Abstract
A major aim of synthetic biology is to program novel cellular behaviors using engineered gene circuits. Early endeavors focused on building simple circuits that fulfill simple functions, such as logic gates, bistable toggle switches, and oscillators. These gene circuits have primarily focused on single-cell behaviors since they operate intracellularly. Thus, they are often susceptible to cell-cell variations due to stochastic gene expression. Cell-cell communication offers an efficient strategy to coordinate cellular behaviors at the population level. To this end, we review recent advances in engineering cell-cell communication to achieve reliable population dynamics, spanning from communication within single species to multispecies, from one-way sender-receiver communication to two-way communication in synthetic microbial ecosystems. These engineered systems serve as well-defined model systems to better understand design principles of their naturally occurring counterparts and to facilitate novel biotechnology applications.
Keywords: synthetic biology, cell-cell communication, quorum sensing, synthetic ecosystems
1. Introduction
At the interface of engineering and biology, a major aim of synthetic biology is to program cells to perform desirable functions in a predictable manner [1, 2]. These programmed functions could have potential applications in biomedicine [3], biofuel [4], bioremediation [5], and cellular computation [6].
We describe here a survey of recent achievements in synthetic biology, highlighting recent trends. Engineered gene circuits have experienced a hierarchical development in terms of circuit complexity, from simple logic gates and regulatory motifs to communication-based circuits. One salient trend is the transition from programming single-cell behaviors to cellular population behaviors, from programming cellular temporal behaviors to spatiotemporal dynamics, and from programming individual microbes to microbial communities.
2. Single-cell synthetic biology
Early studies in synthetic biology primarily focused on the construction of simple genetic devices, such as logic gates, bi-stable switches, and oscillators. These genetic circuits generally operated intracellularly to program single-cell behaviors. Yokobayashi et al. programmed genetic elements to function as logic gates in Escherichia coli [7] (Fig. 1A), in which the LacI/IPTG/Plac parts constitute a logic gate that performs the IMPLIES logic function. They then used a directed evolution approach to optimize the genetic elements in the circuit, making the IMPLIES logic gate match properly to the downstream Inverter logic gate. In an attempt to construct several different types of genetic logic gates, Guet et al. constructed a library of genetic networks with varying connectivity in E. coli by combinatorial synthesis [8], which performed logic gate functions (e.g., NOR, NOT IF, NAND, etc). In addition, Anderson et al. constructed a synthetic AND gate in E. coli that integrates information (salicylate and arabinose induction) from two promoters (Psal and PBAD) as inputs and activates a promoter output (PT7) only when both input promoters are transcriptionally active [9].
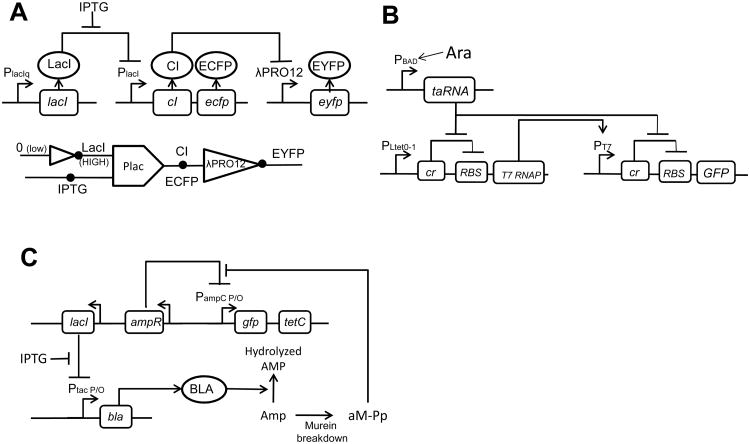
Figure 1.
Engineering genetic circuits at the single-cell level.
(A) A synthetic IMPLIES gate that is interfaced with an Inverter. As shown in the upper panel, the IMPLIES gate involves a constitutively expressed LacI repressor, which inhibits the transcription of CI from the PlacI promoter; IPTG activates the PlacI promoter and induces CI and ECFP expression; CI inhibits the λPRO12 promoter to repress EYFP expression. As shown in the bottom panel, the PlacI promoter comprises an IMPLIES logic gate wherein two inputs, LacI and IPTG, induce the response of the output CI, which acts as the input to the interfaced Inverter based on the λPRO12 promoter. Arrows indicate synthesis. Blunt bars indicate repression. Figure adapted from Yokobayashi et al. [7].
(B) A synthetic riboregulated transcriptional cascade (RTC) counter circuit that can count two pulses of arabinose stimulations. The RTC two-counter includes a transcriptional cascade: promoter PLtet0-1 drives the transcription of T7 RNA polymerase (RNAP), whose protein binds the PT7 promoter and mediates transcription of a green fluorescent protein (GFP). The two genes (T7 RNAP and GFP) are in turn repressed by the riboregulator (cr). Meanwhile, a short, transactivating noncoding RNA (taRNA), driven by the arabinose promoter PBAD, can bind to cr, relieving its repression on RBS and effectively activating the translation of T7 RNAP and GFP proteins. Figure adapted from Friedland et al. [17].
(C) A tunable band-pass filter conferred by a two-arm feed-forward loop (FFL). In one arm, insufficient β-lactamase (BLA) activity causes accumulation of Amp, leading to compromised cell wall synthesis and repressed cell growth. In the second arm, cell wall (murein) breakdown of Amp results in the accumulation of aM-pentapeptide (aM-Pp), which activates the promoter PampC via interacting with the promoter's repressor AmpR. As a result, aM-Pp induces TetC synthesis (which confers cells' Tet resistance) and GFP expression. Alternatively, BLA can be expressed by IPTG induction of Ptac P/O promoter, which is repressed by LacI. Figure adapted from Sohka et al. [18].
Bistable switches often consist of positive-feedback loops mediated by auto-activation or mutual inhibition, which can convert graded inputs into abrupt responses [10]. As an example, Gardner et al. constructed a synthetic bistable toggle switch in E. coli [11], consisting of two mutually repressive transcriptional regulators. Kobayashi et al. further interfaced the toggle switch with a SOS-response signaling pathway and a quorum sensing signaling pathway [12]. An analogous bistable switch based on mutual inhibition has also been implemented in mammalian cells and mice [13]. In addition, there are other synthetic switches that employ different circuit structures. One example is the auto-activation gene circuit [14].
