Abstract
Objectives
Bicuspid aortic valve is the most common congenital cardiac abnormality, occurring in 1% to 2% of the population, and often associates with ascending aortic aneurysm. Based on familial studies, bicuspid aortic valve with aneurysm segregates in an autosomal dominant manner with incomplete penetrance. NOTCH1 mutations have been reported in 6 families with prominent valve calcification and dysfunction and low penetrance of aneurysm. We sought to determine the contribution of NOTCH1 mutations to the more common phenotype of highly penetrant aneurysms with low penetrance of bicuspid aortic valve and with rare valve calcification or dysfunction.
Methods
All exons and splice junctions of NOTCH1 were sequenced in probands from 13 affected families presenting with bicuspid aortic valve with ascending aortic aneurysm in the absence of valve calcification. In addition, mutation analysis was performed on a single individual with aneurysm and calcified tricuspid aortic valve. Sequences were aligned and compared with the reference genomic sequence.
Results
Corroborating previous studies, analysis of the single sporadic patient with calcified aortic valve in the presence of ascending aortic aneurysm revealed a novel heterozygous missense mutation in NOTCH1 resulting in a nonsynonymous amino acid substitution (p.T1090S, c.C3269G) of an evolutionarily conserved residue. This change was not observed in controls. In contrast, we did not identify any pathologic NOTCH1 mutations in the 13 families segregating noncalcified bicuspid aortic valve with highly penetrant aortic aneurysm.
Conclusions
These data suggest that there are phenotypic differences that distinguish families with and without NOTCH1 mutations, indicating a genotype–phenotype correlation with potential implications for patient diagnosis, counseling, and management.
Bicuspid aortic valve (BAV) is the most common congenital cardiac abnormality1,2 affecting about 1% to 2% of individuals in the general population. It is characterized by fusion or incomplete formation of valve commissures during valvulogenesis.
Patients with BAV have varying degrees of valvular dysfunction, ranging from severe (including stenosis and regurgitation) to absent.2 Patients with BAV may also have several additional cardiovascular phenotypes, including valve calcification and ascending aortic aneurysm (AscAA).2-5 However, there is poor correlation between the extent of valve dysfunction and the incidence or severity of aortic aneurysm.6-9 Furthermore, patients with BAV and AscAA have been noted to have dilation of the pulmonary trunk with histologic findings of cystic medial necrosis and elastic fiber fragmentation.4,10 Taken together, these data suggest that the underlying gene defect(s) causing BAV can directly alter vessel wall homeostasis. However, recent studies continue to suggest that eccentric blood flow patterns and/or aortic wall stress, attributed at least in part to perturbation of valve morphology and function, contribute to the increased propensity to aortic dilatation and dissection in patients with BAV.11,12
It has been shown that BAV, either in the presence or absence of these additional clinical features, is highly heritable,13,14 and appears to segregate in an autosomal dominant manner with reduced penetrance.4,15,16 However, the molecular basis of the disorder is not yet well understood.
In 2005, Garg and colleagues17 reported an association between inactivating mutations in NOTCH1 and autosomal dominant aortic valve disease with prominent calcification. Twelve affected individuals in 2 unrelated families were identified to have NOTCH1 mutations. Nine had BAV, with 4 of the 9 also affected by aortic aneurysm. Two other groups have described 4 additional missense NOTCH1 mutations associated with isolated BAV cases, most of whom had valve calcification and aortic aneurysm.18,19 Table E1 presents the clinical information for individuals with previously reported NOTCH1 mutations. Thus, it appears that in some but not all patients with BAV, NOTCH1 mutations are responsible for early valve calcification, significant valvular dysfunction, and aortic aneurysm.
Many patients with BAV have a distinct phenotype, in which the BAV is associated with dilation of the ascending aorta in the absence of early or aggressive valve calcification.3,20 The aneurysms can be observed at the aortic root or, more commonly, just distal to the sinotubular junction.8,21 In such families, the disease is inherited in an autosomal dominant pattern with reduced penetrance. Family members of affected probands can show AscAA in the absence of aortic valve abnormality and vice versa, suggesting that these manifestations share a common etiology but are not interdependent.4 In fact, in these families, AscAA appears to be more highly penetrant when compared with the penetrance of BAV.
NOTCH1, encoded on 9q34.3, is a member of the type 1 transmembrane receptor protein family. Notch signaling is an evolutionarily conserved intercellular pathway that plays a key role in multiple developmental processes by regulating cell fate decisions.22 Given the reported connection between calcific aortic valve disease, aortic aneurysm, and NOTCH1 mutations, we sought to determine the contribution of NOTCH1 mutations to noncalcific BAV with highly penetrant AscAA.
