Abstract
Escherichia coli is used as a model organism for elucidation of menaquinone biosynthesis, for which a hydrolytic step from 1,4-dihydroxy-2-naphthoyl-coenzyme A (DHNA-CoA) to 1,4-dihydroxy-2-naphthoate is still unaccounted for. Recently, a hotdog fold thioesterase has been shown to catalyze this conversion in phylloquinone biosynthesis, suggesting that its closest homolog, YbgC in Escherichia coli, may be the DHNA-CoA thioesterase in menaquinone biosynthesis. However, this possibility is excluded by the involvement of YbgC in the Tol-Pal system and its complete lack of hydrolytic activity toward DHNA-CoA. To identify the hydrolytic enzyme, we have performed an activity-based screen of all nine Escherichia coli hotdog fold thioesterases and found that YdiI possesses a high level of hydrolytic activity toward DHNA-CoA, with high substrate specificity, and that another thioesterase, EntH, from siderophore biosynthesis exhibits a moderate, much lower DHNA-CoA thioesterase activity. Deletion of the ydiI gene from the bacterial genome results in a significant decrease in menaquinone production, which is little affected in ΔybgC and ΔentH mutants. These results support the notion that YdiI is the DHNA-CoA thioesterase involved in the biosynthesis of menaquinone in the model bacterium.
INTRODUCTION
Menaquinone is a lipophilic vitamin (K2) that plays important biological roles in humans and animals (1–4). In bacteria, it serves as a respiration electron transporter through reversible redox cycling between its hydroquinone and quinone forms (5). It is synthesized from chorismate either through a pathway involving o-succinylbenzoic acid (OSB) as a precursor (6) or through a newly discovered pathway using futalosine as an intermediate (7, 8). Due to its absence in humans and animals, the bacterial menaquinone biosynthesis has been an attractive target for development of novel antibiotics against pathogenic microbes such as Mycobacterium tuberculosis (9–11).
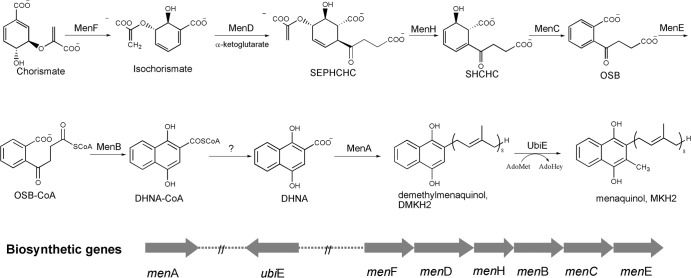
The facultative anaerobe Escherichia coli has been used as a model bacterium for elucidation of the classical menaquinone biosynthetic pathway (12). Early studies of this pathway focused on the genetics of the biosynthesis, leading to identification of eight biosynthetic genes located at three loci, namely, menA, ubiE, and the menFDHBCE cluster. In biochemical characterization of the gene products, MenD was found to synthesize (1R,2S,5S,6S)-2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxy-late (SEPHCHC) as a new intermediate (13, 14), rather than the previously proposed (1R,6R)-2-succinyl-6-hydroxyl-2, 4-cyclohexadiene-1-carboxylate (SHCHC). Subsequently, MenH was identified as the genuine SHCHC synthase, which catalyzes a stepwise elimination reaction with a serine-histidine-aspartate catalytic triad (15, 16). These findings have significantly reshaped the classical pathway as shown in Fig. 1, which requires nine enzymatic conversions but has only eight known enzymes. To fully account for the biosynthesis of menaquinone, a thioesterase is needed for the hydrolytic conversion from 1,4-dihydroxy-2-naphthoyl-coenzyme A (DHNA-CoA) to 1,4-dihydroxy-2-naphthoate (DHNA), which was once thought to be catalyzed by the preceding enzyme, MenB. This possibility, however, has been ruled out by the recent characterization of the enzyme as a dedicated DHNA-CoA synthase (17–19).
Fig 1.
Biosynthesis of menaquinone (vitamin K2) in Escherichia coli. SHCHC, (1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate; SEPHCHC, (1R,2S,5S,6S)-2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate; OSB, o-succinylbenzoate; DHNA, 1,4-dihydroxy-2-naphthoate, AdoMet: S-adenosylmethionine; AdoHcy, S-adenosylhomocysteine.
Through a homolog search for the known men genes, the classic menaquinone biosynthetic pathway has been found to operate in a large number of bacteria (20) as well as in plants, algae, and cyanobacteria in the biosynthesis of phylloquinone (21–23), which shares the same naphthenoid core structure as menaquinones and relays electrons in photosynthesis. Recently, a cyanobacterial hotdog fold thioesterase and its plant homologs have been shown to catalyze DHNA-CoA hydrolysis in phylloquinone biosynthesis (24, 25); the closest homolog to this thioesterase in E. coli is YbgC. However, it is still unknown whether YbgC is indeed involved in menaquinone biosynthesis in the bacteria.
In this study, we expressed YbgC to test its potential involvement in menaquinone biosynthesis but found that it has no detectable DHNA-CoA thioesterase activity. Activity-based screen of all eight other hotdog thioesterases in E. coli found that YdiI, whose function is unknown, and YbdB (or EntH), involved in biosynthesis of the siderophore enterobactin as a type II thioesterase, are active toward DHNA-CoA. Through analysis of menaquinone production in mutants with deletions of the corresponding genes, we have obtained evidence that YdiI is involved in biosynthesis of menaquinone in E. coli.