The first artificial genetic oscillator (the “repressilator”) was engineered by Elowitz and Leibler [15]. The repressilator comprises three transcriptional repressors that form a negative feedback loop. By adopting a relaxation-type circuit architecture with interlocked positive and negative feedback loops, Atkinson et al. constructed a genetic oscillator that generates multiple cycles of damped oscillations [14]. Using a similar architecture but different implementation, Stricker et al. recently constructed a genetic network that can exhibit robust and tunable oscillatory behaviors in single cells [16].
Synthetic gene circuits can also perform a variety of biocomputing functions. For example, Friedland et al. constructed a cellular counter in E. coli that can count up to three induction events [17]. Figure 1B illustrates a riboregulated transcriptional cascade (RTC) two-counter that responds to two pulses of arabinose (Ara) stimulation but does not significantly respond to only one pulse of Ara stimulation [17]. Sohka et al. engineered a bacterial band-pass filter via the construction of a feed-forward loop, which is externally tunable by IPTG and Ampicillin (Amp) concentrations (see Fig. 1C) [18]. Upon plating cells on agar plates with certain concentrations of Amp, IPTG, and tetracycline (Tet), rings or other cellular fluorescence patterns can form.
The above-mentioned gene circuits have focused on programming single cells. That is, they act intracellularly. One common difficulty encountered when dealing with these circuits is that the programmed behavior may vary dramatically from cell to cell, even in an isogenic population. Such heterogeneity is largely due to the stochastic nature of cellular reactions, including gene expression. This stochasticity represents a major hurdle to achieve reliable and coherent behaviors in a population of cells and often makes the quantitative prediction of circuit dynamics difficult. Several strategies may enable more reliable, programmed cellular behavior. One is to develop a library of well-quantified, standardized “biobricks” [19], which can streamline construction of more complex gene circuits. Another way may use proper feedback regulation or cell-cell communication to reduce noise. Negative feedback offers a basic noise reduction mechanism [20]. Despite the stabilization effect of negative feedback on circuit function, one must caution against a long time delay, which can result in oscillations [15, 21-23]. Recent studies also suggest cell-cell communication offers an alternative strategy to alleviate the stochastic nature of cellular processes, thereby facilitating the achievement of robust gene expression dynamics and the engineering of reliable system behaviors in a population of cells [24].
3. Population-level synthetic biology via engineered cell-cell communication
In nature, a wide variety of biological processes are regulated by cell-cell communication, ranging from sophisticated embryogenesis and tissue patterning [25] to bacterial biofilm formation [26]. In multicellular organisms, cells can perceive their microenvironment and make fate decisions to coordinate their collective behaviors in a population, leading to normal physiology and tissue homeostasis. Unicellular organisms (e.g., bacteria) were once perceived as isolated, with individual cells in a population acting independently of one another. It is now recognized that communication exists universally within and across bacterial species [27]. Such communication coordinates behaviors between individual cells to enable a population to act as if it were a multicellular organism [28]. For example, the marine bacterium Vibrio fischeri produces an extracellular factor, called an autoinducer, which coordinates the expression of genes responsible for light production. In this regulatory network, the population must reach a threshold density, or “quorum”, before light is produced. This autoinducer was identified as N-3-(oxohexanoyl) homoserine lactone (3OC6HSL), an acylated homoserine lactone (AHL) molecule broadly adopted as a communication signal by Gram-negative bacteria. The term “quorum sensing” (QS) was coined to specifically refer to density-dependent, coordinated gene expression in bacterial populations. QS is a ubiquitous mechanism in a wide range of Gram-negative and -positive bacteria to regulate their phenotypes and multicellular behaviors, such as pathogenicity, bioluminescence, and biofilm formation [26, 29, 30]. Since the small QS signaling molecules can diffuse freely in the environment and across cell membranes, they can trigger responses in cells nearby or at a distance. This QS-mediated bacterial communication is analogous to the endocrine and paracrine communication mechanisms in multicelluar organisms. Two systems of natural QS genetic components, the LuxR/LuxI QS system from V. fischeri and the LasR/LasI and RhlR/RhlI QS systems from Pseudomonas aeruginosa, have been widely adopted to engineer synthetic cell-cell communication in bacteria [31-34].
In addition to these naturally occurring QS components, there are alternative, engineered components that can fulfill cell-cell communication function. The first class involves engineered LuxR which was constructed using a directed evolution approach [35]. Such evolved variants of LuxR offer either a high degree of specificity in response to a specific AHL molecule or a broad range of specificities for a wide variety of AHLs [36, 37]. LuxR variants with narrow specificities to different AHL signals may be useful in avoiding potential crosstalk in programmed multichannel cell-cell communication circuits. Another class of synthetic cell-cell communication components consists of metabolites synthesized and secreted by cells. For example, Butler et al. reported using a common metabolite, acetate, to engineer tunable, artificial cell-cell communication [38, 39]; Shou et al. reported using two metabolites, adenine and lysine, to construct mutualistic cooperation between two yeast cells [40]. These two examples will be discussed in more detail in the following sections.
3.1 Programming single populations
Engineering assemblies of cells which work collectively cannot be accomplished by simply programming individual cells' behaviors alone. Rather, the utilization of an engineered cell-cell communication module is essential to coordinate individual cells' activities and achieve coherent behavior in cell populations. Such a strategy was implemented by You et al. in the form of a population controller circuit engineered in E. coli by interfacing cell-cell communication with regulated cellular killing [41]. In this circuit, the LuxR/LuxI system is used to broadcast and detect cell density, which autonomously regulates downstream cell death once a threshold cell density has been reached (Fig. 2A). Thus, a sufficiently high cell density results in increased production of killer protein, which ultimately causes cell death. Taken together, this circuit acts as a population density controller. Balagadde et al. invented a microfluidic device, called a microchemostat, which allows for long-term characterization of the nonlinear dynamics of this population control circuit [22]. The microchemostat can maintain continuous operation of micro bioreactors with chip-based control, enabling long-term culturing and real-time monitoring of extremely small populations of bacteria with single-cell resolution and reduced gene mutation. Using a similar circuit design strategy, Anderson et al. linked the capacity to invade mammalian cells with bacterial density by controlling an invasin gene with a quorum sensing module (Fig. 2B) [42]. Here, high bacterial density triggers expression of the invasin gene, enabling E. coli to invade malignant cells. This work further demonstrated that QS-controlled cell-cell communication can control diverse biological processes as outputs.