Materials and Methods
Study Subjects
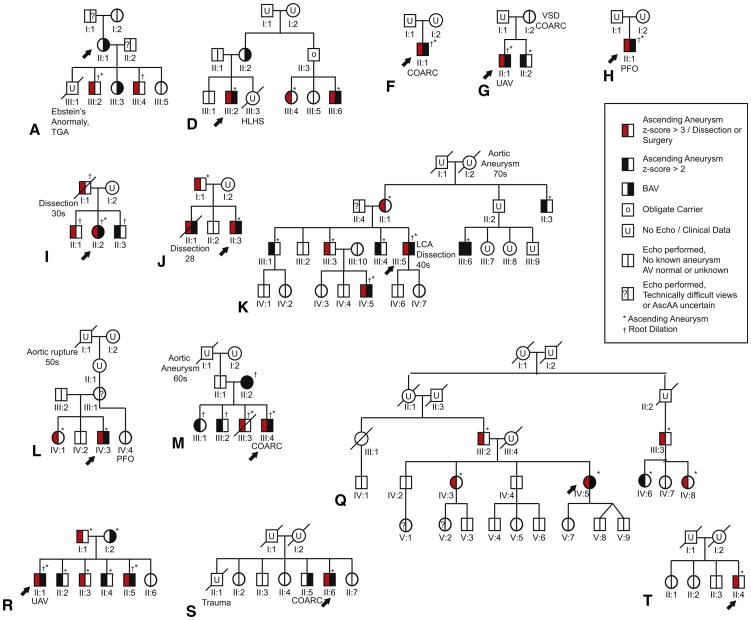
This study was approved by the Institutional Review Board of the Johns Hopkins University School of Medicine. Individuals were recruited, enrolled, and evaluated at the Medical Genetics Clinic of the Johns Hopkins Hospital as previously described.4 Comprehensive clinical analysis for the proband and the extended family was completed before genotyping. History, physical examination, and echocardiograms were obtained for all individuals who gave informed consent. Figure 1 presents the pedigrees of families analyzed.
Figure 1.
Pedigrees of 14 families: 13 with bicuspid aortic valve (BAV)/ascending aortic aneurysm (AscAA), and 1 with calcified tricommissural aortic valve/AscAA. Arrows denote the family member selected for NOTCH1 sequence analysis. AscAA includes both aortic root and ascending aortic dilation. TGA, Transposition of the great arteries; HLHS, hypoplastic left heart syndrome; COARC, coarctation of the aorta; VSD, ventricular septal defect; UAV, unicommissural aortic valve; PFO, patent foramen ovale; LCA, left cerebral artery. Adapted with permission from Loscalzo and colleagues.4
Criteria for Inclusion
Each person was evaluated based on clinical history, examination, and echocardiography.4 Individuals were considered affected with BAV if cardiovascular imaging showed complete or partial fusion of any aortic valve commissure. Individuals were assigned affected status for aneurysm if either the aortic root or more distal ascending aorta showed a dimension with a z score>2 when standardized to age and body size. The z scores were calculated using the Wave Form Echo Program from Boston Children's Hospital for pediatric patients or standard normograms for patients aged 19 years and older.23 In addition, family members with a history of aortic dissection, rupture, or surgery were coded as affected. The individual from each family selected for sequencing is indicated by an arrow in Figure 1.
Criteria for Exclusion
Families demonstrating dysmorphic features or manifestations of a connective tissue disorder were excluded from this study.
NOTCH1 Mutation Analysis
Genomic DNA was isolated from ethylenediaminetetraacetic acid-treated peripheral blood using the DNeasy Blood Kit according the manufacturer's protocol (Qiagen, Valencia, Calif). Mutation analysis of the entire NOTCH1 coding sequence was performed using direct DNA sequencing of polymerase chain reaction-derived amplicons. Amplification of all 34 exons and splice junctions of NOTCH1 was achieved using 42 primer pairs derived from previously published sequencing studies (sequences available upon request)17,18 and the REDExtract-N-Amp PCR Ready Mix (Sigma-Aldric, St Louis, Mo) per manufacturer's instructions. An ABI capillary sequencer (AB, Foster City, Calif) was used for bidirectional sequencing of each amplicon. The resulting sequences were analyzed for variants using MacVector software (MacVector, Inc, Cary, NC) by alignment to the NOTCH1 human reference genome sequence (NG_007458.1).
If a putative mutation was identified, all available family members in that pedigree were genotyped. Sequence conservation was assessed by protein alignment in ClustlW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The accession numbers of each aligned protein are as follows: Homo sapiens NP_060087, Pan troglodytes XP_001171581, Equus caballus XP_001498632, Bos taurus DAA24217, Canis familiaris XP_537795, Rat-tus norvegicus NP_001099191, Mus musculus NP_032740, Gallus gallus XP_415420, Xenopus laevis NP_001081074, Oncorhynchus mykiss NP_00112916, Danio rerio NP_571516, and Drosophila melanogaster NP_476859.