MATERIALS AND METHODS
Chemicals.
The following chemical reagents were purchased from Sigma: dithio-bisnitrobenzoic acid (DTNB), acetyl-CoA, palmitoyl-CoA, salicylic acid, 3,4-dihydroxybenzoic acid, 3,5-dihydroxybenzoic acid, 1-hydroxyl-2-naphthoic acid, 1,4-dihydroxy-2-naphthoic acid (DHNA), benzoic acid, 3-hydroxybenzoic acid, and 4-hydroxybenzoic acid. Biochemicals, including menaquinone-4, coenzyme A, ATP, isopropyl-β-d-thiogalactopyranoside (IPTG), buffers, and other salts, were also purchased from Sigma.
The following aryl-CoA substrates were prepared from the corresponding carboxylic acids and coenzyme A as described previously (15, 26, 27): 1,4-dihydroxy-2-naphthoyl-CoA (DHNA-CoA), 1-hydroxy-2-naphthoyl-CoA (1-HNA-CoA), salicylyl-CoA (SA-CoA), 3,4-dihydroxybenzoyl-CoA, 3,5-dihydroxybenzoyl-CoA, benzoyl-CoA, 3-hydroxybenzoyl-CoA, and 4-hydroxybenzoyl-CoA. All the synthesized thioester substrates were purified by reverse-phase high-pressure liquid chromatography (RP-HPLC). Reagents used in the synthesis, such as N-hydroxysuccinimide (NHS) and N,N′-dicyclohexylcarbodiimide (DCC), were also purchased from Sigma without further treatment.
Expression and purification of proteins.
Overexpression and purification of E. coli EntH or YbdB have been described previously (15, 26). The genes of other hotdog fold proteins in E. coli were amplified from the genomic DNA of Escherichia coli K-12 substrain MG1655 using primers listed in Table 1 and subcloned into pETM (Promega) for expression of the proteins with an N-terminal hexahistidine tag. The recombinant proteins were expressed in BL21(DE3) in Luria broth containing 0.2 mM IPTG at 18°C for 16 h and purified to >95% purity by a combination of metal-chelating chromatography and size exclusion chromatography. The purified proteins were quantified by a Coomassie blue protein assay kit (Pierce) using bovine serum albumin as the standard and stored in 50 mM Tris-HCl buffer (pH 7.8) containing 10% glycerol and 50 mM NaCl at −20°C until use. Protein concentrations from this Bradford assay are consistent with those determined by UV absorption of the proteins at 280 nm using extinction coefficients calculated with ProtParam at ExPASy (http://web.expasy.org/protparam/).
Table 1.
Oligodeoxynucleotide primers used in subcloning of the E. coli hotdog fold thioesterases
| Target gene | Forward primer | Reverse primer |
|---|---|---|
| ybgC | GGCGGATCCATGAATACAACGCTGTTTCGATGGCCGG | GCGGAATTCACTGCTTAAACTCCGCGACAATAGACTTG |
| ybdB (entH) | GCGCGGATCCATGATCCTGCACGCGCAGGCAAAACACG | CCGGAATTCTCAGAAACGCAAGATCTGCGCCAGACTTGC |
| ydiI | GGCGGATCCATGATATGGAAACGGAAAATCACCCTGG | GCGGAATTCTCACAAAATGGCGGTCGTCAATCGTGACG |
| ybaW | GGCGGATCCATGCAAACACAAATCAAAGTTCGTGGATATCATCTCG | CCGGAATTCTTACTTAACCATCTGCTCCAGCTTTTCGCGCAATTCCCC |
| yciA | GGCGGATCCATGTCTACAACACATAACGTCCCTCAGGGCG | GCGGAATTCTTACTCAACAGGTAAGGCGCGAGGTTTTCCTTCAGG |
| yigI | GGCGGATCCATGTCTGCCGTACTGACCGCTGAACAAG | GCGGAATTCTCAACCTACCATATAGGTGGCGGTGGCACTG |
| yiiD | GGCGGATCCATGAGCCAGCTTCCAGGGTTGTCACGGG | GCGGAATTCCTACTCTTCTTCGTTCCCGCCCTCTTCATACGG |
| tesB | GCGGAATTCATGAGTCAGGCGCTAAAAAATTTACTGACATTG | GCGAAGCTTTTAATTGTGATTACGCATCACCCCTTCCTGAACGG |
| paaI | GGCGGATCCATGAGTCATAAGGCCTGGCAAAATGCCCATGC | GCGGAATTCTCAGGCTTCTCCTGTAATGGTGCCGCCGATGCGG |
Thioesterase activity assay.
For aryl-CoA substrates, the thioesterase activity was determined on the basis of the difference in UV-visible light absorption between the substrate and the hydrolytic product as described previously (15, 27). All kinetic measurements were carried out in 200 mM sodium phosphate buffer (pH 7.0) in triplicate at 25 ± 0.5°C. The concentration of the enzyme was adjusted to ensure that consumption of the substrate was less than 5% within the first 3 min of the reaction, during which the initial velocity (v) was measured. In determination of the DHNA-CoA thioesterase activity, the reactions were initiated by adding 10 nM EntH or YdiI and monitored in real time for disappearance of DHNA-CoA by the decrease of the absorbance at 396 nm to determine the initial velocity at six different concentrations of the substrate ranging from 0.3 to 20 μM. When alternative aryl thioester substrates were used, their concentration was adjusted to ensure accurate determination of the Km value, while the enzyme concentration was lowered to 2 nM. The initial velocity was measured at six different concentrations in the ranges of 2.0 to 40 μM for 1-hydroxy-2-naphthoyl-CoA, 50 to 500 μM for salicylyl-CoA, and 10 to 250 μM for 3,4-dihydroxybenzoyl-CoA and 3,5-dihydroxybenzoyl-CoA. The kinetic parameters of maximum velocity (vmax) and Km were determined through the nonlinear regression method from the initial velocity data measured as a function of substrate concentration, using the Michaelis-Menten equation v = vmax [S]/([S] + Km), where [S] is the substrate concentration, v is the initial velocity, and Km is the Michaelis constant. The kcat value was calculated from the ratio of vmax and the concentration of the thioesterase monomer (all known hotdog fold thioesterases contain one active site per monomer).