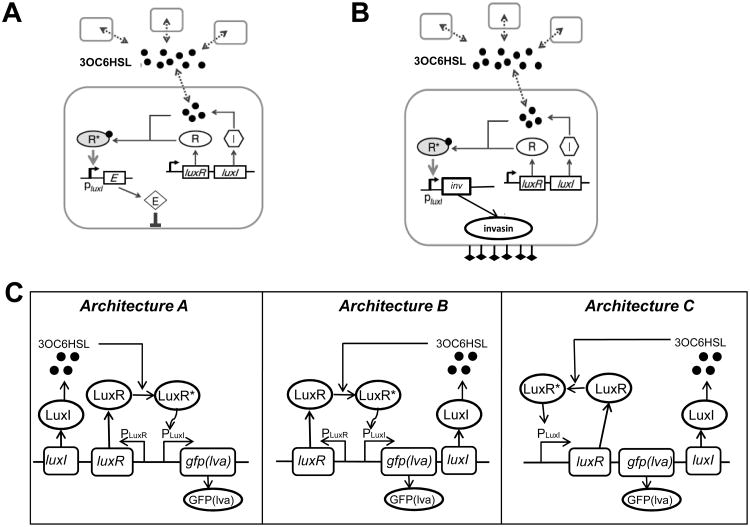
Figure 2.
Programming single-species' populations via engineered cell-cell communication.
(A) A population-control circuit which modulates population density of a single species. The circuit uses the LuxR/LuxI QS circuit, wherein both LuxR and LuxI genes are under the control of the Plac promoter, inducible by IPTG. LuxI produces 3OC6HSL, which can bind and activate LuxR at a sufficiently high concentration. Activated LuxR-3OC6HSL complex (R*) in turn binds to the PluxI promoter to induce expression of a killer gene, ccdB. High expression of ccdB leads to cell killing, thus reducing population density. Figure adapted from You et al. [41].
(B) Synthetic QS circuit to control bacterial invasion of malignant cancer cells. Employing a similar network structure as (A), the killer gene (LacZα-ccdB) in (A) is replaced by an invasion gene inv. Thereby, the invasion of mammalian cells by the programmed bacteria is under the control of QS. Figure adapted from Anderson et al. [42].
(C) Three network architectures for QS gene circuits synthesized by circuit reshuffling. Architecture A: LuxI and LuxR are constitutively expressed, GFP(lva) is under the control of the PluxI promoter, which can be activated by the LuxR-autoinducer complex (LuxR*). Architecture B: LuxR is constitutively expressed; LuxI synthesizes 3OC6HSL and autoactivates expression of itself upon binding with LuxR, forming a positive-feedback loop. Architecture C: Expression of LuxR and LuxI are all under the control of the positive-feedback loop consisting of LuxR-autoinducer and the PluxI promoter. Figure adapted from Haseltine and Arnold [43].
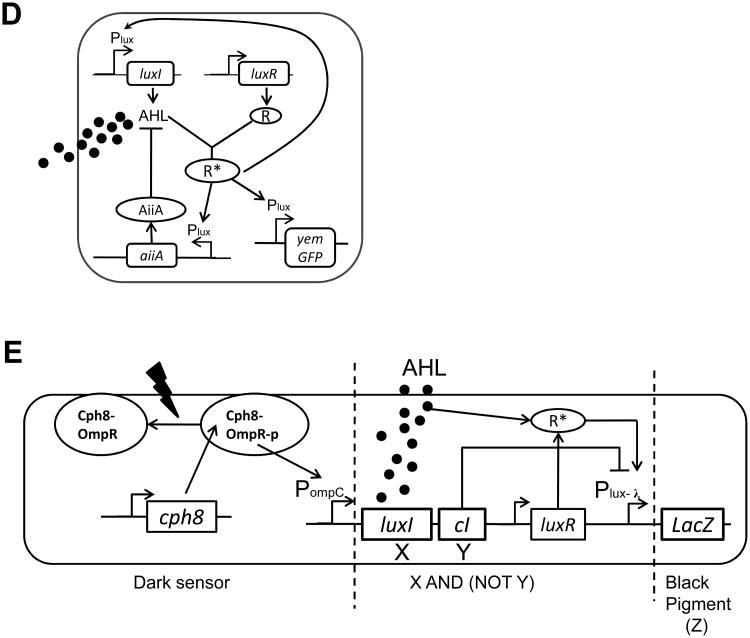
(D) The genetic diagram of a synchronized genetic oscillator. The Plux promoter induces the production of three genes (LuxI, aiiA and yemGFP). The activated LuxR-AHL complex (R*) binds the Plux promoter and activates gene expression. AiiA is an effective protease that catalyzes the cleavage of AHL. The overall circuit functions as coupled negative and positive feedback loops, wherein AHL activates synthesis of itself (positive feedback), and induces AiiA expression, which in turn represses AHL's accumulation by catalyzing AHL's degradation (negative feedback). Figure adapted from Danino et al. [44].
(E) The genetic circuit dictating an edge detection algorithm. The genetic cascade includes three sequential parts: First, the dark sensor part includes a light-sensitive kinase protein Cph8. In the presence of red light, the Cph8 kinase activity is inhibited, precluding the transfer of a phosphoryl group to the response regulator OmpR and subsequent transcriptional activation of the OmpC promoter (PompC). The dark sensor therefore functions as a NOT logic gate. Second, in the X and (NOT Y) logic gate, the PompC promoter drives the synthesis of LuxI and CI. LuxI produces a quorum-sensing signal AHL. CI inhibits the Plux- λ promoter. LuxR-AHL complex (R*) activates the Plux- λ promoter. Therefore, Plux- λ functions as an X and (NOT Y) gate. Lastly, the Plux- λ promoter drives the synthesis of LacZ, which produces β-galactosidase, an enzyme that subsequently cleaves a substrate in the media to produce black pigment. Figure adapted from Tabor et al. [45].