Results
Clinical Findings
Pedigrees for the 13 families and the sporadic case included in this study are shown in Figure 1. Ten of the families described in our study (families A, D, G, I, J, K, L, M, Q, and R) were previously reported4; 3 new families (families F, H, and S) were also studied, as well as the individual with sporadic calcified tricuspid aortic valve (TAV) and aneurysm of the ascending aorta (pedigree T). Three previously described families (families N, O, and P) were excluded due to the lack of sufficient DNA.
All the selected probands had BAV with root or ascending aortic aneurysm, with the exception of the proband in family A. This proband (AII:1) presented with BAV, and was considered affected because she had 2 children with AscAA and another with BAV.
Detailed clinical descriptions for families A, D, G, I, J, K, L, M, Q, and R were previously reported.4 These families include multiple generations of individuals segregating AscAA, BAV, or the combination, in addition to other left ventricular outflow track abnormalities. Three additional pedigrees (F, G, and S) consisted of families with affected members within a single generation. Two of these (F and G) were trios with the offspring presenting with BAV and AscAA.
Proband demographic and clinical data are provided in Table 1. Of the BAV/AscAA probands included in the familial cohort (n = 13), 10 (77%) were male and the median age was 22 years. Two of the probands (15%) were reported to have unicommissural aortic valves, and 2 (15%) had BAV with unknown morphology. Of the 9 probands with known BAV morphology, 8 (62%) had fusion of the right/left coronary commissure whereas 1 (8%) showed fusion of the right/noncoronary commissure. Twelve (92%) of the probands lacked calcification of the aortic valve, whereas the most senior proband at 64 years old (SII:6), was shown to have moderate asymmetric aortic valve calcification with severe aortic stenosis in addition to mitral annular calcification. However, his age (64 years) is a potential confounding variable because aortic valve calcification is commonly observed in individuals aged 60 years and older.1,2
Table 1. Proband demographic and clinical data.
| Proband | ID | Gender | Age (y) | Valve status | Cusp fusion | Aortic stenosis | Aortic regurgitation | Calc | AscAo dilation (age of surgery [y]) |
|---|---|---|---|---|---|---|---|---|---|
| A | II:1 | F | 42 | BAV | R-L | – | – | – | No AscAA |
| D | III:2 | M | 26 | BAV | R-L | Mild | Mild | – | AscAo z>3 |
| F | II:1 | M | 25 | BAV | R-L | – | – | – | Valve-sparing AR replacement (25); coarc |
| G | II:1 | M | 20 | UAV | N/A | Moderate | Moderate | – | Combined AV/AscAo replacement (15) |
| H | II:1 | M | 9 | BAV | R-L | Very mild | Mild | – | Valve-sparing AR replacement (9) |
| I | II:2 | F | 15 | BAV | R-L | – | Mild | – | AR z>2; STJ z>3; AscAo z>3 |
| J | II:3 | M | 22 | BAV | R-L | – | Moderate to severe | – | AR z>3; AscAo z>3 |
| K | III:5 | M | 49 | BAV | Unk | Mild | – | – | Valve-sparing AR replacement (49) |
| L | IV:3 | M | 7 | BAV | R-non | – | – | – | Combined AV/AscAo replacement (5) |
| M | III:4 | M | 16 | BAV | R-L | – | – | – | AR z>3; STJ z>3; AscAo z>3; coarc |
| Q | IV:5 | F | 49 | BAV | R-L | – | Mild | – | Valve-sparing AR replacement (44) |
| R | II:1 | M | 22 | UAV | N/A | Mild | Mild | – | Valve-sparing AR replacement (15) |
| S | II:6 | M | 64 | BAV | Unk | Severe | Mild | Moderate | Combined AV/AscAo replacement (64) |
| T | II:4 | M | 56 | TAV | – | – | + | AscAo z>3 |
Calc, Calcification; AscAo, ascending aorta; F, female; BAV, bicuspid aortic valve; R-L, right-left commissural fusion; AscAA, ascending aortic aneurysm; M, male; AR, aortic root; coarc, coarctation of the aorta, N/A, not applicable; AV, aortic valve; z, z score; STJ, sinotubular junction; Unk, unknown; R-non, right noncommissural fusion; UAV, unicommisural aortic valve; TAV, tricuspid aortic valve.
Amongst these probands, 1 individual (JII:3) was reported to have moderate to severe aortic regurgitation but no aortic valve stenosis. Moderate aortic stenosis and regurgitation was observed in the proband of family G (GII:1). Severe aortic stenosis with mild aortic regurgitation was seen in the proband of family S. Of the remaining 10 probands, 4 had no stenosis or regurgitation, 3 had mild stenosis and regurgitation, 2 had mild regurgitation alone, and 1 had mild stenosis alone. Eight of 13 probands (62%) had surgery to repair an AscAA with a majority (n = 5) undergoing a valve-sparing procedure.