A reported method was modified for the measurement of the thioesterase activity toward acetyl-CoA and palmitoyl-CoA (28). The newly released free thiol from the acyl-CoA substrates was monitored in real time at 412 nm in the presence of excess Ellman's reagent (dithiobis-nitrobenzoic acid [DTNB]). The assay was carried out in 200 mM phosphate buffer (pH 7.0) containing 0.8 to 6 mM acetyl-CoA and a 15 μM concentration of YdiI or other proteins.
Bacterial strains and determination of quinone levels in E. coli mutants.
The E. coli strains used in this study are listed in Table 2. For anaerobic culturing of the strains, 300 ml of Luria broth supplemented with 40 mM dimethyl sulfoxide was inoculated with 4.5 ml of overnight cell culture in a sealed 500-ml flask, which was degassed by vacuum and flushed with nitrogen for three cycles before inoculation. The cells were grown at 37°C and 180 rpm. For aerobic culturing, 300 ml of Luria broth was inoculated with 4.5 ml of overnight cell culture in a 1-liter flask with a cotton stopper. The cells were grown at 37°C and 250 rpm.
Table 2.
E. coli strains obtained from the Coli Genetic Stock Center
| Description | Strain | CGSC no. | Genotype |
|---|---|---|---|
| Parent strain | BW25113 | 7636 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| ΔmenB | JW2257-2 | 11787 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔmenB744::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| ΔmenH | JW2258-1 | 9820 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔyfbB745::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| ΔpaaI | JW1391-1 | 9230 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔpaaI760::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| ΔybgC | JW0726-1 | 11618 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) ΔybgC785::kan λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| ΔentH | JW0589-1 | 8705 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) ΔentH735::kan λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| ΔydiI | JW1676-1 | 11724 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔydiI761::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 |
| ΔyiiD | JW3859-1 | 10787 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 ΔyiiD750::kan Δ(rhaD-rhaB)568 hsdR514 |
| ΔyigI | JW5588-1 | 11477 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 ΔyigI765::kan Δ(rhaD-rhaB)568 hsdR514 |
A reported method was slightly modified for extraction and quantitation of quinones from the cells by reverse-phase high-pressure liquid chromatography (RP-HPLC) (29). The wet cells from a 300-ml culture were harvested at an A600 of 0.7∼1.2, weighed, and suspended in 15 ml of methanol. The cell suspension was then extracted four times, each time with 15 ml of petroleum ether. The extracts were combined and evaporated to dryness under reduced pressure, and the residue was finally dissolved in ethanol. The quinone contents were analyzed and quantified by HPLC on an XTerra RP18 analytical column (10-μm particle size, 19 by 150 mm) using water-methanol (1/99) as the mobile phase for isocratic elution at a flow rate of 1 ml/min. The quinones were separated in the order menaquinone-4 (4.0 min), ubiquinone-8 (UQ-8) (12.8 min), demethylmenaquinone-8 (DMK-8) (19.6 min), and menaquinone-8 (MK-8) (24.0 min), which was authenticated by UV spectroscopy. The elution was monitored at both 270 nm (for both naphthoquinone and ubiquinone) and 245 nm (for naphthoquinone) with a Waters model 2487 dual λ absorbance detector. Menaquinone-4 was used as an internal standard to quantify the content of the quinones assuming that menaquinone-8, demethylmenaquinone-8, and ubiquinone-8 have the same molar absorption coefficient as menaquinone-4 at 245 nm. The content of each quinone in a bacterial strain was averaged from at least three independent measurements.
RESULTS AND DISCUSSION
Assay of YbgC for DHNA-CoA thioesterase activity.
YbgC is one of the hotdog fold proteins in E. coli that shares the highest (28%) sequence identity with the hotdog fold thioesterase Slr0204, shown to be the DHNA-CoA thioesterase in phylloquinone biosynthesis of Synechocystis (24). To test the probability of YbgC as a DHNA-CoA thioesterase, the protein was readily expressed and purified to homogeneity as a soluble protein with an N-terminal hexahistidine tag (Fig. 2). However, it was found to possess no detectable hydrolytic activity toward DHNA-CoA or its aryl-CoA analogs, which included 1-hydroxy-2-naphthoyl-CoA (1-HNA-CoA), 3,4-dihydroxybenzoyl-CoA (3,4-DHB-CoA), and 3,5-dihydroxybenzoyl-CoA (3,5-DHB-CoA). It exhibited only negligible thioesterase activity toward salicylyl-CoA and acetyl-CoA, with a catalytic efficiency smaller than 6.0 M−1 · s−1. The recombinant YbgC with its N-terminal hexahistidine tag removed by thrombin was also found to be inactive toward DHNA-CoA or other aryl-CoA substrates.
