Abstract
Intramembrane proteases regulate diverse processes by cleaving substrates within a transmembrane segment or near the membrane surface. Bacillus subtilis SpoIVFB is an intramembrane metalloprotease that cleaves Pro-σK during sporulation. To elucidate features of Pro-σK important for cleavage by SpoIVFB, coexpression of the two proteins in Escherichia coli was used along with cell fractionation. In the absence of SpoIVFB, a portion of the Pro-σK was peripherally membrane associated. This portion was not observed in the presence of SpoIVFB, suggesting that it serves as the substrate. Deletion of Pro-σK residues 2 to 8, addition of residues at its N terminus, or certain single-residue substitutions near the cleavage site impaired cleavage. Certain multiresidue substitutions near the cleavage site changed the position of cleavage, revealing preferences for a small residue preceding the cleavage site N-terminally (i.e., at the P1 position) and a hydrophobic residue at the second position following the cleavage site C-terminally (i.e., P2′). These features appear to be conserved among Pro-σK orthologs. SpoIVFB did not tolerate an aromatic residue at P1 or P2′ of Pro-σK. A Lys residue at P3′ of Pro-σK could not be replaced with Ala unless a Lys was provided farther C-terminally (e.g., at P9′). α-Helix-destabilizing residues near the cleavage site were not crucial for SpoIVFB to cleave Pro-σK. The preferences and tolerances of SpoIVFB are somewhat different from those of other intramembrane metalloproteases, perhaps reflecting differences in the interaction of the substrate with the membrane and the enzyme.
INTRODUCTION
Regulated intramembrane proteolysis (RIP) was discovered a little more than 10 years ago and has rapidly become a subject of intense investigation (1). RIP governs diverse processes that impact human health, plant productivity, and bacterial survival (2–5). RIP involves cleavage of a substrate within a transmembrane segment (TMS) or near the membrane surface by an intramembrane protease (IP). IPs have active-site residues within TMSs and are classified into three types based on their catalytic residues and mechanism of action: intramembrane metalloproteases (IMMPs) were the first to be identified, serine IPs comprise a large family commonly called rhomboids, and aspartyl IPs include presenilins and signal peptide peptidases (6–10).
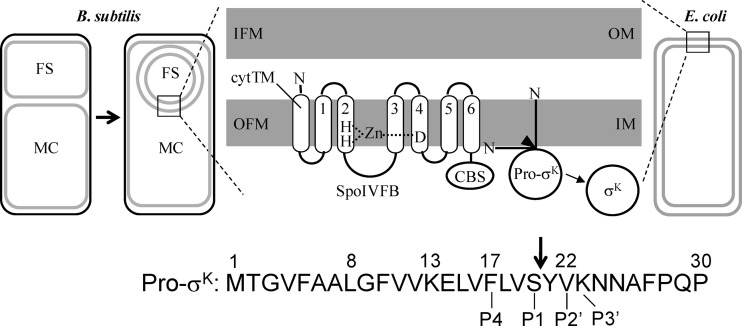
SpoIVFB is an IMMP that cleaves Pro-σK during Bacillus subtilis sporulation (11–13). B. subtilis is a Gram-positive bacterium that forms endospores when nutrients become limiting. During the process of endospore formation, an asymmetrically positioned septum divides the rod-shaped cell into a larger mother cell compartment and a smaller forespore (Fig. 1, left). The mother cell membrane then engulfs the forespore, pinching it off as a free protoplast within the mother cell. Hence, the forespore is surrounded by two membranes, the outer of which is exposed to the mother cell, where SpoIVFB and Pro-σK are synthesized (14–17). SpoIVFB localizes to the outer forespore membrane and is held inactive in a complex that includes SpoIVFA, BofA, and several other proteins (18–21). Serine proteases synthesized in the forespore and secreted into the space between the two membranes surrounding the forespore target SpoIVFA and BofA, releasing SpoIVFB from inhibition (22–25). SpoIVFB cleaves Pro-σK after residue 20 (26), which is referred to as the P1 position in standard substrate nomenclature (i.e., residues preceding the cleavage site N-terminally are designated …P3, P2, and P1 and residues following the cleavage site C-terminally are designated P1′, P2′, P3′…, as shown for several residues at the bottom of Fig. 1). Cleavage releases active σK from the outer forespore membrane into the mother cell (27) (Fig. 1, middle), where it directs expression of more than 100 genes whose products help complete the sporulation process (28).
Fig 1.
Model of intramembrane proteolysis of Pro-σK by SpoIVFB during B. subtilis sporulation and when expressed in E. coli. (Left) During endospore formation, asymmetric septation produces the mother cell (MC) and the forespore (FS) compartments, and then the MC membrane engulfs the FS, pinching it off as a protoplast surrounded by the inner forespore membrane (IFM) and the outer forespore membrane (OFM), within the MC. (Middle) An expanded view of the membranes surrounding the FS or of the E. coli inner membrane (IM) and outer membrane (OM) in the case of heterologous expression. SpoIVFB and Pro-σK expressed in the B. subtilis MC or in the E. coli cytosol insert into the OFM or IM, respectively. SpoIVFB is depicted with an extra transmembrane segment (cytTM) at its N terminus. The transmembrane segments of SpoIVFB are numbered, and their topology is from reference 49. Histidine and aspartate residues likely to coordinate zinc are shown as well as the C-terminal CBS domain. Residues 1 to 27 of Pro-σK are sufficient for membrane association (31), but whether the pro-sequence spans the membrane or associates peripherally was unknown (both possibilities are shown). An arrowhead indicates the cleavage site in Pro-σK. Cleavage of Pro-σK by SpoIVFB releases σK into the B. subtilis MC or into the E. coli cytosol. (Right) Heterologous expression in E. coli. The left, middle, and right portions are adapted from reference 13 with permission. (Bottom) The first 30 residues of Pro-σK are shown with some numbered above, an arrow indicating the cleavage site, and standard substrate position numbering relative to the cleavage site below.
Although RIP of Pro-σK by SpoIVFB normally occurs during B. subtilis sporulation, coexpression of pathway components in Escherichia coli (Fig. 1, middle and right) has contributed greatly to understanding their functions. Coexpression of Pro-σK and SpoIVFB in E. coli resulted in production of a small amount of σK, and it was noted that SpoIVFB contains a motif (HEXXH) found in metalloproteases, suggesting that SpoIVFB directly cleaves Pro-σK (29). Coexpression of BofA with Pro-σK and SpoIVFB in E. coli partially inhibited cleavage of Pro-σK, suggesting that BofA is the primary inhibitor of SpoIVFB (30).
In addition to elucidating functions of components involved in RIP of Pro-σK by SpoIVFB, coexpression in E. coli of SpoIVFB and variants of Pro-σK has been used previously to define features of the substrate that are important for cleavage (31). C-terminal truncations of Pro-σK revealed that residues 1 to 117 are sufficient for cleavage, but residues 1 to 126 allowed more abundant cleavage, comparable to that seen with full-length Pro-σK, and a very similar pattern of results was observed in sporulating B. subtilis. Addition or deletion of 5 residues near the N terminus of Pro-σK allowed abundant cleavage in E. coli or in sporulating B. subtilis. A K13E substitution in Pro-σK prevented cleavage in both organisms. Not only did coexpression in E. coli yield results similar to those seen with sporulating B. subtilis in terms of cleavability of Pro-σK variants, the cleavage was accurate. Just as Pro-σK was cleaved after residue 20 in sporulating B. subtilis (26), full-length Pro-σK(1-241) or Pro-σK(1-126) (13, 30) and variants of Pro-σK(1-126) with 5 residues added or deleted near the N terminus (31) were cleaved exclusively after residue 20, based on N-terminal amino acid sequencing of the purified (C-terminally His-tagged) cleavage products.
Here, coexpression in E. coli of SpoIVFB and variants of Pro-σK was employed to discover additional features of Pro-σK that are important for cleavage by SpoIVFB. These features were examined by sequence comparison in substrates of other IMMPs in order to discern whether particular features might be recognized by all IMMPs, while other features might be unique to SpoIVFB recognition of Pro-σK or apply to their orthologs. SpoIVFB is the founding member of a large subfamily of IMMPs that contain a C-terminal cystathionine-β-synthase (CBS) domain (32). CBS domains appear to regulate the activity of metabolic enzymes, kinases, and channel proteins in response to cellular energy status by binding adenosine-containing ligands such as ATP (33, 34). The CBS domain of SpoIVFB interacts with ATP and with Pro-σK, and ATP is required for purified SpoIVFB to cleave purified Pro-σK (13). This biochemical study utilized cytTM-SpoIVFB-FLAG2-His6, which cleaved Pro-σK(1-126)–His6 accurately upon coexpression in E. coli. The cytTM portion of the SpoIVFB fusion protein is an extra TMS from rabbit cytochrome P450 2B4 that enhances accumulation of the protein in the E. coli inner membrane (Fig. 1, middle). Since cytTM-SpoIVFB-FLAG2-His6 was used throughout this study (with some exceptions; see Fig. 6), we refer to it simply as SpoIVFB here.
Fig 6.

Effects of single-residue substitutions in Pro-σK(1-126)–His6 on cleavage by SpoIVFB in sporulating B. subtilis. Wild-type (WT) Pro-σK(1-126)–His6 or derivatives (abbreviated Pro-σK) with the indicated change were expressed from the native sigK promoter from an ectopic site (amyE) in B. subtilis cells induced to sporulate. All strains had a sigK null mutation to prevent production of native Pro-σK. Strains with only the sigK mutation produce native SpoIVFB (+). Strains that also have a spoIVF null mutation fail to produce SpoIVFB (-). Cells were harvested 5 h after induction of sporulation, and whole-cell extracts were subjected to immunoblot analysis with antibodies that recognize the His6 tag. The product of cleavage is indicated. The arrowhead marks a faint band indicative of cleavage. Above, the wild-type Pro-σK sequence (residues 1 to 30) is shown, with the arrow indicating the cleavage site.
Other subfamilies of IMMPs do not contain a CBS domain. E. coli RseP is the founding member of a large subfamily of mainly bacterial IMMPs that contain at least one PDZ domain (32, 35, 36). RseP cleaves the anti-σ factor RseA after it is first cleaved by DegS in response to extracytoplasmic stress (37). RseP cleaves RseA in its TMS, as well as model proteins unrelated to RseA, provided that their TMS has α-helix-destabilizing residues (38). The importance of residues with a low propensity to form an α-helix, in the vicinity of the cleavage site, was also demonstrated for sterol regulatory element-binding protein-2 (SREBP-2) and ATF6, which are cleaved by human Site-2 protease (S2P) after initial cleavage by Site-1 protease (S1P) (39, 40). S2P is the founding member of the eukaryotic IMMP subfamily (41), most of whose members have a PDZ domain with a cysteine-rich insert of unknown function (32). Helix-destabilizing residues have been proposed to facilitate substrate unwinding that is necessary for cleavage, not only by IMMPs (39), but by rhomboids (42) and signal peptide peptidases (43).