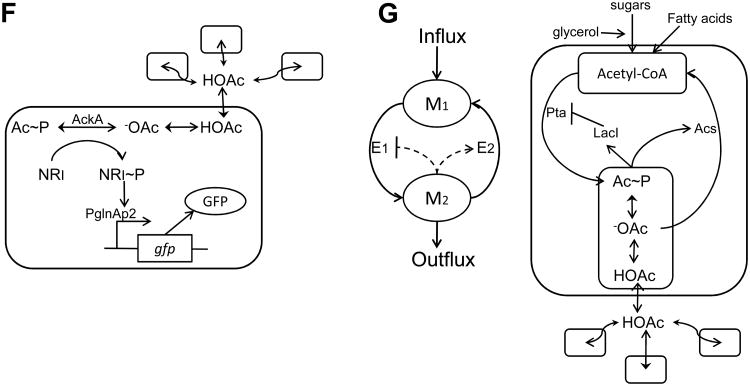
(F) Artificial cell-cell communication using a metabolite signal. Amino acid metabolism results in a metabolite, acetate, as a byproduct. Acetic acid (HOAc), diffuses freely across the cell membrane, playing the role of an artificial cell-cell communication signal. In the cytoplasm, -OAc can be phosphorylated to acetyl phosphate (Ac∼P), which can transfer its phosphate group to NR1. Upon phosphorylation, NR1∼P becomes a transcriptional regulator of the glnAp2 promoter which activates expression of GFP. Therefore, the GFP level reflects the acetate concentration, which in turn is correlated with cell density. Figure adapted from Butler et al. [38].
(G) Metabolator, a synthetic gene-metabolic oscillator that integrates transcriptional regulation with metabolism. The left panel shows the conceptual diagram of the circuit design in which the two metabolite pools (M1 and M2) are controlled by two enzymes (E1 and E2). The right panel illustrates the biological realization of the conceptual design. Acetyl-CoA is converted to acetyl phosphate (Ac∼P) by phosphate acetyltransferase (Pta), which converts to acetate (-OAc) by acetate kinase (Ack). Meanwhile, the enzyme acetyl-CoA synthetase (Acs) is induced in the presence of acetate (-OAc). When Ac∼P reaches a critical concentration, it represses the expression of Pta. Figure adapted from Fung et al. [46].
The basic QS lux module consists of three regulatory components: LuxR, LuxI, and the lux promoter. However, the native lux operon represents one possible combination of these components. To examine the consequences of reshuffling these components, Haseltine and Arnold rewired the lux system and implemented three network architectures (Fig. 2C) [43]. In architecture A, luxI and luxR are constitutively expressed from the PluxR promoter, which leads to a graded response, wherein fluorescence per cell increases linearly with cell density until the fluorescence saturates. In architecture B, luxR is constitutively expressed from the PluxR promoter, and luxI and gfplva are expressed from the PluxI promoter, forming a LuxI auto-activation positive feedback loop. This architecture results in an ultra-sensitive response characteristic of a switch. Finally, in architecture C, luxI, luxR and gfplva are all expressed from the PluxI promoter. This architecture introduces a higher level of nonlinearity to the circuit by the double positive feedback loops of LuxR and LuxI, which results in bistability over a wide range of cellular densities.
Synthetic oscillators that function intracellularly often lead to unsynchronized oscillations in individual cells [14, 15]. A combination of the individual cellular oscillations over the entire population may result in a loss of overall oscillatory behavior. Exploiting cell-cell communication, Danino et al. engineered a synchronized genetic oscillator that operates in a population (Fig. 2D) [44]. The underlying mechanism of the genetic construct (Fig. 2D) is coupled positive and negative feedback loops under the control of diffusible AHL across cell membranes, in which AHL activates its own synthesis (positive feedback), and the synthesis of AiiA, which enzymatically degrades AHL (negative feedback). The synchronized oscillations arise because the AHL signal can both activate the genes necessary for intracellular oscillations and mediate coupling between cells.
Engineered cell-cell communication also enables sophisticated signal processing functions. For example, Tabor et al. constructed a parallel and spatially distributed edge detection algorithm in E. coli, where each bacterium within a spatially distributed population functions as an independent signal processor (Fig. 2E) [45]. The genetic edge detection algorithm allows a lawn of bacteria to identify light-dark boundaries (i.e., edges). To accomplish this logic, the gene circuit in each bacterium requires four components: a dark sensor, a cell-cell communication module, X AND (NOT Y) genetic logic, and a synthesizer of black pigment. Such logic only allows cells in the light but proximal to dark areas to activate LacZ to produce black pigment. Thus, a population of cells programmed with this edge detection gene circuit can produce black pigment only at the boundary between light and dark areas.
The above-mentioned efforts in coordinating individual cells' activities were achieved by taking naturally existing QS components from their native host (e.g., V. fischeri) and integrating them into a target host, such as E. coli. Alternatively, Butler et al. reported the engineering of artificial cell-cell communication based on the synthesis and sensing of metabolites [38, 39]. They demonstrated the design and construction of a gene-metabolic circuit that uses a common metabolite, acetate, to achieve tunable artificial cell-cell communication (see Fig. 2F). Since acetate is constantly produced and secreted by the cell, GFP expression via the promoter glnAp2 depends on the population density, and high levels of transcription only occur when a threshold cell density is reached, satisfying the fundamental characteristic of QS.
The use of the primary metabolite acetate as a signal molecule enables the communication of metabolic states. By taking advantage of the integration of transcriptional regulation with metabolism, Fung et al. constructed an oscillatory circuit, termed the “metabolator”, using glycolytic flux to regulate the signaling metabolite acetyl phosphate (Fig. 2G) [46]. The conceptual design of the metabolator consists of a flux-carrying network with two interconverting metabolite pools (M1 and M2) catalyzed by two enzymes (E1 and E2), the expression of which are negatively and positively regulated by M2, respectively (left panel, Fig. 2G). The biological circuit structure to realize this conceptual design is based on programming the acetate pathway in E. coli (right panel, Fig. 2G). Consistent with model predictions, this system was able to generate sustained oscillatory dynamics.