Fig 2.
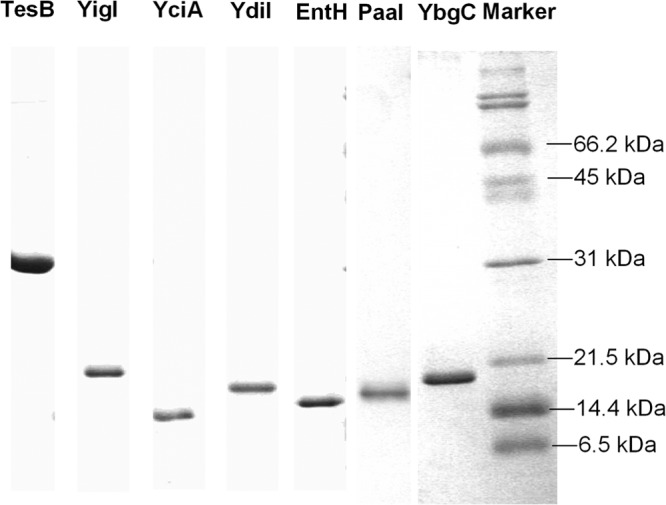
SDS-PAGE of the recombinant hotdog fold thioesterases.
The lack of thioesterase activity of the recombinant YbgC shown here is consistent with previous characterization of its activity in which the protein was found to be moderately active toward acyl-CoA substrates, with the highest catalytic efficiency of 40 M−1 · s−1 for propionyl-CoA (30). Actually, ybgC is part of the Tol-Pal operon involved in maintenance of cell envelope integrity and material transport through the periplasm. Although the exact role of this protein in the Tol-Pal system is still unknown, YbgC is known to affect the virulence and survival of Erwinia chrysanthemi (31). Due to its involvement in the Tol-Pal system and its lack of hydrolytic activity for DHNA-CoA, YbgC is very unlikely to be involved in the menaquinone biosynthesis in spite of its sequence homology to the committed DHNA-CoA thioesterase (Slr0204) in Synechocystis (24).
Assay of various hotdog fold proteins for DHNA-CoA thioesterase activity.
Thioesterases are a large family of proteins in the hotdog fold superfamily that encompasses a large number of proteins involved in diverse biological processes. Despite the conserved hotdog fold, these enzymes exhibit a high degree of sequence diversity, with only one conserved carboxylate residue, Asp or Glu, as the catalytic base or nucleophile (32). In consideration of this low sequence conservation, it is possible that another hotdog fold thioesterase is involved in menaquinone biosynthesis even though it is less homologous to Slr0204 of Synechocystis phylloquinone biosynthesis than YbgC.
There are all together 12 hotdog fold proteins encoded in the E. coli genome (33), of which FabA and FabZ are β-hydroxyacyl-acyl carrier protein (ACP) dehydratases involved in fatty acid biosynthesis (34) and MaoC is an enoyl-CoA dehydratase in polyhydroxyalkanoate biosynthesis (35). The remaining hotdog fold proteins, namely, PaaI, TesB, YbaW, YbdB (or EntH), YbgC, YciA, YdiI, YigI, and YiiD, are all thioesterases. Among them, YdiI, YigI, and YiiD do not have a known physiological role, whereas YciA and TesB are suggested to be involved in lipid metabolism (36–38) and YbgC is involved in the Tol-Pal system without a clearly defined role. The physiological functions of other thioesterases are more certain: YbdB (EntH) has been shown to serve as a type II thioesterase in enterobactin biosynthesis (26, 33), PaaI is involved in the phenylacetate degradation pathway (39, 40), and YbaW has recently been shown to be the third thioesterase (thioesterase III) in fatty acid β-oxidation (41). To test whether any of these thioesterases is involved in menaquinone biosynthesis, they were all overexpressed and purified to homogeneity as a hexahistidine-tagged protein at the N terminus and screened for DHNA-CoA hydrolytic activity.
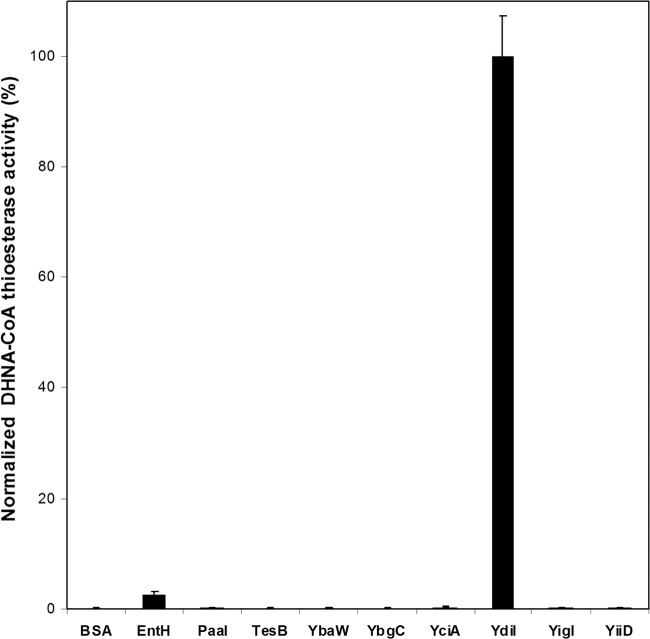
Using DHNA-CoA at 100 μM as the substrate, the specific thioesterase activities of the hotdog fold proteins were determined; they are shown in Fig. 3. The enzyme YdiI was shown to exhibit the highest level of DHNA-CoA thioesterase activity, and EntH was found to also be active in DHNA-CoA hydrolysis, with an activity significantly lower than that of YdiI. Compared to YdiI, all other E. coli hotdog fold thioesterases possess a negligible level of the hydrolytic activity toward DHNA-CoA. Noticeably, neither YdiI nor EntH shares a sequence identity higher than 20% with Slr0204 from Synechocystis or other hotdog fold thioesterases from E. coli. The screening results indeed show that sequence homology is not a good indicator of catalytic functions among hotdog fold thioesterases.