The substrate features that allow IMMPs to cleave a particular peptide bond are not understood. Neither RseP (38) nor S2P (44) appears to recognize a specific sequence near the cleavage site. Here, we report that residues near the cleavage site in Pro-σK, as well as distally N-terminal residues, strongly influence the abundance of cleavage by SpoIVFB. Comparisons of residues near the cleavage sites in Pro-σK, RseA, and SREBP-2, and inferred cleavage sites in their orthologs, suggest that IMMPs share a preference for a medium-size hydrophobic or polar residue at P2′, but SpoIVFB and its orthologs appear to have a stronger preference for a small residue at P1. The arrangement of small and hydrophobic residues near the cleavage site influenced the accuracy with which SpoIVFB cleaved Pro-σK. Unlike RseP (38) and S2P (39), helix-destabilizing residues in Pro-σK were not crucial for cleavage by SpoIVFB. Some of the observed differences between SpoIVFB and other IMMPs likely reflect a difference in the way their substrates interact with the membrane. We provide evidence that a portion of Pro-σK associates peripherally with the membrane and serves as the substrate. In contrast, other IMMP substrates are cleaved in typical TMSs (38, 44). Our report is the most comprehensive analysis of substrate features important for IMMP cleavage to date. The results have broad implications for modulating substrate cleavage by IMMPs similar to SpoIVFB, which are found in both pathogenic and beneficial bacteria.
MATERIALS AND METHODS
Plasmid construction.
Descriptions of plasmid construction and primers are in Tables S1 and S2 in the supplemental material, respectively. DNA sequencing of all cloned PCR products and all genes subjected to mutagenesis (QuikChange kit; Stratagene) confirmed the presence of the desired sequences.
Transformation of plasmids into E. coli and induction of B. subtilis proteins.
For analysis of cleavage and for the cell fractionation experiments whose results are shown in Fig. 2C (see also Fig. S2B in the supplemental material), two plasmids bearing different antibiotic resistance genes and different B. subtilis genes fused to a T7 RNA polymerase promoter were cotransformed into E. coli BL21(DE3) (Novagen), and expression of T7 RNA polymerase and B. subtilis proteins was induced for 2 h with isopropyl-β-d-thiogalactopyranoside (IPTG) (0.25 mM) as described previously (30). For all other cell fractionation experiments, one plasmid designed to produce B. subtilis Pro-σK(1-126)–His6 or its derivative was transformed into E. coli and the strain was induced as described above.
Fig 2.
Fractionation of E. coli expressing Pro-σK(1-126)–His6. (A) Cells induced to express Pro-σK(1-126)–His6 from pZR12 were fractionated, and the supernatant (S) and pellet (P) fractions after the indicated treatments were subjected to immunoblot analysis with antibodies that recognize the His6 tag. The S fraction after low-speed centrifugation (lane 2) was subjected to high-speed centrifugation (lanes 3 to 8). Purified membranes were treated with NaCO3 (lanes 9 and 10). Lane 1 shows protein size markers. (B) Cells induced to express His6-SpoIVFA from pZR74 were fractionated as described for panel A and subjected to immunoblot analysis with antibodies against SpoIVFA, which also recognize a cross-reacting E. coli protein of about 35 kDa. (C) Cells induced to coexpress Pro-σK(1-126)–His6 from pZR12 and SpoIVFB from pZR209 were sonicated to produce a whole-cell lysate (W) and were then fractionated and subjected to immunoblot analysis as described for panel A.
Fractionation of cellular proteins.
Several similar methods of cell fractionation were used. All resulted in 80% to 90% of B. subtilis Pro-σK(1-126)–His6 in the “insoluble fraction” (IF). They differed primarily in the culture volume and the method of cell breakage. For the experiments shown in Fig. S3 in the supplemental material, E. coli cells from cultures (100 ml) induced as described above were fractionated as described previously (30). The same fractionation procedure was used for the experiments showing NaCO3 treatment of purified membranes as presented in Fig. 2A and B except that E. coli cells from cultures (1,000 ml) induced as described above were harvested by centrifugation (12,000 × g) and resuspended in 20 ml of phosphate-buffered saline (PBS) lysis buffer (pH 7.2, containing 100 μM Pefabloc, 0.5 mg/ml lysozyme, 10 μg/ml DNase I, and 10 μg/ml RNase A) and then passed twice through a French pressure cell and centrifuged as described previously (30). In this and the other fractionation procedures described below, a low-speed-centrifugation step (12,000 × g for 10 min at 4°C) was used to sediment and discard proteins in inclusion bodies. The supernatant was then subjected to high-speed centrifugation (200,000 × g for 90 min at 4°C) to sediment proteins associated with membranes. The pellet produced after high-speed centrifugation was rinsed with 10 ml PBS and homogenized in 10 ml PBS containing 5% sucrose, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM EDTA, and 1-ml aliquots (3 ml total) were layered on 4-ml sucrose density gradients to purify membranes as described previously (27). The purified membranes (3 ml) were diluted 10-fold in 100 mM NaCO3 (pH 11) containing 1 mM PMSF and 1 mM EDTA, stirred on ice for 60 min (45), and then centrifuged at 200,000 × g for 90 min at 4°C. Proteins in a sample (500 μl) of the supernatant were precipitated in 10% trichloroacetic acid (TCA), washed with acetone, and resuspended in 100 μl of 62.5 mM Tris·HCl (pH 6.8)–2% SDS–10% glycerol–100 mM dithiothreitol (DTT)–0.1% bromophenol blue and boiled 3 min for immunoblot analysis. The pellet resulting from the high-speed centrifugation was rinsed with 10 ml PBS and homogenized in 3 ml PBS containing 1 mM PMSF and 1 mM EDTA, and a sample (50 μl) was added to 50 μl of 2× sample buffer (50 mM Tris·HCl [pH 6.8], 4% SDS, 20% glycerol, 200 mM DTT, 0.03% bromophenol blue) for immunoblot analysis.
For the experiments whose results are shown in Fig. 3B and C, the fractionation procedure was modified as follows. E. coli from cultures (200 ml) induced as described above were harvested by centrifugation (4,000 × g) and resuspended in 10 ml of PBS lysis buffer. The suspension was incubated at 37°C for 15 min and then passed 3 times through a Nano DeBEE electric benchtop laboratory homogenizer (BEE International) at 14,000 lb/in2. The lysate was centrifuged at 12,000 × g for 10 min at 4°C, and a small sample of the resulting low-speed supernatant was collected for immunoblot analysis, while the rest (10 ml) was centrifuged at 200,000 × g for 90 min at 4°C. A sample of the resulting high-speed supernatant was collected for immunoblot analysis. The pellet was washed gently with 1 ml PBS twice and then ground with a plastic pestle in 10 ml PBS containing 1% Sarkosyl until it was uniformly resuspended, and a sample was collected for immunoblot analysis. For the experiments whose results are shown in Fig. 2 (see also Fig. S1 and S2 in the supplemental material), the procedure was further modified as follows to examine the effects of salt or detergent treatment. To 1 ml of low-speed supernatant, 250 μl PBS (untreated) or 250 μl 5 M NaCl (1 M final concentration) or 237.5 μl PBS plus 12.5 μl Triton X-100 (1% final concentration) was added and the result was mixed by pipetting, stored on ice for 20 min, and then centrifuged at 200,000 × g for 90 min at 4°C. A sample of the resulting high-speed supernatant was collected for immunoblot analysis. The pellet was washed gently with 1 ml PBS and then ground with a plastic pestle in 1.25 ml PBS containing 1% Sarkosyl until it was uniformly resuspended, and a sample was collected for immunoblot analysis.
Fig 3.
Effects of deletions in the pro-sequence of Pro-σK(1-126)–His6 on cleavage and fractionation. (A) Effects on cleavage by SpoIVFB. E. coli cells bearing pZR209 to express cytTM-SpoIVFB-FLAG2-His6 (abbreviated SpoIVFB) and pZR100 (lane 1), pZR247 (lane 2), pZR188 (lane 3), pZR141 (lane 4), or pZR129 (lane 5) to express Pro-σK(1-126)–His6 derivatives (abbreviated Pro-σK) lacking the indicated residues from the pro-sequence were induced with IPTG for 2 h, and whole-cell extracts were subjected to immunoblot analysis using antibodies against FLAG (top) or with antibodies that recognize the His6 tag (bottom). The product of cleavage is indicated. The wild-type pro-sequence (residues 1 to 20) is shown above. (B) Effects on fractionation. E. coli cells bearing pZR12 (lanes 1 to 3), pZR129 (lanes 4 to 6), pZR173 (lanes 7 to 9), or pZR188 (lanes 10 to 12) to express wild-type (WT) Pro-σK(1-126)–His6 or derivatives (abbreviated Pro-σK) lacking the indicated residues from the pro-sequence were induced with IPTG for 2 h, and the cells were fractionated. The supernatant (S) and pellet (P) fractions after the indicated treatments were subjected to immunoblot analysis with antibodies that recognize the His6 tag. Protein size markers are shown at left. (C) Quantification of cell fractionation experiments. The relative intensities of the immunoblot signals in the supernatant (S) and pellet (P) fractions (expressed as a percentage of the total signal) after high-speed centrifugation of the indicated samples in two experiments are shown. Representative immunoblots are shown in panel B. Error bars are 1 standard deviation.
For the experiments whose results are shown in Fig. S8 in the supplemental material, a small-scale fractionation procedure was used. E. coli from cultures (10 ml) induced as described above were harvested by centrifugation (4,000 × g) and resuspended in 500 μl of PBS lysis buffer. The suspension was incubated at 37°C for 15 min, and then cells were disrupted by sonication (Digital Sonifer; Branson) (30% amplitude, 4 times for 10 s each time, with 5 s on an ice-water bath between the sonication procedures). The lysate was centrifuged at 12,000 × g for 10 min at 4°C, and a small sample of the resulting low-speed supernatant was collected for immunoblot analysis, while the rest was centrifuged at 200,000 × g for 90 min at 4°C. A sample of the resulting high-speed supernatant was collected for immunoblot analysis. The pellet was washed gently with 1 ml PBS and then ground with a plastic pestle in 500 μl PBS containing 1% Sarkosyl until it was uniformly resuspended, and a sample was collected for immunoblot analysis.