3.2 Programming interacting populations
One-way communication
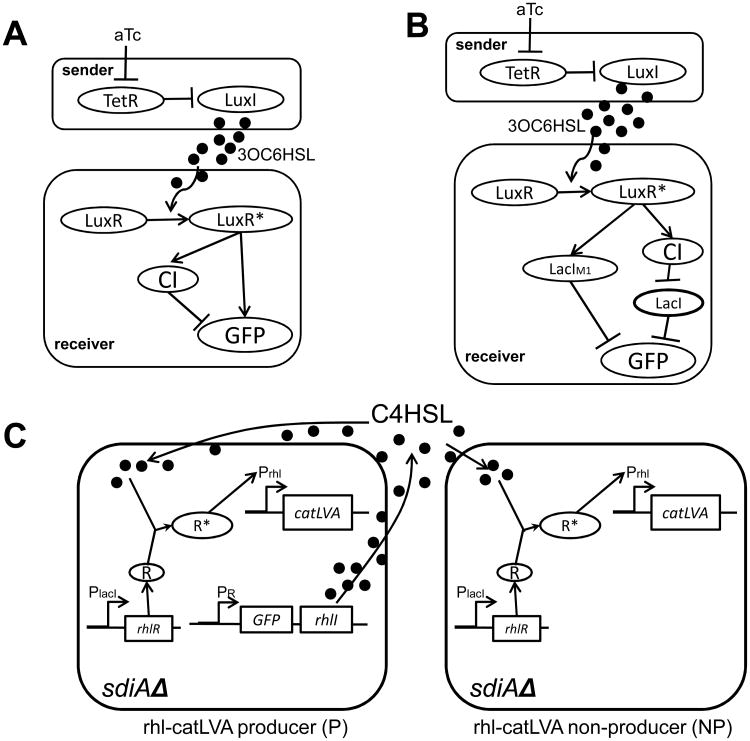
Basu et al. extended engineering cell-cell communication from within a single population to between two populations by constructing two synthetic genetic circuits enabling one-way communication between signal senders and receivers [47, 48]. One example is a pulse-generating network in E. coli [47], in which the receiver cells exhibit transient gene expression in response to a sustained input of AHL signals produced by the sender cells. As depicted in Fig. 3A, upon aTc induction which activates the circuit, the sender cells continuously synthesize an AHL, 3OC6HSL, which freely diffuses to spatially proximate receiver cells to activate their gene expression. The receiver cells contain a gene circuit that functions as a feed-forward motif with two arms of regulation: in the first arm, activated LuxR (LuxR*) induces expression of GFP(lva); in the second, LuxR* also induces CI expression, which in turn inhibits GFP(lva) with a time delay relative to the first arm. This circuit confers a pulse-generating response of the receiver cells to the long-lasting increase of AHL concentration produced by the sender cells. Before induction of the sender circuit, there is a low level of GFP expression in the receiver; upon induction, the GFP level in the receiver increases initially, then decreases before settling to a low level.
Figure 3.
Engineering one-way cell-cell communication.
(A) A synthetic pulse-generator. It consists of one-way communication between the sender and the receiver by a QS mechanism. In the sender cells, upon aTc induction, LuxI synthesizes the autoinducer 3OC6HSL. The receiver cells include a feed-forward network comprised by two arms of regulation: in the first arm, the LuxR-3OC6HSL complex (LuxR*) activates expression of repressor CI, which in turn represses GFP expression; in the second arm, LuxR* induces GFP expression. Figure adapted from Basu et al. [47].
(B) Synthetic self-organized pattern formation. The sender cells produce 3OC6HSL upon induction by aTc. The receiver contains a feed-forward network with two arms: in the first, LuxR* activates expression of a mutated LacI (LacIM1), which then inhibits the output of the circuit, GFP; in the second arm, LuxR* activates expression of the repressor CI, which inhibits expression of LacI; LacI further inhibits GFP expression. The receiver cells are thus conferred with the function of concentration-band detection. Figure adapted from Basu et al. [48].
(C) Simpson's paradox demonstrated by a synthetic microbial ecosystem consisting of QS signal (C4HSL) producers (P) and non-producers (NP). In P, promoter PR drives the synthesis of autoinducer synthase (RhlI), which produces C4HSL. C4HSL diffuses across cell membranes and binds the constitutively expressed RhlR to form an activated complex RhlR-C4HSL (R*). This complex in turn drives expression of catLVA, which confers cells with chloramphenicol resistance. The sdiA null background (sdiAΔ) reduces other Rhl-independent gene expression. In the NP strain, the rhlI gene is absent. The P (with GFP) and NP (no GFP) strains can be differentiated by GFP. Figure adapted from Chuang et al. [51].
(D) An XOR logic gate utilizing cell-cell communication between colonies on a plate. It includes four colonies spatially distributed on an agar plate, in which three colonies contain NOR gates and one contains a buffer gate. The four colonies are spatially arranged such that the computation progresses from left to right; that is, the later layer receives QS signal from the previous layer, thus realizing the function of a XOR logic gate via cell-cell communication. Ara and aTc are inputs. The output of the XOR gate is a yellow fluorescent protein (YFP). Figure adapted from Tamsir et al. [52].
(E) An engineered yeast microbial consortium which utilizes cell-cell communication to achieve the OR logic gate. The microbial consortium includes three cell strains. Two cells receive input stimuli of NaCl and galactose, respectively, and consequently produce the yeast pheromone alpha-factor as a signaling molecule for cell-cell communication in the microbial consortium. The alpha-factor in turn induces GFP expression in the output cell. Figure adapted from Regot et al. [53].
Basu et al. further engineered an analogous feed-forward circuit to program pattern formation (Fig. 3B) [48]. In the solid phase, a gradient of AHL concentration is generated around the sender cells due to AHL diffusion. The receiver cells act as a band detector of the AHL concentration, whereby they maximally respond to an intermediate level of AHL, while they are relatively insensitive to both low and high levels of AHL. That is, receiver cells in both close proximity and far from the sender cells experience a high and low AHL level, respectively, resulting in repressed GFP expression. Only at intermediate distances from sender cells, the receiver cells sense an intermediate level of AHL and maximize their responses with high GFP expression. Therefore, an initially undifferentiated lawn of receiver cells can form a bullseye pattern around a sender colony. More elaborate patterns, such as ellipses and clovers, can be achieved by placing senders at different configurations. One-way communication has also been explored in eukaryotes, such as yeast, by the same group [49].
The maintenance of the “common good” by producers (P) (e.g., of QS signal) in ecosystems is a major question in the evolution of cooperation. Since QS signal non-producers (NP) benefit from the shared resource without paying its cost of producing the QS signal, NP may proliferate faster than P. When P and NP are distributed heterogeneously into subpopulations of varying composition, some start with higher fractions of P, while some start with lower fractions of P. The fraction of P then falls for each subpopulation grown in the co-culture. However, it is possible for the overall fraction of P to rise simultaneously. This phenomenon is generally referred to as “Simpson's paradox” [50]. To test this paradox, Chuang et al. constructed a synthetic microbial ecosystem using two engineered E. coli strains which comprise QS signal AHL producer (P) and non-producer (NP) (Fig. 3C) [51]. The common good in this ecosystem is Rhl autoinducer (C4HSL), which in turn activates chloramphenicol antibiotic (Cm) resistance gene (catLVA) expression (Fig. 3C). The group found that the global proportion of P increased relative to its initial composition, even though within each subpopulation mixture, P decreases in proportion. Their experiments illustrate Simpson's paradox in growing microbial populations.