Fig 3.
Normalized specific DHNA-CoA thioesterase activity of hotdog fold thioesterases from E. coli. The activity of YdiI is set at 100%. The hydrolytic activities were assayed in 200 mM sodium phosphate buffer, pH 7.0, at 25°C, with the DHNA-CoA concentration set at 100.0 μM.
Characterization of the DHNA-CoA thioesterase activities of YdiI and EntH (YbdB).
Both YdiI and EntH (YbdB) were further characterized for their hydrolytic activities. The kinetic parameters of EntH in DHNA-CoA hydrolysis were determined to be 0.33 μM for the Km and 0.13 s−1 for the turnover number (kcat). Surprisingly, the resulting catalytic efficiency (kcat/Km = 4.0 × 105 M−1 · s−1) for this substrate is 1 order of magnitude higher than that for salicylyl-aryl carrier protein, the physiological substrate of this enzyme (26). Despite this high activity, EntH is unlikely to be involved in menaquinone biosynthesis, because both genetic and biochemical studies have provided unambiguous evidence for its involvement in the enterobactin biosynthesis as an editing enzyme for the nonribosomal enterobactin synthase (26, 33). In addition, EntH is also excluded from involvement in menaquinone biosynthesis by the fact that its gene is transcriptionally regulated by the Fur-Fe2+ complex responding to iron deficiency as a component of the enterobactin biosynthetic operon (42).
Consistent with the screening results, YdiI was found to be a much more active DHNA-CoA thioesterase, with a Km of 2.5 μM, a kcat of 6.2 s−1, and a second-ordered rate constant of 2.5 × 106 M−1 · s−1. It was also found to possess a broad substrate spectrum by exhibiting a lower but significant hydrolytic activity toward other aryl-CoA thioesters (Table 3). However, its hydrolytic activity for acyl-CoA thioesters such as acetyl-CoA and palmitoyl-CoA is negligible. These results show that YdiI is a highly efficient thioesterase specific for DHNA-CoA.
Table 3.
Steady-state kinetic parameters for YdiI at 25°Ca
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (μM−1 · s−1) |
|---|---|---|---|
| DHNA-CoA | 2.5 ± 0.3 | 6.2 ± 0.2 | 2.5 ± 0.3 |
| 1-Hydroxy-2-naphthoyl-CoA | 8.0 ± 0.1 | 14.8 ± 0.6 | 1.9 ± 0.2 |
| Salicylyl-CoA | 73 ± 16 | 93.0 ± 9.0 | 1.3 ± 0.3 |
| 3,4-Dihydroxybenzoyl-CoA | 26.9 ± 8.7 | 23.2 ± 3.4 | 0.86 ± 0.28 |
| 3,5-Dihydroxybenzoyl-CoA | 26.5 ± 2.1 | 12.6 ± 0.4 | 0.48 ± 0.04 |
| Acetyl-CoA | 1,559 ± 75 | (4.4 ± 0.2) × 10−3 | (2.8 ± 0.2) × 10−6 |
| Palmitoyl-CoA | ND | ND | ND |
Values are means ± standard deviations. ND, not detected.
YdiI has not been reported to be involved in any physiological processes. Its gene is located between the suf operon and a purR regulon in the E. coli genome and is in the same transcriptional direction as two adjacent genes, ydiH and ydiJ, which encode an uncharacterized protein and a predicted flavin-dependent iron-sulfur oxidoreductase related to biofilm architecture and cell mobility, respectively. No report is available to indicate that the ydiH, ydiI, and ydiJ genes form an operon with related physiological roles. Considering its high DHNA-CoA thioesterase activity, YdiI is likely involved in the biosynthesis of menaquinone in E. coli.
Effect of the ydiI knockout on production of menaquinone.
To test whether YdiI directly contributes to E. coli menaquinone biosynthesis, its gene was deleted from the bacterial genome to examine the effect on the production level of the naphthoquinones. For comparison, the genes for the hotdog fold thioesterases YbgC, EntH (YbdB), YiiD, YigI, and PaaI were also individually knocked out to examine their effects on menaquinone production. The other hotdog fold thioesterases YciA, TesB, YbaW, and MaoC were not included in this test because these proteins are involved in lipid metabolism and their knockout may indirectly affect the biosynthesis of the polyisoprenyl side chain of menaquinone. As positive controls, E. coli mutants without the essential menB gene and the nonessential menH gene of the menaquinone biosynthetic pathway were also included in the test. The knockout mutants were obtained from the Coli Genetic Stock Center (CGSC) along with the parent strain, BW25113 (Table 2). In addition to the targeted gene deletion, all the mutants and the parent strain contain the following genotype: Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ − rph-1 Δ(rhaD-rhaB)568 hsdR514.