Immunoblot analysis.
Except for TCA-precipitated proteins after NaCO3 treatment of purified membranes, all other cell fractions were added to an equal volume of 2× sample buffer and boiled for 3 min and then subjected to immunoblot analysis as described previously (46). Proteins were separated on SDS–14% Prosieve polyacrylamide gels (Lonza) with Tris-Tricine electrode buffer (0.1 M Tris, 0.1 M Tricine, 0.1% SDS) and electroblotted to Immobilon-P membranes (Millipore). Antibodies against pentahistidine (penta-His; Qiagen) and FLAG (Sigma) were used at 1:5,000 dilution. The signal intensities in the immunoblots were measured, and background was subtracted using Quantity One (Bio-Rad) or Multi Gauge (Fujifilm) software.
For analysis of cleavage in E. coli, equivalent amounts of cells from different cultures were collected from 0.5 to 1.0 ml of culture (depending on the optical density at 600 nm) by centrifugation (12,000 × g) and whole-cell extracts were prepared as described previously for immunoblot analysis (30).
Purification of proteins and determination of N-terminal sequences.
To purify the cleaved products of Pro-σK(1-126)–His6 derivatives, plasmids were cotransformed into E. coli as described above and cultures (500 ml) were induced as described previously (30) to coexpress cytTM-SpoIVFB-FLAG2-His6 and a Pro-σK(1-126)–His6 derivative. Cells were harvested by centrifugation (12,000 × g), resuspended in 20 ml of PBS lysis buffer lacking lysozyme, and then passed twice through a French pressure cell and centrifuged as described previously (30). The supernatant remaining after ultracentrifugation was mixed with 0.5 ml cobalt resin slurry (Clontech) by rotating for 30 min at 22°C. Purification of His6-tagged proteins was performed according to the manufacturer's instructions except that elution was performed with PBS (pH 7.2) containing 200 mM imidazole and 150 mM NaCl. Proteins were separated by SDS-PAGE as described above for immunoblot analysis but were then electroblotted to Sequi-Blot polyvinylidene difluoride membranes (Bio-Rad), stained with Coomassie solution (0.1% Coomassie brilliant blue R-250, 1.0% acetic acid, 40% methanol), and destained with 50% methanol. The faster-migrating, cleaved products were sequenced by Edman degradation at the Michigan State University Macromolecular Structure Facility. The percentage of minor-abundance sequences was determined from the amounts (after subtraction of background) of the major and minor amino acid residues in the first round of sequencing except in the two cases (LVSY to AAAA and SY to VAA) in which the two sequences began with the same residue; in those cases, the amounts in the second round of sequencing were used.
Construction of B. subtilis strains and induction of sporulation.
The sigK null mutant BK410 (47) and its sigK spoIVF null mutant derivative described previously (31) were transformed with plasmids (48) by selection on LB agar containing chloramphenicol (5 μg/ml). The plasmids were derived from pDG364, which permits gene replacement of amyE in the chromosome by homologous recombination (double crossover). Transformants that represented an amyE mutant were identified by loss of amylase activity on 1% potato starch medium stained with Gram's iodine solution as described previously (48). The strains are listed in Table S3 in the supplemental material. Sporulation was induced by growing cells in the absence of antibiotic and resuspension of cells in SM medium as described previously (48). Samples (1 ml) were collected 5 h after resuspension and centrifuged (12,000 × g), whole-cell extracts were prepared as described previously for E. coli (30), and proteins in extracts were subjected to immunoblot analysis.
RESULTS
A portion of Pro-σK associates peripherally with the membrane in E. coli and appears to serve as the substrate for SpoIVFB.
Previous work showed that for full-length Pro-σK-His6 expressed in E. coli, the majority that is soluble after low-speed centrifugation (12,000 × g for 10 min to sediment and discard proteins in inclusion bodies) is insoluble after high-speed centrifugation (200,000 × g for 90 min to sediment proteins associated with membranes), which was interpreted to mean that the majority of Pro-σK-His6 not present in inclusion bodies is membrane associated (30). Using a similar cell fractionation method, we obtained similar results for Pro-σK(1-126)–His6 expressed in E. coli (Fig. 2A, lanes 2 to 4). To further investigate the apparent association with the inner membrane, the soluble material present after low-speed centrifugation was incubated with NaCl or Triton X-100 to solubilize peripheral or integral membrane proteins, respectively, followed by high-speed centrifugation. The two treatments solubilized similar amounts of Pro-σK(1-126)–His6 (Fig. 2A, lanes 5 to 8), suggesting that a portion of the protein is peripherally membrane associated. Quantification of several experiments suggested that the detergent solubilizes slightly more protein than the salt, but nearly half of the protein was insoluble even after treatment with 1% Triton X-100 (see Fig. S1 in the supplemental material). In contrast, the integral membrane protein His6-SpoIVFA was solubilized by detergent but not salt (Fig. 2B; see also Fig. S1 in the supplemental material), as expected (20). SpoIVFA has a single TMS (49). Since Pro-σK(1-126)–His6 did not behave like a protein with a single TMS, it is unlikely that the pro-sequence inserts into the membrane like a typical TMS.
Since nearly half of the Pro-σK(1-126)–His6 was insoluble even after treatment with 1% Triton X-100 (see Fig. S1 in the supplemental material), we could not be certain whether the 1 M NaCl treatment solubilized the protein from the membrane or from associations with itself or other cellular components. Therefore, we purified membranes using sucrose step gradients and extracted the membranes with NaCO3. The NaCO3 treatment creates planar membranes and solubilizes peripheral membrane proteins (45). The majority of Pro-σK(1-126)–His6 was solubilized, whereas the majority of His6-SpoIVFA remained insoluble (Fig. 2A and B, lanes 9 and 10). These results further distinguish Pro-σK(1-126)–His6 from a single-pass integral membrane protein and provide additional evidence that a portion of Pro-σK(1-126)–His6 associates peripherally with the membrane in E. coli.
In the experiments reported below, Pro-σK(1-126)–His6 or variants of it were coexpressed with SpoIVFB in E. coli and whole-cell extracts were analyzed by immunoblotting. A typical result for wild-type Pro-σK(1-126)–His6 is shown in lane 2 of Fig. 2C. The lower band was shown previously to be σK(21-126)–His6, demonstrating that SpoIVFB cleaves Pro-σK(1-126)–His6 accurately (13). While cleavage was abundant, cell fractionation revealed why a considerable amount of Pro-σK(1-126)–His6 remained uncleaved. Most of the Pro-σK(1-126)–His6 sedimented upon low-speed centrifugation (compare Fig. 2C, lanes 2 and 3), suggesting that it accumulates in inclusion bodies. Importantly, very little of the remaining Pro-σK(1-126)–His6 was solubilized by salt or detergent (Fig. 2C, lanes 4 to 9). This strongly suggests that the portion of Pro-σK(1-126)–His6 that associates peripherally with the membrane serves as the substrate for SpoIVFB and that the remaining uncleaved Pro-σK(1-126)–His6 is not membrane associated and is therefore presumably not accessible to SpoIVFB. As expected, nearly all of the cleavage product, shown previously to be σK(21-126)–His6 (30), was soluble after high-speed centrifugation (Fig. 2C, lane 4).
We also tested Pro-σK(1-126)–His6 with an S20G substitution at the P1 position preceding the cleavage site, which was shown previously to enhance cleavage by SpoIVFB in vivo and in vitro (13). E. coli expressing Pro-σK(1-126)–His6 S20G yielded fractionation results similar to those seen with the wild-type protein (see Fig. S1 and S2A in the supplemental material). As expected, upon coexpression with SpoIVFB, cleavage of the S20G variant appeared to be enhanced relative to cleavage of the wild-type protein (compare the whole-cell extracts in lane 2 in Fig. S2B in the supplemental material and lane 2 in Fig. 2C). Like the uncleaved wild-type protein (Fig. 2C), the uncleaved S20G variant was not appreciably solubilized by salt or detergent (see Fig. S2B in the supplemental material), indicating that it was not membrane associated. We conclude that the S20G variant is very similar to the wild-type protein in terms of its membrane association characteristics. In both cases, a portion of the pro-protein appears to associate peripherally with the membrane and serve as the substrate for SpoIVFB, but the S20G variant appears to be a slightly better substrate in vivo, and this interpretation is supported by reactions in vitro with purified proteins in detergent or with SpoIVFB proteoliposomes (13).
Taken together, our cell fractionation experiments show that when Pro-σK(1-126)–His6 is expressed in E. coli, a portion associates peripherally with the membrane, and about half is detergent insoluble. The combined material, which was soluble after low-speed centrifugation but insoluble after high-speed centrifugation, is here called the insoluble fraction (IF) (see, e.g., Fig. 2A, lane 4; see also Fig. S1 in the supplemental material, second bar [both labeled “P” for “pellet”]). Coexpression with SpoIVFB results in less Pro-σK(1-126)–His6 in the IF and release of soluble σK(21-126)–His6. Very little of the Pro-σK(1-126)–His6 remaining in the IF can be solubilized by salt, so it is likely that the portion of Pro-σK(1-126)–His6 that was peripherally membrane associated, and perhaps the Pro-σK(1-126)–His6 that associates directly with membrane-embedded SpoIVFB, is cleaved by the enzyme and released to the soluble fraction.
In the experiments reported below, we used coexpression with SpoIVFB in E. coli and immunoblot analysis of whole-cell extracts to screen for Pro-σK(1-126)–His6 variants that result in accumulation of less cleavage product. Variants accumulating less cleavage product were analyzed further by expressing the Pro-σK(1-126)–His6 variant (in the absence of SpoIVFB) and using differential centrifugation to measure the soluble fraction and the IF in order to distinguish features of Pro-σK(1-126)–His6 that might affect its interaction with SpoIVFB from features that, in the absence of SpoIVFB, change the proportion of protein in the IF and therefore affect its interaction with the membrane, itself, or other cellular components.
Deletion of residues 2 to 8 from Pro-σK nearly eliminates cleavage.