In contrast, the sporadic individual (TII:4) with an extensively calcified TAV and a diffuse fusiform AscAA measuring 4.3 cm distal to the STJ was 56 years old at ascertainment. His 3 older siblings had normal echocardiograms. The patient's father (TI:1) lived to 81 years of age and died from pulmonary failure; he was known to have significant arthrosclerosis. The patient's mother (T:I:2) died at age 87 years. Although there was a family history of obesity, there was no known history of aortic aneurysm or BAV.
NOTCH1 Sequencing Results
After sequencing all 34 exons and splice junctions of NOTCH1 in probands from each of 13 affected families, all known polymorphic variants were cataloged (Table 2). Minor allele frequencies were ascertained from the 1000 Genomes Project European ancestry population because this best represents the demographics of our families.
Table 2. Genetic variants within the human NOTCH1 gene in bicuspid aortic valve (BAV)/ascending aortic aneurysm (AscAA) probands.
| Genomic position | cDNA position | Minor | MAF | MAF | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||
| SNP ID | (NG_007458.1) | NOTCH1 gene position | (NM_017617.3) | AA change | Allele | Probands | 1000 gen |
| rs7028061 | g.4849 T>C | 5′ UTR - 76 T/C | A | 0.38 | 0.329 | ||
| rs4489420 | g.26979 T>C | Exon 3 | c.312T>C | p.N104N | A | 0.54 | 0.487 |
| rs72775789 | g.27550 C>A | Exon 4 - 49 C/A | T | 0.04 | 0.01 | ||
| rs2229975 | g.31331 G>A | Exon 5 | c.852G>A | p.P284P | T | 0.15 | 0.228 |
| rs9411208 | g.33042 C>T | Exon 8 + 7 C/T | G | 0.50 | 0.513 | ||
| rs9411254 | g.33359 C>T | Exon 9 - 43 C/T | G | 0.35 | 0.636 | ||
| rs11145767 | g.33525 A>G | Exon 9 + 10 A/G | T | 0.38 | 0.636 | ||
| rs10781498 | g.33617 C>T | Exon 9 + 102 C/T | G | 0.40 | 0.636 | ||
| rs11574887 | g.33620 C>T | Exon 9 + 105 C/T | A | 0.08 | 0.075 | ||
| rs4880100 | g.34560 A>G | Exon 10 - 133 A/G | T | 0.62 | 0.636 | ||
| rs4880099 | g.34650 T>C | Exon 10 - 43 T/C | A | 0.38 | 0.387 | ||
| rs3124603 | g.35062 A>G | Exon 11 - 9 A/G | T | 0.64 | 0.612 | ||
| rs62579232 | g.35591 C>T | Exon 12 + 94 C/T | A | 0.31 | 0.168 | ||
| rs3812609 | g.36347 G>A | Exon 13 + 70 G/A | T | 0.15 | 0.165 | ||
| rs2229971 | g.37307 T>C | Exon 14 | c.2265T>C | p.N755N | A | 0.69 | 0.66 |
| rs3812608 | g.37787 G>A | Exon 15 + 21 G/A | C | 0.58 | 0.483 | ||
| Novel | g.39655 G>A | Exon 16 + 20 G/A | Novel | 0.04 | No data | ||
| rs11574894 | g.39709 G>A | Exon 16 + 74 G/A | T | 0.08 | 0.056 | ||
| rs9411206 | g.39738 T>C | Exon 16 + 103 T/C | A | 0.38 | 0.413 | ||
| rs3125002 | g.39878 T>C | Exon 17 - 104 T/C | A | 0.54 | 0.466 | ||
| rs3125001 | g.39978 G>A | Exon 17 - 4 G/A | C | 0.67 | 0.575 | ||
| rs36119806 | g.40146 C>T | Exon 17 + 12 C/T | A | 0.08 | 0.109 | ||
| rs3124598 | g.41685 A>G | Exon 19 - 31 A/G | T | 0.27 | 0.429 | ||
| rs11145765 | g.41959 G>A | Exon 19 42 G/A | T | 0.08 | 0.097 | ||
| rs3125000 | g.41971 A>G | Exon 19 + 54 A/G | T | 0.31 | 0.429 | ||
| rs35987207 | g.41998_41999ins C | c.3171+81_3171+82insC | No data | 0.50 | No data | ||
| rs3812603 | g.42331 A>G | Exon 20 - 71 A/G | T | 0.33 | 0.429 | ||
| rs3124597 | g.42576 A>G | Exon 20 + 21 A/G | T | 0.38 | 0.429 | ||
| rs4880098 | g.44335 G>A | Exon 24 + 75 G/A | C | 0.50 | 0.549 | ||
| rs11145764 | g.44833 G>A | Exon 25 - 73 G/A | C | 0.52 | 0.55 | ||
| rs61751542 | g.45020 C>T | Exon 25 | c.4129C>T | p.P1377S | A | 0.04 | 0.016 |
| rs13300218 | g.45598 C>T | Exon 26 - 85 C/T | A | 0.08 | 0.1 | ||
| rs11574903 | g.46169 C>T | Exon 26 + 55 C/T | A | 0.23 | 0.219 | ||
| rs10521 | g.47532 C>T | Exon 27 | c.5094C>T | p.D1698D | G | 0.54 | 0.596 |
| rs12344155 | g.48256 T>C | Exon 28 - 43 T/C | G | 0.08 | 0.1 | ||
| rs3124594 | g.48831 T>C | Exon 30 - 43 T/C | A | 0.42 | 0.472 | ||
| rs11574906 | g.49126 A>T | Exon 30 + 87 A/T | A | 0.08 | 0.098 | ||
| rs11574908 | g.49928 C>T | Exon 31 - 12 C/T | A | 0.10 | 0.083 | ||
| rs9632944 | g.51932 T>C | Exon 33 + 44 T/C | G | 0.08 | 0.098 | ||
| rs2229974 | g.53603 C>T | Exon 34 | c.6555C>T | p.D2185D | G | 0.40 | 0.423 |
SNP ID, Single nucleotide polymorphism identification; cDNA, complementary DNA; AA, amino acid; MAF, minor allele frequency.
