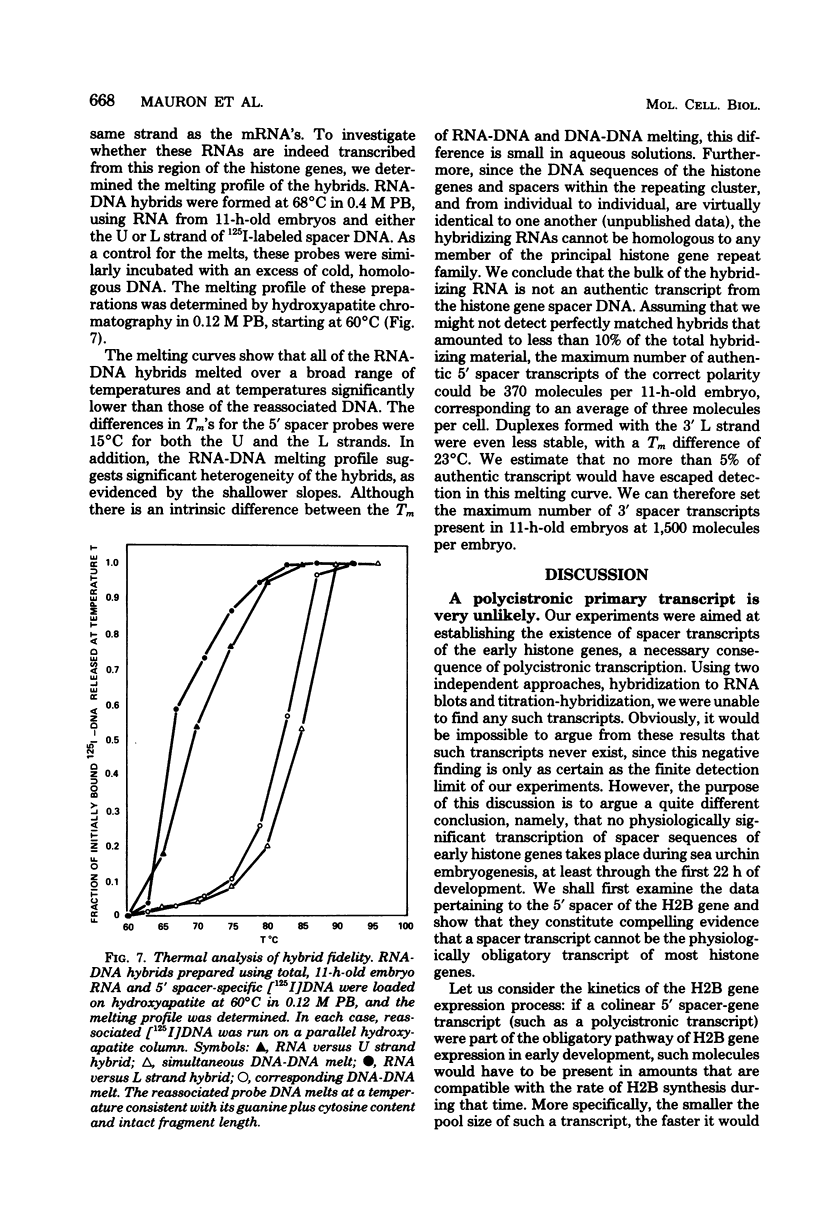
Abstract
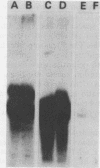
We have examined histone gene expression during the early stages of sea urchin embryogenesis. The five histone genes expressed at that time are contained in tandem repetitive segments. It has been suggested that adjacent coding regions and their intervening spacer sequences are transcribed into large polycistronic messenger ribonucleic acid (RNA) precursors. We have subcloned into pBR322 deoxyribonucleic acid (DNA) sequences mapping either in the coding region, the 5' spacer, or the 3' spacer of the H2B histone gene. These clones were used to produce radioiodinated hybridization probes. We measured the steady-state quantity of H2B messenger RNA as well as spacer-specific RNA in the total RNA from embryos taken at various stages of development from fertilization to hatching of blastulae (0 to 22 h post-fertilization). Small amounts of RNA hybridizing to both spacer probes could be found. However, we show that these RNAs form mismatched hybrids with the spacer DNA and therefore cannot originate from the spacers present in the histone genes. We conclude that there is no detectable transcription of the spacer regions on either side of the H2B histone gene. The detection limit for RNA complementary to the 5' spacer sequence corresponds to a maximum of about three RNA molecules per cell, an amount shown to be far less than the projected steady-state pool size of a putative polycistronic transcript, if such a precursor were to be the obligatory transcript of the histone genes. (This conclusion was derived by using the known rates of production of H2B mRNA throughout early development [R. E. Maxson and F. H. Wilt, Dev. Biol., in press].) The physiologically relevant transcript of the histone genes in early development is therefore monocistronic and probably identical to the messenger RNA itself.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson A. I., Chen K. Rates of RNA chain growth in developing sea urchin embryos. Dev Biol. 1977 Aug;59(1):39–48. doi: 10.1016/0012-1606(77)90238-x. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Schaffner W., Smith H. O. DNA sequences coding for the H2B histone of Psammechinus miliaris. Nature. 1977 Apr 14;266(5603):603–607. doi: 10.1038/266603a0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Birnsteil M. L. A regulatory sequence near the 3' end of sea urchin histone genes. Nucleic Acids Res. 1979 Jul 11;6(9):2997–3008. doi: 10.1093/nar/6.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Levy S., Kedes L. H. Rapid purification of biologically active individual histone messenger RNAs by hybridization to cloned DNA linked to cellulose. Biochemistry. 1979 Jan 9;18(1):208–213. doi: 10.1021/bi00568a032. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Kedes L. H. Histone gene expression during sea urchin embryogenesis: isolation and characterization of early and late messenger RNAs of Strongylocentrotus purpuratus by gene-specific hybridization and template activity. Dev Biol. 1979 Nov;73(1):153–173. doi: 10.1016/0012-1606(79)90144-1. [DOI] [PubMed] [Google Scholar]
- Cohn R. H., Lowry J. C., Kedes L. H. Histone genes of the sea urchin (S. purpuratus) cloned in E coli: order, polarity, and strandedness of the five histone-coding and spacer regions. Cell. 1976 Sep;9(1):147–161. doi: 10.1016/0092-8674(76)90060-x. [DOI] [PubMed] [Google Scholar]
- Costantini F. D., Scheller R. H., Britten R. J., Davidson E. H. Repetitive sequence transcripts in the mature sea urchin oocyte. Cell. 1978 Sep;15(1):173–187. doi: 10.1016/0092-8674(78)90093-4. [DOI] [PubMed] [Google Scholar]
- Galau G. A., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: rate of hybridization of excess RNA with DNA, compared to the rate of DNA renaturation. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1020–1023. doi: 10.1073/pnas.74.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Gronenborn B., Messing J. Methylation of single-stranded DNA in vitro introduces new restriction endonuclease cleavage sites. Nature. 1978 Mar 23;272(5651):375–377. doi: 10.1038/272375a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Schedl P., Kedes L. Sequence analysis and evolution of sea urchin (Lytechinus pictus and Strongylocentrotus purpuratus) histone H4 messenger RNAs. J Mol Biol. 1976 Jun 25;104(2):351–369. doi: 10.1016/0022-2836(76)90276-x. [DOI] [PubMed] [Google Scholar]