Previous work showed that deletion of residues 2 to 6 of Pro-σK(1-126)–His6 did not diminish cleavage by SpoIVFB when the two proteins were coexpressed in E. coli (31) (Fig. 3A, lane 1). We extended the analysis and found that deletion of residues 2 to 7 reduced cleavage, deletion of residues 2 to 8 or 2 to 9 nearly eliminated cleavage, and deletion of residues 2 to 12 abolished cleavage (Fig. 3A, lanes 2 to 5). The deletion of residues 2 to 8 had very little effect on the proportion of protein in the IF (Fig. 3B, lane 12, and Fig. 3C). The deletion of residues 2 to 12 reduced the proportion of protein in the IF by 20%, whereas deletion of residues 2 to 20 (i.e., the entire pro-sequence) reduced it by about 60% (Fig. 3B and C). The protein lacking residues 2 to 20 was included as a control since previous fractionation studies with sporulating B. subtilis suggested that the pro-sequence is required for membrane association of Pro-σK (27) or of a fusion protein in which the first 27 residues of Pro-σK were fused to the green fluorescent protein (GFP) (31). The marked effect of deleting residues 2 to 20 on the proportion of protein in the IF demonstrates that the assay is sensitive to loss of the pro-sequence. Since the deletion of residues 2 to 12 changed the proportion of protein in the IF, the deletion appears to alter associations with the membrane, itself, or other cellular components. On the other hand, the deletion of residues 2 to 8 did not change the proportion of protein in the IF, suggesting that a feature important for interaction with SpoIVFB might be lacking. Of course, we cannot rule out the possibility that loss of residues 2 to 8, for example, changes how the protein interacts with the membrane in a subtle fashion that is not detectable by differential centrifugation. Further work would be needed to distinguish between several possible explanations. Here, we conclude that residues 8 to 20 (for a total of 14 residues, including the N-terminal Met) are sufficient for substantial cleavage. We used this information to guide Ala-scanning mutagenesis (see below).
Previous work showed that insertion of 5 residues (TGVFA) after the N-terminal Met of Pro-σK(1-126)–His6 did not interfere with cleavage in E. coli engineered to coexpress SpoIVFB (31). Addition of the 14-residue S-short tag or the 27-residue S tag prevented cleavage (see Fig. S3A in the supplemental material). The S tag greatly reduced the proportion of protein in the IF, possibly explaining the lack of cleavage, but the S-short tag did not appear to change the proportion in the IF (see Fig. S3B in the supplemental material). We conclude that a 14-residue addition to the N terminus of Pro-σK(1-126)–His6 prevents cleavage. Further work would be needed to determine, for example, whether the extension prevents interaction with SpoIVFB or interferes with a subsequent step in catalysis.
Alanine substitutions at P4, P2′, and P3′ of Pro-σK impair cleavage.
Since deletion of residues 2 to 7 of Pro-σK(1-126)–His6 still permitted substantial cleavage (Fig. 3A, lane 2), we focused on residues 8 to 23 to identify those important for cleavage. Of 12 single-Ala and 1 triple-Ala substitutions tested, F17A, V22A, and K23A greatly reduced cleavage (Fig. 4A). We did not test E14A since E14K was shown previously to allow normal cleavage (31). We did not measure SpoIVFB in some cases where cleavage appeared to be normal (Fig. 4A).
Fig 4.
Effects of single-residue substitutions in Pro-σK(1-126)–His6 on cleavage by SpoIVFB in E. coli. (A) Effects of Ala substitutions for residues 8 to 23. E. coli cells bearing pZR209 to express cytTM-SpoIVFB-FLAG2-His6 (abbreviated SpoIVFB) and pZR12 (lanes 1, 3, 5, 10, 12, 17, 18, and 20), pZR152 (lane 2), pZR153 (lane 4), pZR151 (lane 6), pZR110 (lane 7), pZR273 (lane 8), pZR274 (lane 9), pZR271 (lane 11), pZR186 (lane 13), pZR187 (lane 14), pZR139 (lane 15), pZR138 (lane 16), pZR146 (lane 19), or pZR104 (lane 21) to express wild-type (WT) Pro-σK(1-126)–His6 or derivatives (abbreviated Pro-σK) in which the indicated residue(s) was replaced by A were induced with IPTG for 2 h. Whole-cell extracts were subjected to immunoblot analysis using antibodies against FLAG (top) or with antibodies that recognize the His6 tag (bottom). The product of cleavage is indicated. Above, the wild-type Pro-σK sequence (residues 1 to 23) is shown, with the arrow indicating the cleavage site. (B) Effects of other substitutions for F17, V22, and K23. E. coli cells bearing pZR209 in combination with pZR12 (lanes 3 and 13), pZR289 (lane 1), pZR300 (lane 2), pZR146 (lane 4), pZR199 (lane 5), pZR205 (lane 6), pZR207 (lane 7), pZR197 (lane 8), pZR213 (lane 9), pZR215 (lane 10), pZR220 (lane 11), pZR269 (lane 12), pZR136 (lane 14), pZR107 (lane 15), pZR108 (lane 16), or pZR339 (lane 17) were induced, and extracts were subjected to immunoblot analysis as described for panel A.
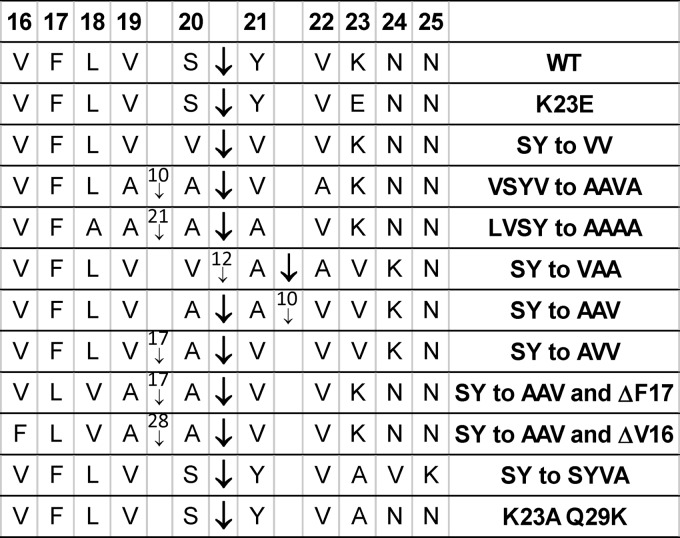
To explore the effects of residues other than F, V, and K at the P4, P2′, and P3′ positions, respectively, other single-residue substitutions were tested. The results of these and other substitutions are summarized in Table 1. At P4, F can be replaced by L or V (Fig. 4B, lanes 1 and 2), indicating that medium-size hydrophobic residues are tolerated, although the smaller hydrophobic residue A is not well tolerated (Fig. 4A, lane 11). An alignment of Pro-σK orthologs revealed primarily F, L, or V at the putative P4 position, and A is rarely observed at this position (see Fig. S4 in the supplemental material), suggesting that SpoIVFB orthologs that likely cleave Pro-σK in other bacteria prefer F or medium-size hydrophobic residues at P4. This preference is not shared by more distantly related IMMPs that cleave other types of substrates. E. coli RseP cleaves the anti-σ RseA and an artificial substrate with TMS 1 from the galactoside permease LacY (38), both of which have G at P4 (see Fig. S5 in the supplemental material), and RseA orthologs have G or A at the putative P4 position (see Fig. S6 in the supplemental material). Human S2P cleaves the transcription factor SREBP-2, which has R at P4 (see Fig. S5 in the supplemental material), and R is perfectly conserved at the putative P4 position of SREBP-2 orthologs (see Fig. S7 in the supplemental material).
Table 1.
Effects of single-residue substitutions on cleavage of Pro-σK(1-126)-His6 by coexpressed SpoIVFB and on the proportion of protein in the insoluble fraction in E. coli
At P2′, V can be replaced by I, T, L, or M, which have side chains of similar sizes, but not by F, which has a larger side chain, and not by P, G, and N, which have a low propensity to form an α-helix (Fig. 4B, lanes 5 to 12). Pro-σK orthologs have V, L, or I at the putative P2′ position (see Fig. S4 in the supplemental material). Interestingly, E. coli RseA and human SREBP-2 also have V at P2′ (see Fig. S5 in the supplemental material), and their orthologs have V, M, or T at the putative P2′ position (see Fig. S6 and S7 in the supplemental material), so a preference for a medium-size hydrophobic or polar residue at P2′ appears to be a general feature of IMMPs. However, E. coli LacY TMS 1 has F at P2′ (38) (see Fig. S5 in the supplemental material), so RseP can tolerate a larger side chain at this position, in contrast to SpoIVFB (Fig. 4B, lane 5). Like RseP, S2P appeared to tolerate F at the P2′ position of SREBP-2, although the cleavage site was not mapped (44).
At P3′, K can be replaced by E, R, S, or Q, which have a variety of side chain types (Fig. 4B, lanes 14 to 17). To assess the accuracy of cleavage of the K23E variant, the cleavage product was purified and subjected to N-terminal amino acid sequencing by Edman degradation. A single sequence, YVENNAF, was observed, indicating that cleavage occurred at the normal position between S20 and Y21 (Fig. 5). Pro-σK orthologs have primarily K, S, or T at the putative P3′ position, but residues with a variety of side chain types are found (see Fig. S4 in the supplemental material). RseA has S at P3′ (see Fig. S5 in the supplemental material), and S is highly conserved at the putative P3′ position of RseA orthologs (see Fig. S6 in the supplemental material), but LacY TMS 1 has Y at P3′ (see Fig. S5 in the supplemental material), indicating that E. coli RseP can tolerate a large side chain at this position. Likewise, SREBP-2 has L at P3′ (see Fig. S5 in the supplemental material), and SREBP-2 orthologs have L, F, or M at the putative P3′ position (see Fig. S7 in the supplemental material), suggesting that human S2P prefers a large hydrophobic or aromatic residue at P3′.
Fig 5.
Cleavage sites in Pro-σK(1-126)–His6 derivatives. Cleavage products were purified and subjected to N-terminal amino acid sequencing by Edman degradation. Wild-type (WT) Pro-σK(1-126)–His6 was cleaved exclusively after residue 20, as indicated by a large arrow. Some derivatives yielded two sequences, and the percentage of the minor cleavage product is indicated by the number over the smaller arrow.
Each variant protein that exhibited reduced cleavage upon coexpression with SpoIVFB was expressed alone in E. coli to compare the IFs. F17A at P4 reduced the proportion of protein in the IF by about 20% (see Fig. S8 in the supplemental material), which is comparable to the reduction in the IF observed for the deletion of residues 2 to 12 (Fig. 3B and C), suggesting that both substitutions alter the association of Pro-σK(1-126)–His6 with the membrane, itself, or other cellular components. Substituting A, F, P, N, or G for V22 at P2′, or A for K23 at P3′, did not change the proportion of protein in the IF significantly (see Fig. S8 in the supplemental material), so there was no indication that these substitutions alter the membrane association of Pro-σK(1-126)–His6.