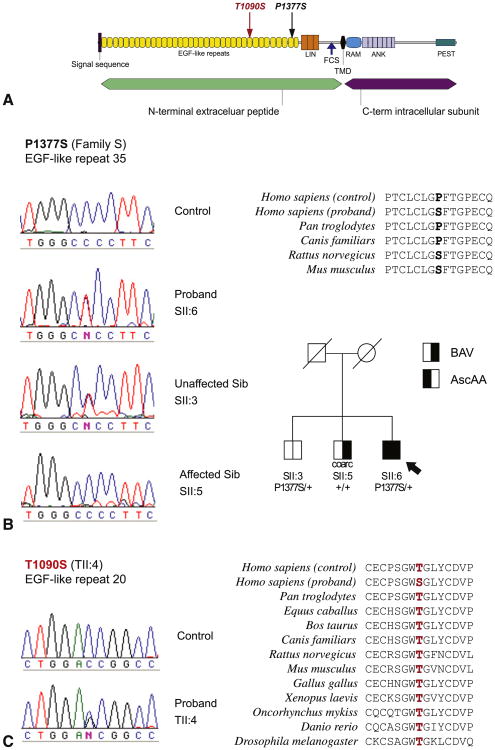
A single novel intronic variant was identified, exon 16+20G>A in the proband (SII:6) from family S. This variant, although noncoding, could theoretically have an effect on splicing; however, it does not have an intuitive effect on messenger RNA processing or stability. Additionally, this variant was identified in his unaffected brother (SII:3), further supporting its neutrality. The proband from family S (SII:6) was also heterozygous for a nonsynonymous variant in exon 25 p.P1377S (c.C4129T) (Figure 2). This variant was absent in his affected brother (individual SII:5) with BAV and coarctation of the aorta in the absence of AscAA and present in his unaffected brother (SII:3) (Figure 2, B). Protein alignment analysis showed that this amino acid is not highly conserved, because rat and mouse NOTCH1 show serine at this position whereas humans have proline (Figure 2, B). This variant was previously observed in BAV cases and was rejected as pathogenic in those studies because it was also detected in similar frequencies in control individuals.18,19 Hence, this variant is also unlikely to be pathogenic.
Figure 2.
Characterization of coding variants identified within NOTCH1. A, Schematic of NOTCH1 protein domains with the location of each variant indicated. B, Variant P1377S in family S is present in proband SII:6 and unaffected sibling (Sib) SII:3, but absent in a control and affected sibling SII:5. Protein conservation reveals proline at position 1377 is not conserved in rat and mouse. C, Variant T1090S in proband TII:4. This substituted amino acid is conserved down to fruit fly. EGF, Epidermal growth factor; LIN, LIN-12/Notch repeats; FCS, furin cleavage site; TMD, transmembrane domain; RAM, RAM domain; ANK, ankyrin repeats; PEST, proline, glutamate, serine, threonine-rich degradation motif; BAV, bicuspid aortic valve; AscAA, ascending aortic aneurysm.
A coding variant was also identified in the proband from family T (TII:4). This individual had AscAA and early and severe calcification and dysfunction of a TAV. He was heterozygous for a threonine to serine (p.T1090S;c.C3269G) substitution in exon 20 (Figure 2, C). This mutation is located within an epidermal growth factor-like domain and involves a residue that is highly conserved through evolution. It was not observed in 180 human control chromosomes and was also absent from publicly available databases, including the 1000 genomes project. No additional family members were available for segregation analysis.
No other variants were identified in the probands from the other pedigrees.