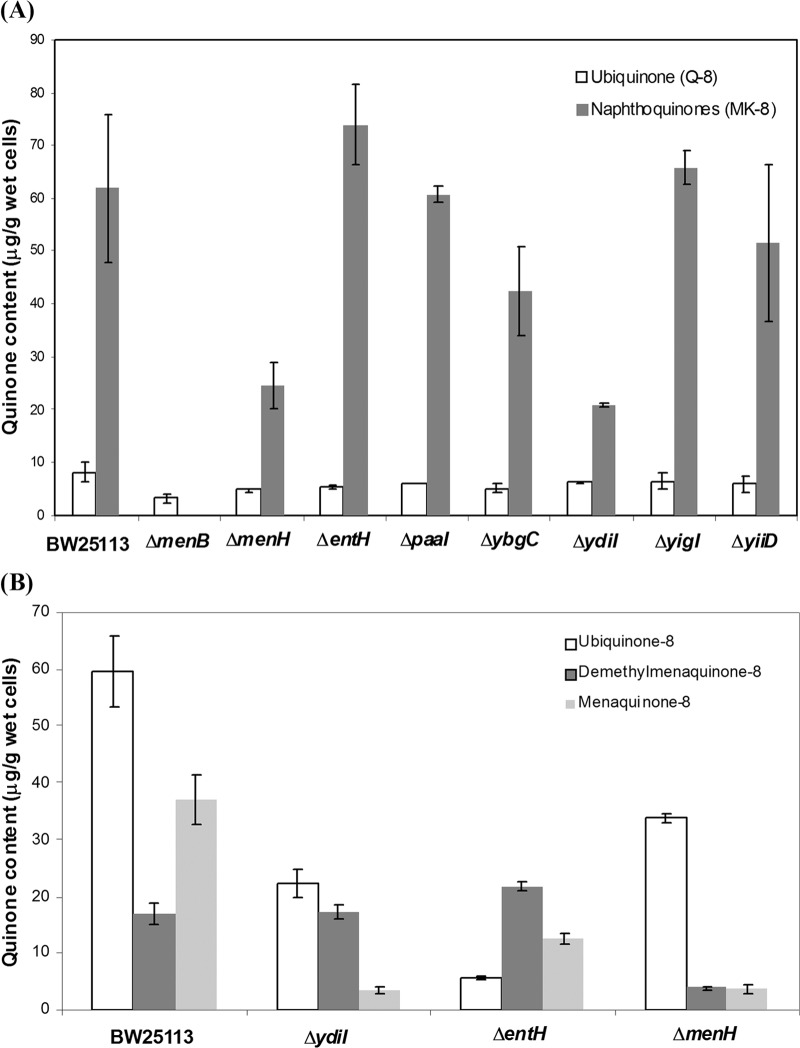
E. coli predominantly uses ubiquinone-8 (UQ-8) in respiration under aerobic conditions (6, 12) but produces menaquinone-8 (MK-8) and demethylmenaquinone-8 (DMK-8) as an obligatory electron carrier with fumarate, dimethylsufoxide, or trimethylamine N-oxide as the electron acceptor under anaerobic conditions (43, 44). To examine the mutational effects on menaquinone biosynthesis, the mutants along with the parent strain were grown under anaerobic conditions in the presence of dimethyl sulfoxide for compositional analysis of the quinone pool. Under these conditions, the production of DMK-8 is negligible. The results for the production of MK-8 and UQ-8 are shown in Fig. 4A. As expected, the production level of UQ-8 was little affected by deletion of the investigated genes, and deletion of the essential menB gene in menaquinone biosynthesis completely eliminated the production of both MK-8 and DMK-8.
Fig 4.
Contents of ubiquinone and naphthoquinones in the deletion mutants of menaquinone biosynthetic genes or hotdog fold thioesterase genes under anaerobic (A) and aerobic (B) conditions. BW25113 is the parent strain of the deletion mutants. Note that the amount of both naphthoquinones and ubiquinone is arbitrary, assuming that all these quinones are the same as menaquinone-4 in molar absorption coefficient at 245 nm.
The parent strain, BW25113, and most of the hotdog fold thioesterase deletion mutants, including the ΔyiiD, ΔyigI, ΔybgC, ΔentH, and ΔpaaI mutants, produce similar levels of MK-8 (Fig. 4A). This result clearly indicates that the deleted genes are not directly involved in the menaquinone biosynthesis, consistent with the known physiological functions of EntH, YbgC, and PaaI. In contrast, the ΔydiI mutant produces 67% less MK-8 than the parent strain, similar to the ΔmenH mutant with a defective menaquinone biosynthetic pathway. This similarity in naphthoquinone decrease strongly supports that YdiI is directly involved in the biosynthesis of menaquinone like MenH, as suggested by its high DHNA-CoA thioesterase activity. The residual naphthoquinone produced by both mutants likely results from the partial complementation of the gene functions by factors of the intracellular environment. In the ΔmenH mutant, the MenH substrate SEPHCHC is known to undergo spontaneous 1,4 elimination of pyruvate to form the SHCHC product and therefore to be capable of partial complementation of the MenH activity (13). In the ΔydiI mutant, the DHNA-CoA thioesterase activity may be partially complemented by slow spontaneous hydrolysis of DHNA-CoA due to its labile thioester function or by hydrolysis of the thioester by the thioesterases or hydrolases of other pathways, which are generally known to have broad substrate specificities as exemplified by YdiI and EntH. The inability of these cellular factors to fully complement the activity of MenH or YdiI is actually a good indicator of their direct involvement in menaquinone biosynthesis.