We engineered B. subtilis to express wild-type Pro-σK(1-126)–His6 or key Ala-substituted variants during sporulation. The proteins were expressed from the native sigK promoter. The constructs were integrated into the chromosome ectopically (replacing amyE) in sigK or sigK spoIVF null mutant B. subtilis. Wild-type Pro-σK(1-126)–His6 accumulated poorly in the sigK spoIVF null mutant, but a cleavage product was observed in the sigK null mutant with an intact spoIVF locus (Fig. 6, lanes 1 and 2). Surprisingly, the F17A variant at P4 was cleaved (lane 4), indicating that coexpression in E. coli does not always faithfully predict results in sporulating B. subtilis. On the other hand, as expected from the E. coli results, the V22A variant at P2′ and the K23A variant at P3′ exhibited little or no cleavage product, although, like wild-type Pro-σK(1-126)–His6, the uncleaved protein accumulated poorly and appeared to be susceptible to degradation even in the absence of SpoIVFB (lanes 7 to 11).
Substitutions at P1 of Pro-σK can impair cleavage.
At the P1 position of B. subtilis Pro-σK(1-126)–His6, an S20G substitution enhances accumulation of cleavage product upon coexpression with SpoIVFB in E. coli (13) (see lanes 2 and 3 in Fig. S2B in the supplemental material) and an S20A substitution appeared to have no effect on cleavage (Fig. 4A, lane 15). We tested the effects of other residues at the P1 position. As with the S20A substitution (Fig. 7A, lane 3), there was little or no effect on cleavage when C, L, V, or N replaced S at P1 (lanes 5, 7, 18, and 19). On the other hand, D, with a negatively charged side chain, reduced cleavage slightly (lane 16); W and Y, with aromatic side chains, abolished and greatly reduced cleavage, respectively (lanes 17 and 22); and P, which is incompatible with α-helical structure (although it is not known whether the pro-sequence is normally α-helical), greatly reduced cleavage (lane 20). When expressed alone in E. coli, the S20D variant showed about 20% less protein in the IF, but the S20W, S20Y, and S20P variants had the normal proportion of protein in the IF (see Fig. S8 in the supplemental material). The S20W variant failed to be cleaved in sporulating B. subtilis (Fig. 6, lane 6). We conclude that aromatic or Pro residues are unfavorable at P1. In agreement, orthologs of Pro-σK do not have such residues at the putative P1 position but have S, G, or A (see Fig. S4 in the supplemental material), suggesting that a small residue is preferred. RseA and SREBP-2 have A and L, respectively, at P1 (see Fig. S5 in the supplemental material). RseA orthologs have A at the putative P1 position (see Fig. S6 in the supplemental material). SREBP-2 orthologs have L, I, or M at the putative P1 position (see Fig. S7 in the supplemental material), suggesting that S2P has less preference for a small residue at P1. RseP tolerates the aromatic residue F at P1 when it cleaves LacY TMS 1 (see Fig. S5 in the supplemental material). Likewise, S2P appeared to tolerate the double substitution, LC to FF, at P1 and P1′ of SREBP-2, although the cleavage site was not mapped (44).
Fig 7.
Effects of substitutions in Pro-σK(1-126)–His6 on cleavage by SpoIVFB in E. coli. (A) Effects of one- and two-residue substitutions near the cleavage site. E. coli cells bearing pZR209 to express cytTM-SpoIVFB-FLAG2-His6 (abbreviated SpoIVFB) and pZR12 (lanes 1, 6, 14, and 21), pZR138 (lane 2), pZR139 (lane 3), pZR92 (lane 4), pZR150 (lane 5), pZR335 (lane 7), pZR334 (lane 8), pZR337 (lane 9), pZR336 (lane 10), pZR340 (lane 11), pZR179 (lane 12), pZR279 (lane 13), pZR329 (lane 15), pZR328 (lane 16), pZR333 (lane 17), pZR226 (lane 18), pZR315 (lane 19), pZR316 (lane 20), or pZR314 (lane 22) to express wild-type (WT) Pro-σK(1-126)–His6 or derivatives (abbreviated Pro-σK) in which one or two residues around the cleavage site were replaced as indicated were induced with IPTG for 2 h. Whole-cell extracts were subjected to immunoblot analysis using antibodies against FLAG (top) or with antibodies that recognize the His6 tag (bottom). The product of cleavage is indicated. We did not measure SpoIVFB in some cases where cleavage appeared to be normal (lanes 1 to 13). Above, the wild-type Pro-σK sequence (residues 1 to 30) is shown, with the arrow indicating the cleavage site. (B) Effects of multiresidue substitutions near the cleavage site. E. coli cells bearing pZR209 to express cytTM-SpoIVFB-FLAG2-His6 (abbreviated SpoIVFB) and pZR195 (lane 1), pZR206 (lane 2), pZR208 (lane 3), pZR12 (lanes 4 and 9), pZR263 (lane 5), pZR264 (lane 6), pZR303 (lane 7), pZR217 (lane 8), pZR179 (lane 10), pZR177 (lane 11), pZR185 (lane 12), pZR190 (lane 13), pZR291 (lane 14), pZR293 (lane 15), pZR295 (lane 16), pZR304 (lane 17), or pZR298 (lane 18) to express wild-type (WT) Pro-σK(1-126)–His6 or derivatives (abbreviated Pro-σK) in which residues in the vicinity of the cleavage site were replaced as indicated were induced with IPTG for 2 h. Whole-cell extracts were subjected to immunoblot analysis as described for panel A. (C) Effects of changes C-terminal of the cleavage site. E. coli cells bearing pZR209 to express cytTM-SpoIVFB-FLAG2-His6 (abbreviated SpoIVFB) and pZR12 (lanes 1, 3, and 7), pZR301 (lane 2), pZR312 (lane 4), pZR163 (lane 5), pZR155 (lane 6), or pZR280 (lane 8) to express wild-type (WT) Pro-σK(1-126)–His6 or derivatives (abbreviated Pro-σK) in which residues C-terminal of the cleavage site were added or substituted as indicated were induced with IPTG for 2 h. Whole-cell extracts were subjected to immunoblot analysis as described for panel A.
Although Y is highly conserved at the putative P1′ position of Pro-σK orthologs (see Fig. S4 in the supplemental material), as noted above the Y at P1′ of B. subtilis, Pro-σK(1-126)–His6 tolerated a substitution to A (Fig. 4A, lane 16, and Fig. 7A, lane 2). Not surprisingly, a conservative substitution to F was also tolerated at P1′ (Fig. 7A, lane 4). Since a fairly broad range of residue types appeared to be tolerated at P1 and P1′, we replaced SY at these positions with different combinations of residues, creating double substitutions. All combinations were tolerated except YS (lanes 8 to 13 and 15). Since the single S20Y substitution greatly reduced cleavage (lane 22), it is not surprising that the YS double substitution was not tolerated (lane 15). The YS variant showed the normal proportion of protein in the IF (see Fig. S8 in the supplemental material). Perhaps most surprising was that the GP combination was cleaved abundantly (Fig. 7A, lane 11), indicating that Pro is tolerated at P1′, even though Pro at P1 impairs cleavage (lane 20). The VV combination was cleaved abundantly (lane 13), but we wondered whether the position of cleavage was ambiguous, since the double substitution creates the sequence VVVV at residues 19 to 22. Purification of the cleavage product and determination of its N-terminal amino acid sequence yielded a single sequence indicative of cleavage between residues 20 and 21 (Fig. 5).
Multiresidue substitutions near the cleavage site in Pro-σK affect the abundance and position of cleavage.
To test the limits of the ability of SpoIVFB to properly cleave Pro-σK(1-126)–His6, several residues near the cleavage site were changed simultaneously. Changing VSY to AAA allowed cleavage, but changing VSYV to AVAA severely impaired cleavage (Fig. 7B, lanes 1 and 2), without changing the proportion of protein in the IF (see Fig. S8 in the supplemental material), perhaps due to substitution of A for V at P2′ as shown above (Fig. 4A, lane 19). However, changing VSYV to AAVA allowed cleavage, despite substitution of A for V at P2′, although cleavage was reduced compared with the wild-type sequence results (Fig. 7B, lanes 3 and 4). Nevertheless, sufficient cleavage product from the AAVA-substituted Pro-σK(1-126)–His6 was produced to permit purification and determination of the N-terminal amino acid sequence. Two sequences were obtained. The more abundant (90%) sequence began with VA, and a minor-abundance (10%) sequence began with AVA (Fig. 5). These results show that SpoIVFB mainly cleaves AAVA after residue 20, tolerating A at the P2′ position, but about 10% of the time cleaves after residue 19, placing V at the P2′ position. This result is remarkable for two reasons. First, the single-residue substitution V22A greatly reduced cleavage (Fig. 4A, lane 19, and Fig. 6, lane 8), so A is not well tolerated at the P2′ position in the context of the otherwise wild-type sequence. Second, ambiguity in the position of cleavage was not observed for the wild-type sequence or when SY was changed to VV (creating VVVV at residues 19 to 22) (Fig. 5). Changing LVSY to AAAA also allowed substantial cleavage (Fig. 7B, lane 6) but caused greater ambiguity in the cleavage site, with SpoIVFB cleaving after residue 19 about 21% of the time (Fig. 5). We conclude that effects of particular residues near the cleavage site in Pro-σK depend on the context of other residues and that this context influences both the abundance and position of cleavage. Taken together with our other results, we infer that residues near the active site of SpoIVFB interact with several residues spanning at least from S20 at P1 to K23 at P3′ of Pro-σK.
Effects of adding a residue near the cleavage site in Pro-σK depend on the arrangement of small and hydrophobic residues and on residues in an N-terminal position with respect to F17.
The effects of changing residues near the cleavage site in Pro-σK suggested that the arrangement of small (Ala) and hydrophobic (Val) residues can profoundly influence the abundance and position of cleavage. To examine this further, we tested the effects of changing SY to AAA, VAA, AAV, or AVV in Pro-σK(1-126)–His6. Interestingly, AAA nearly eliminated cleavage (Fig. 7B, lane 11) without changing the proportion of protein in the IF (see Fig. S8 in the supplemental material), VAA and AAV allowed cleavage, albeit reduced (lanes 12 to 13), and AVV allowed abundant cleavage (lane 14). For the three substitutions that allowed substantial cleavage, N-terminal amino acid sequencing of the cleavage products revealed ambiguity in the position of cleavage in each case (Fig. 5). To our surprise, SpoIVFB cleaved mainly after residue 21 in the case of VAA and mainly after residue 20 for AAV and AVV. In each case, cleavage occurred mainly with an A residue at P1 and a V residue at P2′, consistent with the notions mentioned above that SpoIVFB and its orthologs prefer a small residue at P1 and that IMMPs in general prefer a medium-size hydrophobic or polar residue at P2′. However, the minor cleavage products confirm that SpoIVFB can tolerate V at P1 (in the case of the VAA and AVV substitutions of SY, as in the case of the main cleavage product when SY was changed to VV) and A at P2′ (in the case of the VAA substitution of SY, as in the case of the main cleavage product when VSYV was changed to AAVA) (Fig. 5).