Discussion
The results of this study indicate that NOTCH1 mutations do not contribute significantly to the more common phenotype of noncalcific BAV and AscAA. The underlying genetic cause for families with highly penetrant AscAA and low penetrance of BAV in the absence of aggressive valve calcification or dysfunction has yet to be elucidated.
BAV with AscAA
BAV with AscAA is a phenotype that shows autosomal dominant inheritance with reduced penetrance and variable expression; however, the disease mechanism has not been fully illustrated.4,15,16 Thus we maintain high incentive to identify a molecular marker for disease predisposition. Mutations in NOTCH1 have been associated with aortic valve disease in familial as well as sporadic cases.17-19 Garg and colleagues17 compellingly demonstrated the link between inactivating mutations in NOTCH1 and autosomal dominant aortic valve disease with pronounced calcification in 2 unrelated families. Nine individuals had BAV and amongst these, dilated ascending aorta was noted in four individuals. One individual with AscAA, TAV, no valve calcification, and normal valve function was negative for the familial NOTCH1 mutation.17 Four additional missense mutations were subsequently reported in sporadic BAV cases.18,19 Each of these cases had a phenotype of dysfunctional BAV with stenosis/insufficiency and coexisting AscAA and all but 1 also showed prominent calcification. Taken together, these patient phenotypes indicate that valve malformation, calcification, and dysfunction are the predominant phenotype caused by altered NOTCH1 signaling (Table E1).
Because the contribution of NOTCH1 mutations to BAV with AscAA in the absence of valve calcification has not been established, we chose to approach this question by sequencing families with highly penetrant AscAA and low penetrance of aortic valve dysmorphology and dysfunction.
The absence of NOTCH1 mutations in our cohort, although identifying a potentially pathogenic NOTCH1 mutation in a sporadic individual with AscAA and prominent calcification, suggests that there are phenotypic differences that distinguish families with and without NOTCH1 mutations. We suggest that at least 2 distinct disease mechanisms exist: a NOTCH1-dependent mechanism that produces stenotic, insufficient and/or calcified aortic valve with rare aneurysm, and a NOTCH1-independent mechanism that produces highly penetrant AscAA in the presence of a noncalcified and often normally functioning BAV. Identification of the causative gene for BAV patients is important not only for understanding the mechanism of disease, but also from the perspective of managing the disease, especially given that aortic valve calcification, dysfunction, and aortic aneurysm show an age-dependent onset.
Screening for NOTCH1 Mutations and Follow-up
In general we show that NOTCH1 mutations are exceedingly rare, with now a total of 7 reported to date (Table E1). Therefore, it would be low yield and cost-ineffective to genotype all families with BAV and AscAA, in particular families with decreased penetrance of BAV and highly penetrant AscAA who have a low incidence of early valve calcification. When screening family members of patients with NOTCH1 mutations, the presence of BAV, aortic valve calcification, and/or aortic valve dysfunction suggests these individuals are affected, and hence require continual care and monitoring. On the other hand, the absence of BAV does not exclude the presence of a pathologic NOTCH1 mutation or the predisposition for aneurysm, because data from our lab and others show variable clinical expression of NOTCH1 mutations in the context of TAV. More study is necessary to determine if valve calcification can be a robust read out for NOTCH1 mutations. It is also notable that a full phenotypic description of children with NOTCH1 mutations is lacking.
For patients with the more common presentation of non-calcific BAV with aortic aneurysm, due to the variable penetrance, variable expression and age-dependent onset of AscAA, we recommend that family members of affected individuals be regularly monitored by echocardiogram for AscAA regardless of their aortic valve morphology. First-degree as well as more distantly related individuals should have regular echocardiograms and imaging studies that include the ascending aorta distal to the sinotubular junction.
Based on our observations and other reported studies,17-19 we suggest that patient management should differ between patients with BAV and families with and without NOTCH1 mutations.
Surgical Management
Individuals with BAV have an increased risk for aortic aneurysms and dissections. Although aneurysms and dissections can be observed in individuals in the absence of aortic valve abnormalities, the incidence of such abnormalities in patients with BAV is 8- to 10-fold greater than in patients with no aortic valve abnormalities.2,3,14,24,25 BAV with AscAA carries a significant burden of disease with important surgical implications. We propose that there may be different etiologies of different subclasses of BAV. In terms of implications for management, disruption of NOTCH1 expression primarily affects valve calcification, morphology, and function, and AscAA generally occurs in the setting of significant hemodynamic perturbations. For those patients with BAV with unequivocally pathologic NOTCH1 mutations, there should be significant concern regarding the long-term durability of the native aortic valve. Such patients will likely require valve replacement within their lifetime, and they may benefit from early valve replacement because this may reduce their risk of secondary aortic aneurysms.