Cell-cell communication signals can also function as chemical wires, which can carry out complex and distributed biological computations in genetically programmed bacteria placed at proper spatial configurations. By spatially seeding a library of simple logic gates carried out by different strains and manipulating their one-way interactions via AHLs, Tamsir et al. were able to construct all 16 possible two-input Boolean logic gates, including NOR, NAND, XOR, etc. [52]. As an example, Fig. 3D illustrates a XOR gate which functions through programmed communication between four colonies on an agar plate. The XOR gate was built with three NOR gates and a buffer gate, and each of the four colonies was composed of a strain containing a single logic gate (either NOR or buffer). By spotting the four colonies onto an agar plate with the spatial configuration shown in Fig. 3D, the computation progresses from left to right in response to the stimulation of two inputs (Ara and aTc). The overall microbial consortium (including cells 1 to 4) behaved as a digital XOR gate (see its truth table, inset, Fig. 3D).
Using engineered yeast cells and a cell-cell communication strategy, Regot et al. were also able to build a set of Boolean logic gates with diverse input/output functions [53]. As an example, Fig. 3E illustrates an OR gate using three engineered cell strains, wherein Cells 1 and 2 respond to the presence of NaCl (input 1) and galactose (input 2) to produce the wiring molecule pheromone, which subsequently induces the output of Cell 3 (expression of GFP). The microbial consortium was co-cultured in liquid phase, and cells expressing GFP were counted by flow cytometry. More complicated biological computations (such as a multiplexer and a 1-bit adder with carrier) can be achieved by employing four or five engineered strains, demonstrating the flexibility of this method [53].
Two-way communication
The examples above involve primarily one-way communication between sender and receiver cells. However, microbial ecosystems, consisting of communities with multiple microbial populations interacting reciprocally, are much more ubiquitous in nature [54]; thus, engineering two-way communication interactions has recently received increasing attention.
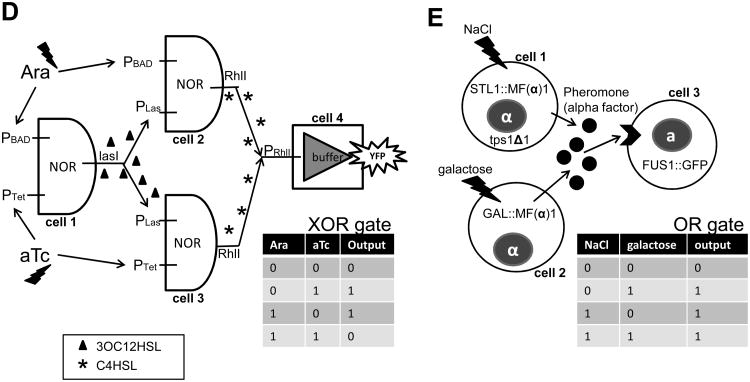
Brenner et al. described a synthetic microbial consortium implemented in E. coli wherein its members communicate bi-directionally to achieve “consensus” gene expression [31]. Guided by mathematical modeling, they designed a microbial consensus consortium (MCC) using the QS components LasI/LasR and RhlI/RhlR (Fig. 4A). The two populations communicate reciprocally via the exchange of two QS signals, generating gene expression responses if and only if the two populations are simultaneously present at sufficiently high cell densities. Therefore, the MCC functions as a logic AND gate in which the two inputs are the population densities of the two types of cells containing Circuit A and B, respectively.
Figure 4.
Engineering two-way cell-cell communication.
(A) A microbial consensus consortium (MCC) circuit. It implements two-way cell-cell communication using two QS modules, LasR/LasI and RhlR/RhlI, to achieve a consensus response. In Circuit A, LasI produces 3OC12HSL, which diffuses into cells containing Circuit B and binds LasR, thereby inducing expression of RhlI. RhlI in Circuit B synthesizes C4HSL, which diffuses into cells containing Circuit A and binds RhlR to activate LasI expression, closing the positive feedback loop. Figure adapted from Brenner et al. [31].
(B) A synthetic obligatory cooperative system achieved by exchanging metabolites in a two-way communication system. This system consists of two auxotrophic yeast strains that depend on each other for survival. One strain, is unable to synthesize adenine but overproduces lysine. In contrast, another strain, is unable to synthesize lysine but overproduces adenine. Figure adapted from Shou et al. [40].
(C) An engineered cooperative microbial consortium, in which only the mixture of the two bacteria can chemotax toward the combined gradient of asparagine and phenylacetyl glycine (PAG). One strain synthesizes penicillin acylase (Pac), which enzymatically hydrolyzes phenylacetyl glycine (PAG) to produce phenylacetic acid (PAA), a chemoattractant to E. coli cells with Tar chemoreceptors. The other strain expresses AnsB gene, which encodes Asparaginase. Asparaginase enzymatically converts Asparagine to Aspartate, functioning as a ligand (or chemoattractant) to E. coli cells with TarPA chemoreceptors. This engineered mutualistic consortium is not able to chemotax toward a gradient consisting of only one of the two compounds (Asparagine and PAA). Figure adapted from Goldberg et al. [55].
(D) Schematic of the gene circuit in a synthetic predator-prey ecosystem. The system consists of two types of engineered E. coli. Each controls the other's survival and death via two QS circuits, LuxR/LuxI from V. fisheri and LasR/LasI from P. aeruginosa. The predator synthesizes 3OC12HSL, which diffuses into the prey and binds to LasR to induce expression of the ccdB killer protein, thereby killing the prey. On the other hand, the prey synthesizes 3OC6HSL, which diffuses into the predator and binds to LuxR to induce expression of the ccdA antidote protein. This protein neutralizes the toxicity of the ccdB killer protein, thereby rescuing the predator. Figures adapted from Balagadde et al. [32].
Metabolites represent another type of exchangeable agent that can dictate intercellular communication [38]. Shou et al. reported engineering a synthetic obligatory cooperative ecosystem, which consists of a pair of genetically engineered, auxotrophic Saccharomyces cerevisiae strains (Fig. 4B) [40]. The first strain, can produce an amino acid, lysine, essential to the survival of the second strain, but its own survival relies on another amino acid, adenine, which is produced by the second strain, The two populations thus form a mutualistic ecosystem— in the absence of one partner, the other cannot maintain its survival, and the two species reach their maximal viability only in the presence of one another.