- Hackett P. B., Traub P., Gallwitz D. The histone genes in HeLa cells are on individual transcriptional units. J Mol Biol. 1978 Dec 25;126(4):619–635. doi: 10.1016/0022-2836(78)90012-8. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr, Rosbash M., Kaback D. B. Isolation of yeast histone genes H2A and H2B. Cell. 1979 Dec;18(4):1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Kunkel N. S., Hemminki K., Weinberg E. S. Size of histone gene transcripts in different embryonic stages of the sea urchin, Strongylocentrotus purpuratus. Biochemistry. 1978 Jun 27;17(13):2591–2598. doi: 10.1021/bi00606a021. [DOI] [PubMed] [Google Scholar]
- Lev Z., Thomas T. L., Lee A. S., Angerer R. C., Britten R. J., Davidson E. H. Developmental expression of two cloned sequences coding for rare sea urchin embryo messages. Dev Biol. 1980 May;76(2):322–340. doi: 10.1016/0012-1606(80)90382-6. [DOI] [PubMed] [Google Scholar]
- Levy S., Sures I., Kedes L. H. Sequence of the 5'-end of Strongylocentrotus purpuratus H2b histone mRNA and its location within histone DNA. Nature. 1979 Jun 21;279(5715):737–739. doi: 10.1038/279737a0. [DOI] [PubMed] [Google Scholar]
- Lifton R. P., Goldberg M. L., Karp R. W., Hogness D. S. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Maxwell I. H., Maxwell F., Hahn W. E. Removal of RNase activity from DNase by affinity chromatography on agarose coupled aminophenylphosphoryl-uridine-2' (3')-phosphate. Nucleic Acids Res. 1977 Jan;4(1):241–246. doi: 10.1093/nar/4.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G. P., Costantini F. D., Posakony J. W., Davidson E. H., Britten R. J. Evolutionary conservation of repetitive sequence expression in sea urchin egg RNA's. Science. 1980 May 30;208(4447):1046–1048. doi: 10.1126/science.6154974. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Cohen L. H., Hendricks M. B., Donnelly R. J., Weinberg E. S. Stage-specific mRNAs coding for subtypes of H2A and H2B histones in the sea urchin embryo. Cell. 1978 Jun;14(2):327–336. doi: 10.1016/0092-8674(78)90118-6. [DOI] [PubMed] [Google Scholar]