In contrast, in the more common presentation of high penetrance of AscAA with low penetrance of BAV (with minimal calcification or dysfunction), it appears that the underlying gene defect is altering aortic wall homeostasis as a primary manifestation. In this scenario, it seems appropriate to consider sparing the natural aortic valve. In these patients, valve repair or replacement alone would not be predicated to mitigate the risk for aneurysm progression and/or dissection.
Limitations
The limitations of this study must be recognized. There is a possibility of incomplete ascertainment of the phenotype. Whereas BAV is congenital, the onset of both aortic valve calcification and AscAA are age-dependent, leading to the possibility of misclassification. Although our proposed correlation between early valve calcification and the presence of a NOTCH1 mutation is based on (1) the mutation-negative status of 13 of 13 families without significant valve calcification, (2) our identification of a NOTCH1 mutation in the sole individual in our study with early valve calcification, and (3) the documentation of aggressive valve calcification in a vast majority of patients previously reported to have NOTCH1 mutations (Table E1), further prospective work is needed to define the full predictive power of this apparent phenotype–genotype correlation. Furthermore, although we provide significant genetic evidence that the novel missense mutation (T1090S) observed in our case of AscAA in the presence of calcified aortic valve likely contributes to the phenotype, we must be cautious when concluding that this is absolutely the cause of disease in the absence of functional studies. Further analysis was limited by the inability to study additional family members. Another limitation is that we did not analyze the untranslated regions of the NOTCH1 gene or look for duplications/deletions involving NOTCH1.
In conclusion, BAV with the predisposition for aortic aneurysm is not a single disorder. Understanding of disease genes and elucidation phenotype–genotype correlations will inform patient diagnosis, counseling, and management.
Supplementary Material
ID, Identification; Calc, calcification; AscAA, ascending aortic aneurysm; CV, cardiovascular; F, female; TAV, tricuspid aortic valve; N/A, not applicable; AscAo, ascending aorta; AVR, aortic valve repair; BAV, bicuspid aortic valve; Unk, unknown; M, male; VSD, ventral septal defect; MS, mitral stenosis; MV, mitral valve; AoR, aortic root replacement; MA, mitral atresia; HLV; hypoplastic left ventricle; DORV, double outlet right ventricle; R-L, right-left commissural fusion; R-non, right-non commissural fusion. *Carriers assumed to have NOTCH1 mutations but this was not confirmed by sequencing.
Acknowledgments
Supported by National Institutes of Health grants AR41135 and AR49698 and the Howard Hughes Medical Institute.
Abbreviations and Acronyms
- AscAA
ascending aortic aneurysm
- BAV
bicuspid aortic valve
- TAV
tricuspid aortic valve
Footnotes
Supplemental material is available online.
Disclosures: Authors have nothing to disclose with regard to commercial support
References
- 1.Roberts WC. The congenitally bicuspid aortic valve. A study of 85 autopsy cases. Am J Cardiol. 1970 Jul;26:72–83. doi: 10.1016/0002-9149(70)90761-7. [DOI] [PubMed] [Google Scholar]
- 2.Ward C. Clinical significance of the bicuspid aortic valve. Heart. 2000;83:81–5. doi: 10.1136/heart.83.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards WD, Leaf DS, Edwards JE. Dissecting aortic aneurysm associated with congenital bicuspid aortic valve. Circulation. 1978;57:1022–5. doi: 10.1161/01.cir.57.5.1022. [DOI] [PubMed] [Google Scholar]
- 4.Loscalzo ML, Goh DL, Loeys B, Kent KC, Spevak PJ, Dietz HC. Familial thoracic aortic dilation and bicommissural aortic valve: a prospective analysis of natural history and inheritance. Am J Med Genet A. 2007;143:1960–7. doi: 10.1002/ajmg.a.31872. [DOI] [PubMed] [Google Scholar]
- 5.McBride KL, Pignatelli R, Lewin M, et al. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: segregation, multiplex relative risk, and heritability. Am J Med Genet A. 2005;134:180–6. doi: 10.1002/ajmg.a.30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pachulski RT, Weinberg AL, Chan KL. Aortic aneurysm in patients with functionally normal or minimally stenotic bicuspid aortic valve. Am J Cardiol. 1991;67:781–2. doi: 10.1016/0002-9149(91)90544-u. [DOI] [PubMed] [Google Scholar]
- 7.Nistri S, Sorbo MD, Marin M, Palisi M, Scognamiglio R, Thiene G. Aortic root dilatation in young men with normally functioning bicuspid aortic valves. Heart. 1999;82:19–22. doi: 10.1136/hrt.82.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn RT, Roman MJ, Mogtader AH, Devereux RB. Association of aortic dilation with regurgitant, stenotic and functionally normal bicuspid aortic valves. J Am Coll Cardiol. 1992;19:283–8. doi: 10.1016/0735-1097(92)90479-7. [DOI] [PubMed] [Google Scholar]