The parent strain, BW25113, and three deletion mutants were also analyzed for their production of the quinones under aerobic conditions (Fig. 4B). The production of both DMK-8 and MK-8 is much lower in the ΔmenH mutant than in the bacterial strains with an intact menaquinone biosynthetic pathway, including BW25113 and the ΔentH mutant. The ΔydiI mutant generates a similarly low level of MK-8 as the ΔmenH mutant, but its production of DMK-8 is similar to that of the parent strain. Overall, both the ΔmenH and ΔydiI mutants produce a significantly lower level of naphthoquinones than do other bacterial strains, consistent with the fact that they contain a defective menaquinone biosynthetic gene. Noticeably, the chosen bacterial strains produce a highly variable level of ubiquinone without a clear correlation to their genotypes.
Conservation of YdiI in menaquinone biosynthesis.
Results from the thioesterase screen, in vitro biochemical characterization, and in vivo analysis of naphthoquinone production converge to indicate that YdiI is responsible for the hydrolysis of DHNA-CoA in E. coli menaquinone biosynthesis, similar to the hotdog fold DHNA-CoA thioesterase Slr0204 in Synechocystis (24). However, these two proteins share very low sequence identity (6%) in spite of their similar structures. The closest E. coli homolog of Slr0204, YbgC, is excluded from involvement in the biosynthetic pathway, providing an interesting example in which proteins with identical physiological function share a lower level of sequence homology than two functionally irrelevant proteins. This weak correlation of sequence homology with protein function may be characteristic of members in the hotdog fold superfamily, which are well known to share the same structural fold with low sequence homology (32). Another example of this weak relationship is that EntH and YdiI share a high level of sequence identity (59%) with similar catalytic profiles but otherwise perform completely different physiological roles.
A large number of YdiI homologs with a sequence identity greater than 50% are found in a diverse array of microorganisms through a BLAST search of GenBank. Examination of the sequences found that all of these homologs contain the conserved catalytic base or nucleophile identified in the structures of YdiI (PDB code: 1VH5) (45), EntH (PDB code: 1VH9) (45), and the Arthrobacter 4-hydroxybenzoyl-CoA thioesterase (PDB code:1Q4T) (46). Due to the high sequence identity, a large proportion of these homologs, particularly those with a sequence identity of 70% or higher, are believed to be the DHNA-CoA thioesterase in menaquinone or phylloquinone biosynthesis. However, as noted previously, the correlation between sequence homology and biological function is fragile for hotdog fold thioesterases. Further investigation is needed to determine how many of these YdiI homologs are actually involved in biosynthesis of the naphthoquinones.
In summary, we have identified YdiI as the DHNA-CoA thioesterase in menaquinone biosynthesis, which is designated MenI. This leads to complete elucidation of the model primary biosynthetic pathway in Escherichia coli. It will also greatly facilitate identification of the YdiI orthologs in other microorganisms, particularly the menaquinone-dependent pathogenic bacteria. In this regard, complete elucidation of the biosynthetic pathway will also facilitate development of novel antibiotics targeting the biosynthetic pathway.
ACKNOWLEDGMENTS
This work was supported by GRF601209 from the Research Grants Council and the Research Infrastructure Fund FSGRF12SC06 from the University Grants Committee of the Hong Kong Special Administrative Region.
Footnotes
Published ahead of print 5 April 2013
REFERENCES
- 1. Unden G, Dunnwald P. 11 March 2008, posting date Chapter 3.2.2, The aerobic and anaerobic respiratory chain of Escherichia coli and Salmonella enterica: enzymes and energetics. In Böck A, Curtiss R, III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, Ussery D. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. 10.1128/ecosal.3.2.2 [DOI] [PubMed] [Google Scholar]
- 2. Suttie JW. 2009. Vitamin K in health and disease. CRC Press, Boca Raton, FL [Google Scholar]
- 3. Nakagawa K, Hirota Y, Sawada N, Yuge N, Watanabe M, Uchino Y, Okuda N, Shimomura Y, Suhara Y, Okano T. 2010. Identification of UBIAD1 as a novel human menaquinone-4 biosynthetic enzyme. Nature 468:117–121 [DOI] [PubMed] [Google Scholar]
- 4. Vos M, Esposito G, Edirisinghe JN, Vilain S, Haddad DM, Slabbaert JR, Van Meensel S, Schaap O, De Strooper B, Meganathan R, Morais VA, Verstreken P. 2012. Vitamin K2 is a mitochondrial electron carrier that rescues pink1 deficiency. Science 336:1306–1310 [DOI] [PubMed] [Google Scholar]
- 5. Wallace BJ, Young IG. 1977. Role of quinones in electron transport to oxygen and nitrate in Escherichia coli. Studies with a ubiA− menA− double quinone mutant. Biochim. Biophys. Acta 461:84–100 [DOI] [PubMed] [Google Scholar]
- 6. Meganathan R. 2010. Menaquinone/ubiquinone biosynthesis and enzymology, p 411–444 In Mander L, Liu H-W. (ed), Comprehensive natural products II: chemistry and biology, vol 7 Elsevier Science, Oxford, United Kingdom [Google Scholar]