In light of the apparent flexibility of SpoIVFB with respect to cleavage of sequences with A or V at P1 and P2′ and to toleration of an additional residue between V19 and V22 of Pro-σK, it is surprising that changing SY to AAA nearly eliminated cleavage (Fig. 7B, lane 11). In contrast, changing SY to AA allowed abundant cleavage (lane 10), as did changing VSY to AAA (lane 1), LVSY to VAAA or AAAA (lanes 5 and 6), or FLVSY to LLAAA (lane 7). However, like the SY-to-AAA change (lane 11), changing VSY to LAAA resulted in poor cleavage (lane 8) without changing the proportion of protein in the IF (see Fig. S8 in the supplemental material). Both changes create the sequence AAA at residues 20 to 22, the normal P1-P2′ positions, which seems to disfavor cleavage.
We also explored the effects of inserting or deleting a residue N-terminal of F17. Insertion of an A residue between V16 and F17 of otherwise wild-type Pro-σK(1-126)–His6 nearly eliminated cleavage (Fig. 7B, lane 17) without changing the proportion of protein in the IF (see Fig. S8 in the supplemental material). Also, strikingly, deletion of V16 or F17 from the variant in which SY was changed to AAV increased cleavage (Fig. 7B; compare lanes 15 and 16 with lane 13). N-terminal amino acid sequencing of cleavage products revealed that the V16 and F17 deletions shifted the predominant cleavage site within the AAV sequence (Fig. 5). The two deletion variants were cleaved mainly between the A and V residues, whereas the SY-to-AAV variant without deletion was cleaved mainly between the A residues. The minor-abundance cleavage products also differed. We conclude that residues N-terminal of F17 in Pro-σK can influence cleavage by SpoIVFB, at least when the pro-sequence is shortened by 6 or 7 residues (Fig. 3A, lanes 2 and 3) or lengthened by 1 residue (Fig. 7B, lane 17) or when SY is changed to AAV and V16 is deleted (Fig. 7B, lane 16, and Fig. 5). In agreement, a K13E charge-reversal substitution in Pro-σK-His6 resulted in poor accumulation of the protein in sporulating B. subtilis and no cleavage was detected (31). Yet neither an E14K charge reversal (31) nor Ala substitutions for residues 8, 9, 10 to 12, 13, 15, or 16 (Fig. 4A) affected cleavage of Pro-σK(1-126)–His6 upon coexpression with SpoIVFB in E. coli. Also, deleting V16 of Pro-σK(1-126)–His6 did not impair cleavage (Fig. 7B, lane 18). Perhaps several residues N-terminal of F17 in Pro-σK interact with SpoIVFB and/or the membrane to influence cleavage, and certain changes alter the interactions enough to cause an observable cleavage defect.
Changes C-terminal of V22 in Pro-σK can affect cleavage, but helix-destabilizing residues are not crucial.
A K23A substitution at P3′ in Pro-σK(1-126)–His6 greatly reduced cleavage (Fig. 4A, lane 21), but K23 could be replaced by a variety of other residues and little effect on cleavage was observed (Fig. 4B, lanes 14 to 17). K23 is at the P3′ position with respect to cleavage of wild-type Pro-σK but at the P4′ position with respect to the major cleavage site of variants in which SY was replaced with AAV or AVV (Fig. 5). Replacing SY with SYAA greatly reduced cleavage, apparently due in part to loss of V at P2′, since replacing SY with SYVA allowed slightly more cleavage (Fig. 7C, lanes 2 and 4). Sequencing of cleavage products of the SYVA variant showed that cleavage occurred at the normal position, after residue 20, with K in the P5′ position (Fig. 5). To determine whether K could be moved even farther from the cleavage site, a K23A Q29K double substitution was tested, since we knew that Q29A allowed cleavage (data not shown). The double substitution permitted substantial cleavage (Fig. 7C, lane 5), and sequencing revealed that cleavage occurred after residue 20, so K was at the P9′ position (Fig. 5). In contrast, a K23A Y21K double substitution, changing YVK to KVA, nearly eliminated cleavage (Fig. 7C, lane 6). However, the YVK-to-KVA change appeared to alter associations of the protein with the membrane, itself, or other cellular components, since the proportion of protein in the IF was greatly reduced (see Fig. S8 in the supplemental material). Taken together, our results show that SpoIVFB tolerates charged or polar residues at position 23 of Pro-σK but that an A residue at position 23 greatly reduces cleavage unless K is present C-terminally and that the position of the K is flexible (K at P4′, P5′, or P9′ allowed substantial cleavage of different variants).
The importance of α-helix-destabilizing residues in the vicinity of the cleavage site has been documented for the IMMPs S2P (39, 40) and RseP (38) and for other IPs (42, 43). Pro-σK has four residues with a low α-helical propensity C-terminal of the cleavage site (N24, N25, P28, and P30). Changing all four residues simultaneously to F or A reduced cleavage slightly (Fig. 7C, lane 8). The proportion of protein in the IF was unaltered (see Fig. S8 in the supplemental material). The change reduced, but did not abolish, cleavage in sporulating B. subtilis (Fig. 6, lane 14). Apparently, helix-destabilizing residues are not crucial for SpoIVFB to cleave Pro-σK(1-126)–His6, distinguishing SpoIVFB from other IMMPs and IPs studied so far.
DISCUSSION
By measuring the effects on cleavage by SpoIVFB of Pro-σK with deletions in the pro-sequence and a large number of substitutions near the cleavage site, we have discovered features of the substrate that influence the abundance and accuracy of cleavage by an IMMP that is broadly conserved in endospore-forming bacteria. This knowledge can be used to modulate cleavage of Pro-σK orthologs, which govern spore formation (contributing to persistence) and biosynthesis of toxins as well as useful products in Bacilli and Clostridia that are deadly pathogens and beneficial industrial microorganisms (50–53). Most of these bacteria have orthologs of SpoIVFB. Most also have orthologs of RseP, as do most bacteria whose genomes have been sequenced. By comparing our results for SpoIVFB with previous work on RseP and its eukaryotic PDZ domain-containing counterpart S2P, we found that, in general, RseP and S2P do not share with SpoIVFB the same preferences for residues near the cleavage site in the substrate and, in particular, that RseP and S2P appear to tolerate an aromatic residue at the P1 or P2′ position in the substrate, unlike SpoIVFB. We also found that helix-destabilizing residues in the substrate are not crucial for cleavage by SpoIVFB, unlike the results seen with RseP (38) and S2P (39). These differences appear to reflect differences in the ways the substrates interact with the membrane and with their cognate IMMP.
Substrate interaction with the membrane.
Our results provide evidence that a portion of Pro-σK(1-126)–His6 produced in E. coli interacts peripherally with the inner membrane but that, upon coexpression with SpoIVFB, this portion is cleaved. In contrast, RseP (35, 36, 38) and S2P (40, 44, 54, 55) cleave substrates within a TMS after cleavage by another protease. Pro-σK(1-126)–His6 produced in E. coli did not behave like a protein with a TMS (Fig. 2), and it can be purified and cleaved by purified SpoIVFB, demonstrating that prior cleavage by another protease is not required (13).
The cell fractionation behavior of Pro-σK(1-126)–His6 produced in E. coli is consistent with the behavior of full-length Pro-σK in sporulating B. subtilis. The majority of Pro-σK is membrane associated but can be released from membranes with salt (0.5 M NaCl or 0.6 M KCl), suggesting a peripheral association (27). The pro-sequence mediates the membrane interaction during sporulation (27, 31). In agreement, deletion of the pro-sequence (residues 2 to 20) from Pro-σK(1-126)–His6 dramatically reduced the proportion of protein in the IF (Fig. 3). Moreover, purified Pro-σK(1-126)–His6 with the S20G substitution associates readily with preformed liposomes made from E. coli lipids, but σK(21-126)–His6 lacking the pro-sequence fails to associate with liposomes (13).
Since the pro-sequence has two charged residues (K13 and E14) bordered by hydrophobic residues (4 to 12 and 15 to 19), as do many of its orthologs (see Fig. S4 in the supplemental material), we propose that K13 and/or E14 interacts with the membrane surface and that the hydrophobic parts of the pro-sequence loop into the membrane (Fig. 8). The charge-reversal substitution E14K in Pro-σK(1-126)–His6 did not affect cleavage upon coexpression with SpoIVFB in E. coli, but a K13E substitution prevented cleavage (31). The K13E substitution in full-length Pro-σK-His6 resulted in poor accumulation and undetectable cleavage in sporulating B. subtilis. The variant protein appeared to be membrane associated, but its interaction with the membrane might be altered. For example, K13 might normally interact with phosphate groups of membrane phospholipids. With respect to the hydrophobic parts of the pro-sequence, we found that deletion of residues 2 to 8 impaired cleavage (Fig. 3). The proportion of protein in the IF was unchanged, but this does not rule out a subtle change in interaction with the membrane, especially since deletion of residues 2 to 12 reduced the proportion of protein in the IF. The contribution of hydrophobic residues 15 to 19 remains to be fully addressed, but changing two of the five residues to Ala (i.e., residues 18 and 19 in the LVSY to AAAA variant) permitted cleavage (Fig. 7B, lane 6).
Fig 8.

Model for interaction of Pro-σK with the membrane and with SpoIVFB. The pro-sequence interacts peripherally with the membrane, and the hydrophobic parts may loop into the membrane. Pro-σK interacts with the C-terminal part of SpoIVFB, which contains the CBS domain. ATP binding to the CBS domain changes the conformation (or oligomeric state [not depicted]) of the enzyme, allowing entry of the pro-sequence near the membrane surface into the SpoIVFB active site (star), where cleavage occurs, releasing σK and the pro-sequence from the membrane into the mother cell cytosol of sporulating B. subtilis or into the cytosol of E. coli engineered to coexpress SpoIVFB and Pro-σK (see Fig. 1).
