomes using serial analysis of gene expression. Exp Nephrol. 2002;10:82–92. doi: 10.1159/000049903. [DOI] [PubMed] [Google Scholar]
- 60.Cheval L, Duong Van Huyen JP, Bruneval P, Verbavatz JM, Elalouf JM, Doucet A. Plasticity of mouse renal collecting duct in response to potassium depletion. Physiol Genomics. 2004;19:61–73. doi: 10.1152/physiolgenomics.00055.2004. [DOI] [PubMed] [Google Scholar]
- 61.Sengoelge G, Luo W, Fine D, Perschl AM, Fierlbeck W, Haririan A, Sorensson J, Rehman TU, Hauser P, Trevick JS, Kulak SC, Wegner B, Ballermann BJ. A SAGE-based comparison between glomerular and aortic endothelial cells. Am J Physiol Renal Physiol. 2005;288:F1290–F1300. doi: 10.1152/ajprenal.00076.2004. [DOI] [PubMed] [Google Scholar]
- 62.Schena M, Shalon D, Davis R, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 63.Cohen CD, Lindenmeyer MT, Eichinger F, Hahn A, Seifert M, Moll AG, Schmid H, Kiss E, Grone E, Grone HJ, Kretzler M, Werner T, Nelson PJ. Improved elucidation of biological processes linked to diabetic nephropathy by single probe-based microarray data analysis. PLoS One. 2008;3:e2937. doi: 10.1371/journal.pone.0002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naito Y, Uchiyama K, Mizushima K, Kuroda M, Akagiri S, Takagi T, Handa O, Kokura S, Yoshida N, Ichikawa H, Takahashi J, Yoshikawa T. Microarray profiling of gene expression patterns in glomerular cells of astaxanthin-treated diabetic mice: a nutrigenomic approach. Int J Mol Med. 2006;18:685–695. [PubMed] [Google Scholar]
- 65.Thongboonkerd V, Barati MT, McLeish KR, Benarafa C, Remold-O’Donnell E, Zheng S, Rovin BH, Pierce WM, Epstein PN, Klein JB. Alterations in the renal elastin-elastase system in type 1 diabetic nephropathy identified by proteomic analysis. J Am Soc Nephrol. 2004;15:650–662. doi: 10.1097/01.asn.0000115334.65095.9b. [DOI] [PubMed] [Google Scholar]
- 66.Tilton RG, Haidacher S, Lejeune W, Zhang X, Zhao Y, Kurosky A, Brasier A, Denner L. Diabetes-induced changes in the renal cortical proteome assessed with 2-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2007;7:1729–1742. doi: 10.1002/pmic.200700017. [DOI] [PubMed] [Google Scholar]
- 67.Sharma K, Lee S, Han S, Francos B, McCue P, Wassell R, Shaw MA, RamachandraRao SP. Two-dimensional fluorescence difference gel electrophoresis analysis of the urine proteome in human diabetic nephropathy. Proteomics. 2005;5:2648–2655. doi: 10.1002/pmic.200401288. [DOI] [PubMed] [Google Scholar]
- 68.Dihazi H, Muller GA, Lindner S, Meyer M, Asif AR, Oellerich M, Strutz F. Characterization of diabetic nephropathy by urinary proteomic analysis: identification of a processed ubiquitin form as a differentially excreted protein in diabetic nephropathy patients. Clin Chem. 2007;53:1636–1645. doi: 10.1373/clinchem.2007.088260. [DOI] [PubMed] [Google Scholar]
- 69.Rossing K, Mischak H, Dakna M, Zurbig P, Novak J, Julian BA, Good DM, Coon JJ, Tarnow L, Rossing P. Urinary proteomics in diabetes and CKD. J Am Soc Nephrol. 2008;19:1283–1290. doi: 10.1681/ASN.2007091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang YH, Zhang S, Cui JF, Lu B, Dong XH, Song XY, Liu YK, Zhu XX, Hu RM. Diagnostic potential of serum protein pattern in Type 2 diabetic nephropathy. Diabet Med. 2007;24:1386–1392. doi: 10.1111/j.1464-5491.2007.02312.x. [DOI] [PubMed] [Google Scholar]
- 71.Kanwar YS, Wada J, Sun L, Xie P, Wallner EI, Chen S, Chugh S, Danesh FR. Diabetic nephropathy: mechanisms of renal disease progression. Exp Biol Med (Maywood) 2008;233:4–11. doi: 10.3181/0705-MR-134. [DOI] [PubMed] [Google Scholar]
- 72.Nayak B, Xie P, Akagi S, Yang Q, Sun L, Wada J, Thakur A, Danesh FR, Chugh SS, Kanwar YS. Modulation of renal-specific oxidoreductase/myo-inositol oxygenase by high-glucose ambience. Proc Natl Acad Sci USA. 2005;102:17952–17957. doi: 10.1073/pnas.0509089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziyadeh FN, Simmons DA, Snipes ER, Goldfarb S. Effect of myo-inositol on cell proliferation and collagen transcription and secretion in proximal tubule cells cultured in elevated glucose. J Am Soc Nephrol. 1991;1:1220–1229. doi: 10.1681/ASN.V1111220. [DOI] [PubMed] [Google Scholar]
- 74.Lin S, Sahai A, Chugh SS, Pan X, Wallner EI, Danesh FR, Lomasney JW, Kanwar YS. High glucose stimulates synthesis of fibronectin via a novel protein kinase C, Rap1b, and B-Raf signaling pathway. J Biol Chem. 2002;277:41725–41735. doi: 10.1074/jbc.M203957200. [DOI] [PubMed] [Google Scholar]
- 75.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein B, Nusbaum C, Jaffe D, Gnirke A, Jaenisch R, Lander E. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, Raskin P, Zinman B. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med. 2005;353:2643–2653. doi: 10.1056/NEJMoa052187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ebe Y, Kuno A, Uchiyama N, Koseki-Kuno S, Yamada M, Sato T, Narimatsu H, Hirabayashi J. Application of lectin microarray to crude samples: differential glycan profiling of lec mutants. J Biochem. 2006;139:323–327. doi: 10.1093/jb/mvj070. [DOI] [PubMed] [Google Scholar]