Goldberg et al. engineered a cooperative chemotactic microbial consortium [55]. As schematized in Fig. 4C, Strain 1 expresses the ansB gene, which encodes Asparaginase, an enzyme that converts asparagines to aspartate. Aspartate is a chemoattractant to Strain 2 with the TarPA chemoreceptor expressed on its membrane. On the other hand, Strain 2 expresses penicillin acylase (Pac), which converts PAG to PAA. PAA is a chemoattractant to Strain 1 with Tar chemoreceptor expressed on its membrane. Thereby, each strain contains a subset of the components necessary for the chemotaxis to asparagine or PAG and is unable to respond to either attractant alone. The two strains comprise a mutualistic consortium, and contain all of the components required for chemotactic responses. The mixed population showed robust chemotaxis on the combined gradient of both asparagines and PAG. Such synergistic interactions resemble many natural ecosystems [56].
In comparison to other types of ecological interactions, predation often generates richer dynamics, and as such, represents a great challenge to engineer de novo. To achieve the predator-prey interaction logic, Balagadde et al. constructed bi-directional cell-cell communication between the predator and the prey populations using E. coli, wherein the two bacterial populations regulate each other's gene expression and survival via two QS modules, LuxI/LuxR and LasI/LasR [32]. Figure 4D schematically depicts the predator-prey logic: the predator produces 3OC12HSL, which diffuses into the prey and leads to its death following induction by the LacZ-ccdB killer protein; meanwhile, the prey produces another QS signal, 3OC6HSL, which permeates into the predator and rescues the predator by eliciting expression of an antidote protein, ccdA, neutralizing ccdB's toxicity. This synthetic system satisfies the broader definition of the predator-prey ecosystem, where one species (prey) suffers from the prosperity of the other (predator) while the latter benefits from the growth of the former. Oscillatory dynamics of the population densities of the predator and the prey were observed in a microchemostat.
Finally, Weber et al. extended engineering two-way cell-cell communication to mammalian cells. They used a volatile chemical, acetaldehyde, and antibiotics to engineer a few synthetic ecosystems, including mutualism, parasitism, and predation [57].
4. Conclusions and perspectives
Here, we have reviewed recent endeavors in synthetic biology, particularly highlighting the use of engineered cell-cell communication. Such programmed coordination of cellular population dynamics by cell-cell communication cannot be realized by artificial gene circuits acting intracellularly. Engineering cell-cell communication opens a new horizon for the synthetic biology field.
We envision that synthetic ecosystems can serve as well-defined model systems to address ecological and evolutionary questions and elucidate biological design principles that are infeasible to study in natural ecosystems due to their extreme complexity or unidentified interaction modes. As an extension of engineering population behaviors in a single species, one can engineer population behaviors in multiple species with diverse interactions. One advantage of these engineered microbial communities is that such systems involve compositional parts with well-defined genetic properties and interactions, facilitating a quantitative investigation of their spatiotemporal dynamics. In contrast, such analysis is often much more difficult when studying their naturally occurring counterparts, due to much more complex interactions and confounding factors. For example, by programming a synthetic predator-prey ecosystem, Balagadde et al. addressed the occurrence conditions for extinction, steady-state coexistence, and oscillatory coexistence in a predator-prey ecosystem [32]. Song et al. further examined how biodiversity changes in the chemical-mediated ecosystem over time and space [58]. The underlying principles driving these changes differ significantly from contact-based ecosystems [59, 60]. Such investigation of the impact of spatiotemporal interactions (e.g., cellular motility and segregation) on the relative abundance of the predator and the prey species may provide a systematic understanding of spatiotemporal modulation of a predator-prey ecosystem's biodiversity, which is a central element in determining stability and persistence of ecosystems [58, 61].
Synthetic ecosystems have also been used to address evolutionary theory in ecological systems. Shou et al. constructed a synthetic obligatory cooperative system to investigate the adaptation and co-evolution of cooperative species [40]. They demonstrated that two synthetic cooperative populations can co-evolve through time by repeated dilution and regrowth in co-culture. Chuang et al. synthesized an ecosystem consisting of QS signal producer and non-producer to demonstrate Simpson's paradox and the Hamilton rule, which rationalized the existence of cooperation during evolution [51, 62]. Gore et al. constructed an ecosystem that included a wild-type (invertase producer) and a cheater yeast cell (invertase non-producer) to illustrate the plausibility of snowdrift game theory as a underlying mechanism for the origin of cooperation [63]. In the future, we envision that more ecological and evolutionary theories will be addressed by synthetic ecosystems.
Microbial ecosystems are of great importance in a variety of real-world applications, ranging from biocomputing [52, 53] to environmental remediation [64] and the facilitation of human immunities [65]. In addition, microbial ecosystems can perform more complex tasks than individual populations, enabling novel applications, such as drug and fuel production [66, 67]. Given the many advantages of microbial ecosystems, we envision the engineering of such ecosystems for the development of potential biotechnology applications [33, 68] as a future area of significant interest throughout the field of synthetic biology.
Acknowledgments
This work was partially supported by a Start-Up Grant from NTU (HS), a Tier-1 grant from MOE of Singapore (HS), the National Institutes of Health (5R01CA118486), a National Science Foundation CAREER award (LY), a DuPont Young Professorship (LY), a David and Lucile Packard Fellowship (LY), and a Department of Homeland Security Graduate Fellowship (SP).
Footnotes
The authors have declared no conflict of interest.