An attractive feature of our proposed interaction of Pro-σK with the membrane (Fig. 8) is that its orientation (N terminus toward the mother cell for B. subtilis or toward the cytosol for E. coli) would be the same as for other IMMP substrates. Generally, IPs cleave TMSs that span the membrane in one orientation or the other. Since the orientation of highly conserved motifs (e.g., HEXXH and DG) in TMSs of SpoIVFB appears to the same as for other IMMPs (49, 56–58), it has seemed unusual that their substrates would span the membrane in opposite orientations, as would be the case if the pro-sequence of Pro-σK inserts in the membrane like a typical TMS. If the pro-sequence of Pro-σK instead interacts with the membrane surface as we propose (Fig. 8), this conundrum would be solved. As further support, it is worth noting that the unusually long pro-sequences of two B. cereus strains and two B. thuringiensis strains have charged residues that are mostly positive (see Fig. S4 in the supplemental material), consistent with a cytosolic location (59). The unusually short pro-sequences of two other B. cereus strains and B. pseudomycoides (see Fig. S4 in the supplemental material) probably do not permit membrane association or cleavage, based on our deletion results (Fig. 3). These orthologs of Pro-σK likely are active σ factors without being cleaved, since deletion of just five or six residues from the N terminus of Pro-σK produced σ factor activity in vivo (31) or in vitro (60), respectively.
If the pro-sequence interacts with the membrane as depicted in Fig. 8 rather than inserting like a typical TMS, it could explain our findings that helix-destabilizing residues in Pro-σK are not crucial for cleavage by SpoIVFB in E. coli (Fig. 7C, lane 8) or in sporulating B. subtilis (Fig. 6, lane 14), although they appear to facilitate cleavage in both cases. The normal pro-sequence may be partially or fully unwound, and even when all four helix-destabilizing residues are replaced with Ala the pro-sequence may be partially unwound. This would presumably facilitate access of the pro-sequence to the active site of SpoIVFB near the membrane surface and stable interaction until cleavage occurs (Fig. 8).
Substrate access to the active site of the enzyme.
While the interaction of Pro-σK with the membrane appears to be novel, since other IP substrates are integral rather than peripheral membrane proteins, the concept of an IP substrate accessing the active site of the enzyme near the membrane surface is not new. It was previously proposed that S1P cleavage of SREBP causes partial unfolding of the target TMS followed by S2P cleavage near the membrane surface (39). Evidence for this model is a requirement for helix-destabilizing residues in the vicinity of the cleavage site. RseP substrates also exhibit this requirement in vivo, but two observations did not fit the model (38). First, RseP cleaves near the middle of TMSs (see Fig. S5 in the supplemental material). Second, helix-destabilizing residues are required for purified RseP to cleave a purified substrate under detergent-solubilized conditions, and the same peptide bond is cleaved as in vivo (38). Therefore, an intact membrane is not needed for cleavage site selection, and neither does it explain the need for helix-destabilizing residues. Subsequently, helix-destabilizing residues in the TMS of a model substrate were shown to stabilize the substrate-RseP complex, in which substrate residues near the cleavage site are in close proximity to the active site of the enzyme (61). These results by no means rule out a role of helix-destabilizing residues in facilitating substrate access to the RseP active site, although cleavage near the middle of the target TMS suggests lateral entry within the membrane rather than near its surface.
Helix-destabilizing residues in substrates of rhomboid IPs have also been proposed to facilitate substrate access to the active site near the membrane surface (62–64). These enzymes normally require helix-destabilizing residues in the TMS that gets cleaved (42, 62). Recently, studies of rhomboids reconstituted into proteoliposomes or expressed in cells indicated that helix-destabilizing residues in the target TMS of the substrate, as well as the dynamic properties of the enzyme in the membrane, play the primary role in substrate recognition (65).
Importantly, our study involved expression in E. coli or in sporulating B. subtilis, where SpoIVFB is embedded in a membrane and Pro-σK appears to associate peripherally with a membrane, and we found that substrate helix-destabilizing residues facilitate cleavage but are not essential. The unique dispensability of substrate helix-destabilizing residues in vivo could reflect the unique interaction of Pro-σK with the membrane compared with other IP substrates. It could also reflect a unique characteristic of the SpoIVFB subfamily of IMMPs, the CBS domain (32). We propose that binding of ATP to the CBS domain changes the conformation or oligomeric state of SpoIVFB, allowing access of the pro-sequence to the active site near the membrane surface (Fig. 8). This part of the model is based on findings that the CBS domain-containing C-terminal part of SpoIVFB interacts with Pro-σK(1-126)-FLAG2 and that ATP changes the interaction (13). Also, ATP binds to the C-terminal part of SpoIVFB and is required for purified SpoIVFB to cleave purified Pro-σK(1-126)–His6. The CBS domain of SpoIVFB may sense the energy status of the developing mother cell (13), which appears to feed the forespore small molecules through channels that form during engulfment, allowing the forespore to maintain its integrity and σG RNA polymerase to transcribe its regulon (66–69). Subsequent inactivation of the channels has been proposed to cause a rise in ATP in the mother cell that is sensed by the CBS domain of SpoIVFB, coupling cleavage of Pro-σK to channel inactivation (71).
Cleavage site selection.
We found that SpoIVFB prefers a small residue at P1 and a hydrophobic residue at P2′. Sequence comparisons of substrate orthologs imply that SpoIVFB orthologs have similar preferences (see Fig. S5 in the supplemental material). Recently, orthologs of SpoIVFB from Anabaena variabilis were shown to cleave B. subtilis Pro-σK(1-126)–His6 S20G with G at P1 and V at P2′ upon coexpression in E. coli (70), consistent with a preference for a small residue at P1 and a hydrophobic residue at P2′. Typically, such preferences reflect the existence of binding pockets near the active site of the protease for substrate residue side chains; however, in a model of SpoIVFB based on the available IMMP structure (58), we were unable to distinguish between several possible P1 and P2′ binding pockets.
Comparison of the preferences and tolerances of SpoIVFB with those of RseP and S2P revealed similar preferences for a medium-size hydrophobic or polar residue at P2′, but in contrast to SpoIVFB, the other two IMMPs tolerate F at P1 and P2′. RseP tolerates F simultaneously at P1 and P2′ when it cleaves LacY TMS 1 (38) (see Fig. S5 in the supplemental material). S2P appeared to tolerate F simultaneously at P1 and P1′, or singly at P2′, when it cleaves SREBP-2, although the cleavage site was not mapped (44). While RseP and S2P appeared to tolerate an aromatic residue at P1 or P2′, neither these IMMPs nor their orthologs appear to prefer an aromatic residue at these positions as determined on the basis of examining the residues at these positions in RseA and SREBP-2 (see Fig. S5 in the supplemental material) and examining the putative P1 and P2′ residues in substrate orthologs (see Fig. S6 and S7 in the supplemental material). Aromatic residues are not found at P1 or P2′. Rather, RseP appears to prefer A at P1 and V at P2′, consistent with the preferences of SpoIVFB, and S2P appears to prefer L at P1 and V, M, or T at P2′, suggesting a preference for a larger residue at P1 than SpoIVFB, although SpoIVFB tolerated L at P1 of Pro-σK(1-126)–His6 (Fig. 7A, lane 7). SpoIVFB also tolerated the polar residue T at P2′ of Pro-σK(1-126)–His6 (Fig. 4B, lane 7), although a polar residue was not found at the putative P2′ position of Pro-σK orthologs (see Fig. S4 in the supplemental material).
In terms of designing substitutions likely to block cleavage of Pro-σK orthologs, W at the putative P1 position (Fig. 6, lane 6, and Fig. 7A, lane 17) or P, N, or G at the P2′ position (Fig. 4B, lanes 9, 11, and 12) would appear to be a good choice.
The P1 and P2′ residues are not the only features of Pro-σK that influence cleavage by SpoIVFB. We discovered that multiresidue substitutions near the cleavage site in Pro-σK can affect the abundance and position of cleavage (Fig. 5 and 7B). Notably, changing VSYV to AAVA allowed substantial cleavage (Fig. 7B, lane 3) but introduced ambiguity in the cleavage site, and the most abundant cleavage product indicated that A was tolerated at P2′ (Fig. 5), whereas A is not well tolerated at P2′ in the context of the otherwise wild-type sequence (Fig. 4A, lane 19, and Fig. 6, lane 8). This implies that the sequence context influences cleavage site selection and cleavage efficiency rather than simply preferences for a small residue at P1 and a hydrophobic residue at P2′. If only the residues at P1 and P2′ were important, cleavage mainly after residue 19, placing A at P1 and V at P2′, would have been observed, but instead, this was a minor-abundance cleavage product (Fig. 5). We think that cleavage after residue 19 is not more abundant because residues N-terminal of F17 in Pro-σK also interact with SpoIVFB to influence cleavage. Several observations support this notion, but among the most convincing is that insertion of an A residue between F17 and V16 (i.e., changing VF to VAF) greatly impaired cleavage (Fig. 7B, lane 17) without altering the proportion of protein in the IF (see Fig. S8 in the supplemental material).
Interactions between SpoIVFB and residues N-terminal of F17 in Pro-σK might explain the effects of other substitutions that insert or delete a residue near the cleavage site in Pro-σK. For example, the main cleavage product of the variant with the SY-to-VAA substitution was unique in that cleavage occurred after residue 21. This one-residue shift allows A at P1 and V at P2′, favoring cleavage, but it adds one residue between F17 and the cleavage site, which may disfavor interaction of residues N-terminal of F17 with SpoIVFB, accounting for the low overall cleavage efficiency (Fig. 7B, lane 12). The variant with the SY-to-AAV substitution likewise causes low cleavage efficiency (Fig. 7B, lane 13), despite allowing cleavage after residue 20 with A at P1 and V at P2′ (Fig. 5). A possible explanation is suggested by the minor-abundance cleavage product, which indicates that this substrate can bind in an alternate register that allows cleavage after residue 21 with A at P1 and V at P2′. Such binding adds one residue between F17 and the cleavage site, which may disfavor interaction of residues N-terminal of F17 with SpoIVFB, resulting in an inefficient competing reaction that lowers the overall cleavage efficiency. In agreement, deleting F17 or V16 in combination with the SY-to-AAV substitution restored the normal cleavage efficiency (Fig. 7B, lanes 15 and 16) and the location of the cleavage was after residue 19 (minor) or 20 (major), with A at P1 and V at P2′ in both cases (Fig. 5). Taking these results together, it appears that SpoIVFB cleaves Pro-σK more readily after residue 20 than after residue 19 or 21, perhaps due to interactions with residues N-terminal of F17 and with P1 and P2′ near the cleavage site and a constraint on the number of residues in between (i.e., a “molecular ruler” mechanism). More work is needed to define features N-terminal of F17 in Pro-σK that are important for cleavage by SpoIVFB and to understand precisely how the cleavage site is determined.