- 9.Keane MG, Wiegers SE, Plappert T, Pochettino A, Bavaria JE, Sutton MG. Bicuspid aortic valves are associated with aortic dilatation out of proportion to coexistent valvular lesions. Circulation. 2000;102(19 Suppl 3):III35–9. doi: 10.1161/01.cir.102.suppl_3.iii-35. [DOI] [PubMed] [Google Scholar]
- 10.de Sa M, Moshkovitz Y, Butany J, David TE. Histologic abnormalities of the ascending aorta and pulmonary trunk in patients with bicuspid aortic valve disease: clinical relevance to the ross procedure. J Thorac Cardiovasc Surg. 1999;118:588–94. doi: 10.1016/S0022-5223(99)70002-4. [DOI] [PubMed] [Google Scholar]
- 11.Girdauskas E, Borger MA, Secknus MA, Girdauskas G, Kuntze T. Is aortopathy in bicuspid aortic valve disease a congenital defect or a result of abnormal hemodynamics? A critical reappraisal of a one-sided argument. Eur J Cardiothorac Surg. 2011;39:809–14. doi: 10.1016/j.ejcts.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Nathan DP, Xu C, Plappert T, et al. Increased ascending aortic wall stress in patients with bicuspid aortic valves. Ann Thorac Surg. 2011;92:1384–9. doi: 10.1016/j.athoracsur.2011.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emanuel R, Withers R, O'Brien K, Ross P, Feizi O. Congenitally bicuspid aortic valves. Clinicogenetic study of 41 families. Br Heart J. 1978;40:1402–7. doi: 10.1136/hrt.40.12.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol. 2004;44:138–43. doi: 10.1016/j.jacc.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 15.Huntington K, Hunter AG, Chan KL. A prospective study to assess the frequency of familial clustering of congenital bicuspid aortic valve. J Am Coll Cardiol. 1997;30:1809–12. doi: 10.1016/s0735-1097(97)00372-0. [DOI] [PubMed] [Google Scholar]
- 16.Clementi M, Notari L, Borghi A, Tenconi R. Familial congenital bicuspid aortic valve: a disorder of uncertain inheritance. Am J Med Genet. 1996;62:336–8. doi: 10.1002/(SICI)1096-8628(19960424)62:4<336::AID-AJMG2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 17.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–4. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed SA, Aherrahrou Z, Liptau H, et al. Novel missense mutations (p.T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem Biophys Res Commun. 2006;345:1460–5. doi: 10.1016/j.bbrc.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 19.McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM., 3rd Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 2007;134:290–6. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 20.Roberts CS, Roberts WC. Dissection of the aorta associated with congenital malformation of the aortic valve. J Am Coll Cardiol. 1991;17:712–6. doi: 10.1016/s0735-1097(10)80188-3. [DOI] [PubMed] [Google Scholar]
- 21.Nkomo VT, Enriquez-Sarano M, Ammash NM, Melton LJ, 3rd, Bailey KR, Desjardins V, et al. Bicuspid aortic valve associated with aortic dilatation: a community-based study. Arterioscler Thromb Vasc Biol. 2003;23:351–6. doi: 10.1161/01.atv.0000055441.28842.0a. [DOI] [PubMed] [Google Scholar]
- 22.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–47. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Roman MJ, Devereux RB, Kramer-Fox R, O'Loughlin J. Two-dimensional echocardiographic aortic root dimensions in normal children and adults. Am J Cardiol. 1989;64:507–12. doi: 10.1016/0002-9149(89)90430-x. [DOI] [PubMed] [Google Scholar]
- 24.Fedak PW, Verma S, David TE, Leask RL, Weisel RD, Butany J. Clinical and pathophysiological implications of a bicuspid aortic valve. Circulation. 2002;106:900–4. doi: 10.1161/01.cir.0000027905.26586.e8. [DOI] [PubMed] [Google Scholar]
- 25.Sabet HY, Edwards WD, Tazelaar HD, Daly RC. Congenitally bicuspid aortic valves: a surgical pathology study of 542 cases (1991 through 1996) and a literature review of 2,715 additional cases. Mayo Clin Proc. 1999;74:14–26. doi: 10.4065/74.1.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ID, Identification; Calc, calcification; AscAA, ascending aortic aneurysm; CV, cardiovascular; F, female; TAV, tricuspid aortic valve; N/A, not applicable; AscAo, ascending aorta; AVR, aortic valve repair; BAV, bicuspid aortic valve; Unk, unknown; M, male; VSD, ventral septal defect; MS, mitral stenosis; MV, mitral valve; AoR, aortic root replacement; MA, mitral atresia; HLV; hypoplastic left ventricle; DORV, double outlet right ventricle; R-L, right-left commissural fusion; R-non, right-non commissural fusion. *Carriers assumed to have NOTCH1 mutations but this was not confirmed by sequencing.