- Probst E., Kressmann A., Birnstiel M. L. Expression of sea urchin histone genes in the oocyte of Xenopus laevis. J Mol Biol. 1979 Dec 15;135(3):709–732. doi: 10.1016/0022-2836(79)90173-6. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
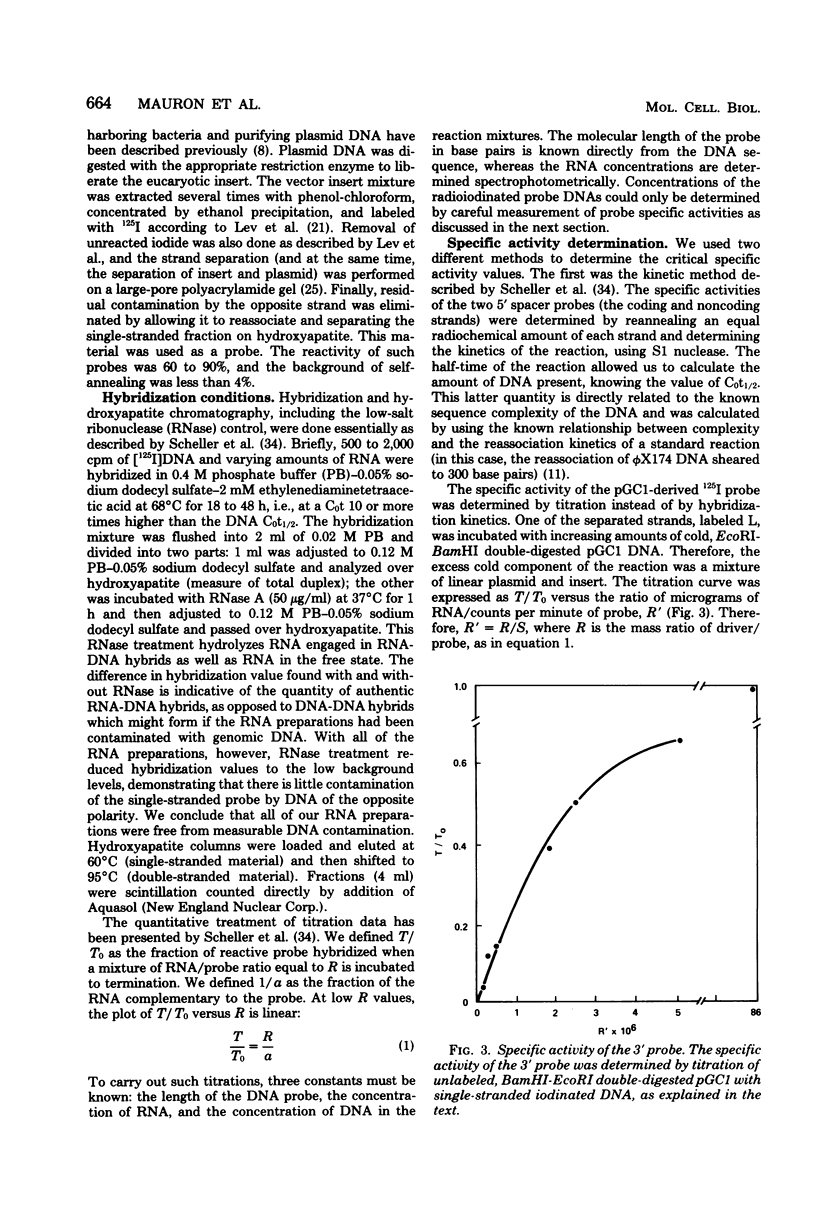
- Scheller R. H., Costantini F. D., Kozlowski M. R., Britten R. J., Davidson E. H. Specific representation of cloned repetitive DNA sequences in sea urchin RNAs. Cell. 1978 Sep;15(1):189–203. doi: 10.1016/0092-8674(78)90094-6. [DOI] [PubMed] [Google Scholar]
- Spinelli G., Melli M., Arnold E., Casano C., Gianguzza F., Ciaccio M. High molecular weight RNA containing histone messenger in the sea urchin Paracentrotus lividus. J Mol Biol. 1980 May 15;139(2):111–122. doi: 10.1016/0022-2836(80)90299-5. [DOI] [PubMed] [Google Scholar]
- Strohman R. C., Moss P. S., Micou-Eastwood J., Spector D., Przybyla A., Paterson B. Messenger RNA for myosin polypeptides: isolation from single myogenic cell cultures. Cell. 1977 Feb;10(2):265–273. doi: 10.1016/0092-8674(77)90220-3. [DOI] [PubMed] [Google Scholar]
- Sures I., Levy S., Kedes L. H. Leader sequences of Strongylocentrotus purpuratus histone mRNAs start at a unique heptanucleotide common to all five histone genes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1265–1269. doi: 10.1073/pnas.77.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Sures I., Maxam A., Cohn R. H., Kedes L. H. Identification and location of the histone H2A and H3 genes by sequence analysis of sea urchin (S. purpuratus) DNA cloned in E. coli. Cell. 1976 Dec;9(4 Pt 1):495–502. doi: 10.1016/0092-8674(76)90031-3. [DOI] [PubMed] [Google Scholar]
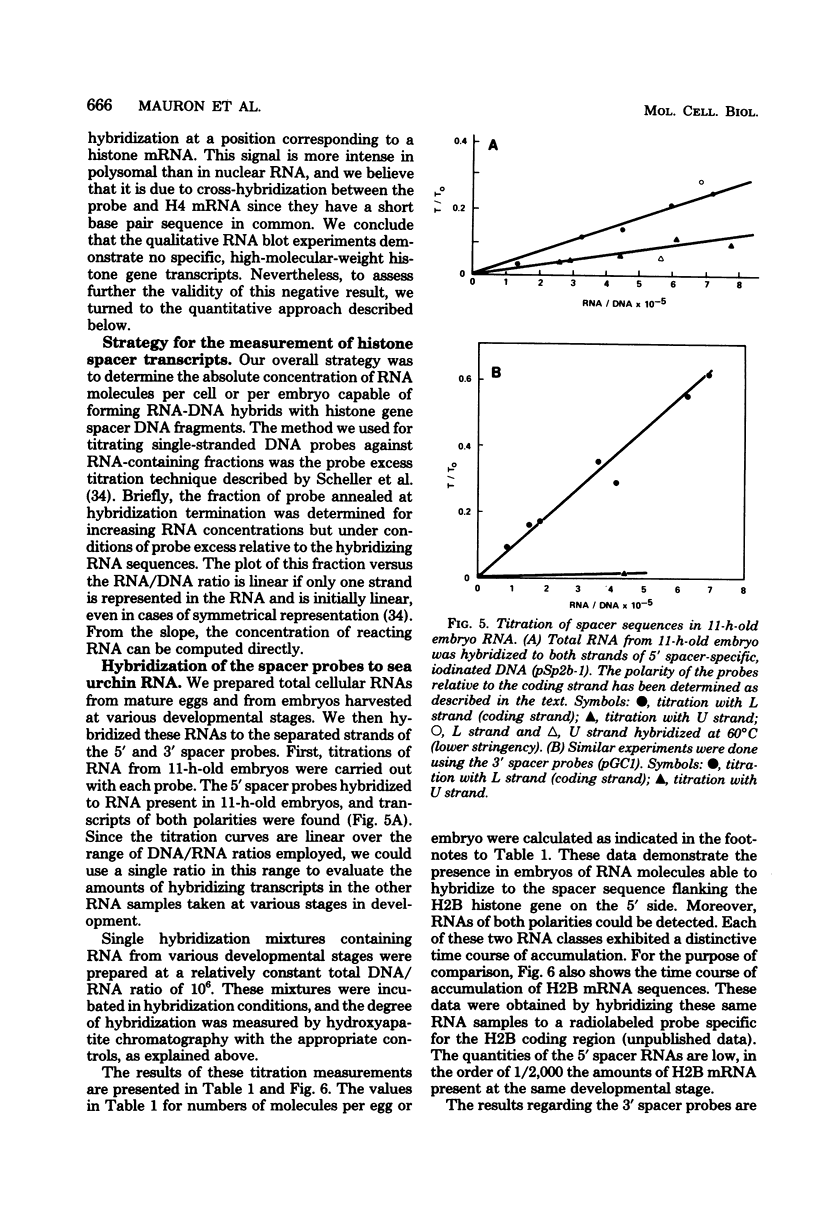
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Wold B. J., Klein W. H., Hough-Evans B. R., Britten R. J., Davidson E. H. Sea urchin embryo mRNA sequences expressed in the nuclear RNA of adult tissues. Cell. 1978 Aug;14(4):941–950. doi: 10.1016/0092-8674(78)90348-3. [DOI] [PubMed] [Google Scholar]