We discovered one feature of Pro-σK C-terminal of P2′ in addition to the helix-destabilizing residues discussed above. We found that replacing K with A at P3′ of Pro-σK(1-126)–His6 greatly impaired cleavage in E. coli (Fig. 4A, lane 21) without changing the proportion of protein in the IF (see Fig. S8 in the supplemental material) and that it greatly impaired cleavage in sporulating B. subtilis (Fig. 6, lane 11). The positively charged K at P3′ could be replaced with negatively charged E or with a polar residue, and SpoIVFB cleaved normally (Fig. 4B, lanes 14, 16, and 17, and Fig. 5). Strikingly, the K23A substitution no longer impaired cleavage of Pro-σK(1-126)–His6 in E. coli when combined with a Q29K substitution (Fig. 7C, lane 5). This variant was cleaved with K at P9′, and other variants were cleaved with K at P4′ or P5′ (Fig. 5). It appears that SpoIVFB requires a charged or polar residue at P3′ or farther in the C-terminal direction; however, the three polar residues at P4′, P5′, and P9′ (i.e., N24, N25, and Q29) are insufficient with A at P3′ (i.e., the K23A variant). This cannot reflect a requirement for at least four polar or charged residues at P3′-P9′, since replacing NNAFPQP at P4′-P9′ with FAAFAQA reduced cleavage only slightly (Fig. 7C, lane 8). The ability of SpoIVFB to tolerate charge reversal or a polar residue at P3′, or movement of K from P3′ to P9′, suggests a highly flexible interaction between residues near the active site of SpoIVFB and residues C-terminal of P2′ in Pro-σK. This feature of the interaction between SpoIVFB and Pro-σK, as well as many other features of our model (Fig. 8), need to be explored with further experiments, including structural studies.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant GM43585 (to L.K.), by Michigan State University AgBioResearch, by National Science Foundation grant MCB0914691 (to R.Z.), and by the South Dakota Agricultural Experiment Station.
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00229-13.
REFERENCES
- 1. Brown MS, Ye J, Rawson RB, Goldstein JL. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391–398 [DOI] [PubMed] [Google Scholar]
- 2. Wolfe MS, Kopan R. 2004. Intramembrane proteolysis: theme and variations. Science 305:1119–1123 [DOI] [PubMed] [Google Scholar]
- 3. Freeman M. 2008. Rhomboid proteases and their biological functions. Annu. Rev. Genet. 42:191–210 [DOI] [PubMed] [Google Scholar]
- 4. Urban S. 2009. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 7:411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen G, Zhang X. 2010. New insights into S2P signaling cascades: regulation, variation, and conservation. Protein Sci. 19:2015–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urban S, Shi Y. 2008. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr. Opin. Struct. Biol. 18:432–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wolfe MS. 2009. Intramembrane proteolysis. Chem. Rev. 109:1599–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ha Y. 2009. Structure and mechanism of intramembrane protease. Semin. Cell Dev. Biol. 20:240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Urban S. 2010. Taking the plunge: integrating structural, enzymatic and computational insights into a unified model for membrane-immersed rhomboid proteolysis. Biochem. J. 425:501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolfe MS. 2010. Structure, mechanism and inhibition of gamma-secretase and presenilin-like proteases. Biol. Chem. 391:839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudner D, Fawcett P, Losick R. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. U. S. A. 96:14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu Y-TN, Kroos L. 2000. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 182:3305–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou R, Cusumano C, Sui D, Garavito RM, Kroos L. 2009. Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP. Proc. Natl. Acad. Sci. U. S. A. 106:16174–16179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. 1990. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell 62:239–250 [DOI] [PubMed] [Google Scholar]
- 15. Cutting S, Roels S, Losick R. 1991. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J. Mol. Biol. 221:1237–1256 [DOI] [PubMed] [Google Scholar]
- 16. Lu S, Halberg R, Kroos L. 1990. Processing of the mother-cell σ factor, σK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc. Natl. Acad. Sci. U. S. A. 87:9722–9726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stragier P, Kunkel B, Kroos L, Losick R. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243:507–512 [DOI] [PubMed] [Google Scholar]
- 18. Doan T, Marquis KA, Rudner DZ. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 55:1767–1781 [DOI] [PubMed] [Google Scholar]
- 19. Jiang X, Rubio A, Chiba S, Pogliano K. 2005. Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control sigma activity. Mol. Microbiol. 58:102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Resnekov O, Alper S, Losick R. 1996. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells 1:529–542 [DOI] [PubMed] [Google Scholar]
- 21. Rudner DZ, Losick R. 2002. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 16:1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Campo N, Rudner DZ. 2006. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol. Cell 23:25–35 [DOI] [PubMed] [Google Scholar]
- 23. Campo N, Rudner DZ. 2007. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor sigmaK in Bacillus subtilis. J. Bacteriol. 189:6021–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong TC, Cutting SM. 2003. SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-σK in Bacillus subtilis. Mol. Microbiol. 49:1425–1434 [DOI] [PubMed] [Google Scholar]
- 25. Zhou R, Kroos L. 2005. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-σK during Bacillus subtilis development. Mol. Microbiol. 58:835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kroos L, Kunkel B, Losick R. 1989. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science 243:526–529 [DOI] [PubMed] [Google Scholar]
- 27. Zhang B, Hofmeister A, Kroos L. 1998. The pro-sequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J. Bacteriol. 180:2434–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328. 10.1371/journal.pbio.0020328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu S, Cutting S, Kroos L. 1995. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor pro-σK in the absence of other sporulation gene products. J. Bacteriol. 177:1082–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou R, Kroos L. 2004. BofA protein inhibits intramembrane proteolysis of pro-σK in an intercompartmental signaling pathway during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U. S. A. 101:6385–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prince H, Zhou R, Kroos L. 2005. Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-σK. J. Bacteriol. 187:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kinch LN, Ginalski K, Grishin NV. 2006. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 15:84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ignoul S, Eggermont J. 2005. CBS domains: structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 289:C1369–C1378 [DOI] [PubMed] [Google Scholar]
- 34. Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. 2004. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Invest. 113:274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 16:2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kanehara K, Ito K, Akiyama Y. 2002. YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σE, RseA. Genes Dev. 16:2147–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ades SE. 2008. Regulation by destruction: design of the σE envelope stress response. Curr. Opin. Microbiol. 11:535–540 [DOI] [PubMed] [Google Scholar]
- 38. Akiyama Y, Kanehara K, Ito K. 2004. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 23:4434–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. 2000. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl. Acad. Sci. U. S. A. 97:5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355–1364 [DOI] [PubMed] [Google Scholar]
- 41. Rawson R, Zelenski N, Nijhawan D, Ye J, Sakai J, Hasan M, Chang T, Brown M, Goldstein J. 1997. Complementation cloning of SP2, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1:47–57 [DOI] [PubMed] [Google Scholar]
- 42. Urban S, Freeman M. 2003. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 11:1425–1434 [DOI] [PubMed] [Google Scholar]
- 43. Lemberg MK, Martoglio B. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735–744 [DOI] [PubMed] [Google Scholar]
- 44. Duncan E, Dave U, Sakai J, Goldstein J, Brown M. 1998. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 273:17801–17809 [DOI] [PubMed] [Google Scholar]
- 45. Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kroos L, Yu YT, Mills D, Ferguson-Miller S. 2002. Forespore signaling is necessary for pro-σK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest. J. Bacteriol. 184:5393–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kunkel B, Sandman K, Panzer S, Youngman P, Losick R. 1988. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J. Bacteriol. 170:3513–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England [Google Scholar]
- 49. Green D, Cutting S. 2000. Membrane topology of the Bacillus subtilis Pro-σK processing complex. J. Bacteriol. 182:278–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de Hoon MJ, Eichenberger P, Vitkup D. 2010. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 20:R735–R745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harry KH, Zhou R, Kroos L, Melville SB. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 191:2728–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jones SW, Paredes CJ, Tracy B, Cheng N, Sillers R, Senger RS, Papoutsakis ET. 2008. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 9:R114. 10.1186/gb-2008-9-7-r114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paredes CJ, Alsaker KV, Papoutsakis ET. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 54. Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. 1996. Sterol-regulated release of SREBP-2 from cell membrane requires two sequential cleavages, one within a transmembrane domain. Cell 85:1037–1048 [DOI] [PubMed] [Google Scholar]
- 55. Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124:587–599 [DOI] [PubMed] [Google Scholar]
- 56. Kanehara K, Akiyama Y, Ito K. 2001. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene 281:71–79 [DOI] [PubMed] [Google Scholar]
- 57. Zelenski N, Rawson R, Brown M, Goldstein J. 1999. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274:21973–21980 [DOI] [PubMed] [Google Scholar]
- 58. Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. 2007. Structure of a site-2 protease family intramembrane metalloprotease. Science 318:1608–1612 [DOI] [PubMed] [Google Scholar]
- 59. von Heijne G. 1992. Membrane protein structure prediction: hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 60. Johnson B, Dombroski A. 1997. The role of the pro-sequence of Bacillus subtilis σK in controlling activity in transcription initiation. J. Biol. Chem. 272:31029–31035 [DOI] [PubMed] [Google Scholar]
- 61. Koide K, Ito K, Akiyama Y. 2008. Substrate recognition and binding by RseP, an Escherichia coli intramembrane protease. J. Biol. Chem. 283:9562–9570 [DOI] [PubMed] [Google Scholar]
- 62. Akiyama Y, Maegawa S. 2007. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol. Microbiol. 64:1028–1037 [DOI] [PubMed] [Google Scholar]
- 63. Maegawa S, Koide K, Ito K, Akiyama Y. 2007. The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyses an extramembrane peptide bond of substrates. Mol. Microbiol. 64:435–447 [DOI] [PubMed] [Google Scholar]
- 64. Wang Y, Maegawa S, Akiyama Y, Ha Y. 2007. The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J. Mol. Biol. 374:1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moin SM, Urban S. 2012. Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. eLife 1:e00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Camp AH, Losick R. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69:402–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Camp AH, Losick R. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23:1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr, Rudner DZ. 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 5:e1000566. 10.1371/journal.pgen.1000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Meisner J, Wang X, Serrano M, Henriques AO, Moran CP., Jr 2008. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. U. S. A. 105:15100–15105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen K, Gu L, Xiang X, Lynch M, Zhou R. 2012. Identification and characterization of five intramembrane metalloproteases in Anabaena variabilis. J. Bacteriol. 194:6105–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kroos L, Akiyama Y. Biochemical and structural insights into intramembrane metalloprotease mechanisms. BBA Biomemb., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.