References
- 1.Marguet P, Balagadde F, Tan C, You L. Biology by design: reduction and synthesis of cellular components and behaviour. J R Soc Interface. 2007;4:607–623. doi: 10.1098/rsif.2006.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drubin DA, Way JC, Silver PA. Designing biological systems. Genes Dev. 2007;21:242–254. doi: 10.1101/gad.1507207. [DOI] [PubMed] [Google Scholar]
- 3.Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol. 2006;2:674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- 4.Fortman JL, Chhabra S, Mukhopadhyay A, Chou H, et al. Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol. 2008;26:375–381. doi: 10.1016/j.tibtech.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Pieper DH, Reineke W. Engineering bacteria for bioremediation. Curr Opin Biotechnol. 2000;11:262–270. doi: 10.1016/s0958-1669(00)00094-x. [DOI] [PubMed] [Google Scholar]
- 6.Tan C, Song H, Niemi J, You L. A synthetic biology challenge: making cells compute. Mol Biosyst. 2007;3:343–353. doi: 10.1039/b618473c. [DOI] [PubMed] [Google Scholar]
- 7.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 9.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 11.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi H, Kaern M, Araki M, Chung K, et al. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 14.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 15.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 16.Stricker J, Cookson S, Bennett MR, Mather WH, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedland AE, Lu TK, Wang X, Shi D, et al. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohka T, Heins RA, Phelan RM, Greisler JM, et al. An externally tunable bacterial band-pass filter. Proc Natl Acad Sci U S A. 2009;106:10135–10140. doi: 10.1073/pnas.0901246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 20.Guido NJ, Wang X, Adalsteinsson D, McMillen D, et al. A bottom-up approach to gene regulation. Nature. 2006;439:856–860. doi: 10.1038/nature04473. [DOI] [PubMed] [Google Scholar]
- 21.Marguet P, Tanouchi Y, Spitz E, Smith C, You L. Oscillations by minimal bacterial suicide circuits reveal hidden facets of host-circuit physiology. PLoS One. 2010;5:e11909. doi: 10.1371/journal.pone.0011909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balagadde FK, You L, Hansen CL, Arnold FH, Quake SR. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 23.Swinburne IA, Silver PA. Intron delays and transcriptional timing during development. Dev Cell. 2008;14:324–330. doi: 10.1016/j.devcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanouchi Y, Tu D, Kim J, You L. Noise reduction by diffusional dissipation in a minimal quorum sensing motif. PLoS Comput Biol. 2008;4:e1000167. doi: 10.1371/journal.pcbi.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–319. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- 26.Parsek MR, Greenberg EP. Sociomicrobiology: the connections between qourum sensing and biofilms. Trends in Microbiology. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Pai A, You L. Optimal tuning of bacterial sensing potential. Mol Syst Biol. 2009;5:286. doi: 10.1038/msb.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 29.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and diseases. Annu Rev Biomed Eng. 2008;10:145–167. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- 31.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci U S A. 2007;104:17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balagadde FK, Song H, Ozaki J, Collins CH, et al. A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenner K, You L, Arnold FH. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Pai A, Tanouchi Y, Collins CH, You L. Engineering multicellular systems by cell-cell communication. Curr Opin Biotechnol. 2009;20:461–470. doi: 10.1016/j.copbio.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 36.Collins CH, Arnold FH, Leadbetter JR. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol Microbiol. 2005;55:712–723. doi: 10.1111/j.1365-2958.2004.04437.x. [DOI] [PubMed] [Google Scholar]
- 37.Collins CH, Leadbetter JR, Arnold FH. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotechnol. 2006;24:708–712. doi: 10.1038/nbt1209. [DOI] [PubMed] [Google Scholar]
- 38.Bulter T, Lee SG, Wong WW, Fung E, et al. Design of artificial cell-cell communication using gene and metabolic networks. Proc Natl Acad Sci U S A. 2004;101:2299–2304. doi: 10.1073/pnas.0306484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerchman Y, Weiss R. Teaching bacteria a new language. Proc Natl Acad Sci U S A. 2004;101:2221–2222. doi: 10.1073/pnas.0400473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shou W, Ram S, Vilar JM. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci U S A. 2007;104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You L, Cox RS, 3rd, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 42.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 43.Haseltine EL, Arnold FH. Implications of rewiring bacterial quorum sensing. Appl Environ Microbiol. 2008;74:437–445. doi: 10.1128/AEM.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, et al. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fung E, Wong WW, Suen JK, Bulter T, et al. A synthetic gene-metabolic oscillator. Nature. 2005;435:118–122. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- 47.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004;101:6355–6360. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 49.Chen MT, Weiss R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat Biotechnol. 2005;23:1551–1555. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- 50.Sober E, Wilson DS. Unto Others: The Evolution and Psychology of Unselfish Behavior. Harvard University Press; Cambridge, MA: 1998. [Google Scholar]
- 51.Chuang JS, Rivoire O, Leibler S. Simpson's paradox in a synthetic microbial system. Science. 2009;323:272–275. doi: 10.1126/science.1166739. [DOI] [PubMed] [Google Scholar]
- 52.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regot S, Macia J, Conde N, Furukawa K, et al. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 54.Atlas RM, Bartha R. Microbial Ecology: Fundamentals and Applications. The Benjamin/Cummings Publishing Co. Inc; 1998. [Google Scholar]
- 55.Goldberg SD, Derr P, DeGrado WF, Goulian M. Engineered single- and multi-cell chemotaxis pathways in E. coli. Mol Syst Biol. 2009;5:283. doi: 10.1038/msb.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schink B. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek. 2002;81:257–261. doi: 10.1023/a:1020579004534. [DOI] [PubMed] [Google Scholar]
- 57.Weber W, Daoud-El Baba M, Fussenegger M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc Natl Acad Sci U S A. 2007;104:10435–10440. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song H, Payne S, Gray M, You L. Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem. Nat Chem Biol. 2009;5:929–935. doi: 10.1038/nchembio.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 60.Reichenbach T, Mobilia M, Frey E. Mobility promotes and jeopardizes biodiversity in rock-paper-scissors games. Nature. 2007;448:1046–1049. doi: 10.1038/nature06095. [DOI] [PubMed] [Google Scholar]
- 61.Loreau M, Naeem S, Inchausti P, Bengtsson J, et al. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science. 2001;294:804–808. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- 62.Chuang JS, Rivoire O, Leibler S. Cooperation and Hamilton's rule in a simple synthetic microbial system. Mol Syst Biol. 2010;6:398. doi: 10.1038/msb.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gore J, Youk H, van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459:253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atlas RM, Philp J. Bioremediation: Applied microbial solutions for real-world environmental cleanup. ASM Press; Herndon, VA: 2005. [Google Scholar]
- 65.Kaper JB, Sperandio V. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect Immun. 2005;73:3197–3209. doi: 10.1128/IAI.73.6.3197-3209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol. 2005;71:7099–7106. doi: 10.1128/AEM.71.11.7099-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McMahon KD, Martin HG, Hugenholtz P. Integrating ecology into biotechnology. Current Opinion in Biotechnology. 2007;18:287–292. doi: 10.1016/j.copbio.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 68.Li B, You L. Synthetic biology: Division of logic labour. Nature. 2011;469:171–172. doi: 10.1038/469171a. [DOI] [PubMed] [Google Scholar]