- 7. Seto H, Jinnai Y, Hiratsuka T, Fukawa M, Furihata K, Itoh N, Dairi T. 2008. Studies on a new biosynthetic pathway for menaquinone. J. Am. Chem. Soc. 130:5614–5615 [DOI] [PubMed] [Google Scholar]
- 8. Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, Dairi T. 2008. An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321:1670–1673 [DOI] [PubMed] [Google Scholar]
- 9. Kurosu M, Narayanasamy P, Biswas K, Dhiman R, Crick DC. 2007. Discovery of 1,4-dihydroxy-2-naphthoate prenyltransferase inhibitors: new drug leads for multidrug-resistant gram-positive pathogens. J. Med. Chem. 50:3973–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dhiman RK, Mahapatra S, Slayden RA, Boyne ME, Lenaerts A, Hinshaw JC, Angala SK, Chatterjee D, Biswas K, Narayanasamy P, Kurosu M, Crick DC. 2009. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol. Microbiol. 72:85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu X, Zhou R, Sharma I, Li X, Kumar G, Swaminathan S, Tonge PJ, Tan DS. 2012. Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. Chembiochem 13:129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meganathan R, Kwon O. 23 December 2009, posting date Chapter 3.6.3.3, Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In Böck A, Curtiss R, III, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, Ussery D. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang M, Cao Y, Guo ZF, Chen M, Chen X, Guo Z. 2007. Menaquinone biosynthesis in Escherichia coli: identification of 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate (SEPHCHC) as a novel intermediate and reevaluation of MenD activity. Biochemistry 46:10979–10989 [DOI] [PubMed] [Google Scholar]
- 14. Jiang M, Chen M, Cao Y, Yang Y, Sze KH, Chen X, Guo Z. 2007. Determination of the stereochemistry of 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylic acid, a key intermediate in menaquinone biosynthesis. Org. Lett. 9:4765–4767 [DOI] [PubMed] [Google Scholar]
- 15. Jiang M, Chen X, Guo ZF, Cao Y, Chen M, Guo Z. 2008. Identification and characterization of (1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase in the menaquinone biosynthesis of Escherichia coli. Biochemistry 47:3426–3434 [DOI] [PubMed] [Google Scholar]
- 16. Jiang M, Chen X, Wu XH, Chen M, Wu Y, Guo Z. 2009. Catalytic mechanism of SHCHC synthase in the menaquinone biosynthesis of Escherichia coli: identification and mutational analysis of the active site residues. Biochemistry 48:6921–6931 [DOI] [PubMed] [Google Scholar]
- 17. Truglio JJ, Theis K, Feng Y, Gajda R, Machutta C, Tonge PJ, Kisker C. 2003. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J. Biol. Chem. 278:42352–42360 [DOI] [PubMed] [Google Scholar]
- 18. Jiang M, Chen M, Guo ZF, Guo Z. 2010. A bicarbonate cofactor modulates 1,4-dihydroxy-2-naphthoyl coenzyme A synthase in menaquinone biosynthesis of Escherichia coli. J. Biol. Chem. 285:30159–30169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song H, Guo Z. 2012. Characterization of the 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in the phylloquinone biosynthesis of Synechocystis sp. PCC 6803. Sci. China B 55:98–105 [Google Scholar]
- 20. Glasner ME, Fayazmanesh N, Chiang RA, Sakai A, Jacobson MP, Gerlt JA, Babbitt PC. 2006. Evolution of structure and function in the o-succinylbenzoate synthase/N-acylamino acid racemase family of the enolase superfamily. J. Mol. Biol. 360:228–250 [DOI] [PubMed] [Google Scholar]
- 21. Lefebvre-Legendre L, Rappaport F, Finazzi G, Ceol M, Grivet C, Hopfgartner G, Rochaix JD. 2007. Loss of phylloquinone in Chlamydomonas affects plastoquinone pool size and photosystem II synthesis. J. Biol. Chem. 282:13250–13263 [DOI] [PubMed] [Google Scholar]
- 22. Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux JP. 2008. Characterization and biological function of the isochorismate synthase 2 gene of Arabidopsis. Plant Physiol. 147:1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gross J, Meurer J, Bhattacharya D. 2008. Evidence of a chimeric genome in the cyanobacterial ancestor of plastids. BMC Evol. Biol. 8:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Widhalm JR, van Oostende C, Furt F, Basset GJC. 2009. A dedicated thioesterase of the hotdog-fold family is required for the biosynthesis of the naphthoquinone ring of vitamin K1. Proc. Natl. Acad. Sci. U. S. A. 106:5599–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Widhalm JR, Ducluzeau AL, Buller NE, Elowsky CG, Olsen LJ, Basset GJC. 2012. Phylloquinone (vitamin K1) biosynthesis in plants: two peroxisomal thioesterases of lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-CoA. Plant J. 71:205–215 [DOI] [PubMed] [Google Scholar]
- 26. Guo ZF, Sun Y, Zheng S, Guo Z. 2009. Preferential hydrolysis of aberrant precursors suggests an active proofreading mechanism for the type II thioesterase in Escherichia coli enterobactin biosynthesis. Biochemistry 48:1712–1722 [DOI] [PubMed] [Google Scholar]
- 27. Chen M, Jiang M, Sun Y, Guo ZF, Guo Z. 2011. Stabilization of the second oxyanion intermediate by 1,4-dihydroxy-2-naphthoyl coenzyme A synthase of the menaquinone pathway: spectroscopic evidence of the involvement of a conserved aspartic acid. Biochemistry 50:5893–5904 [DOI] [PubMed] [Google Scholar]
- 28. Zhou Y, Guo XC, Yi T, Yoshimoto T, Pei D. 2000. Two continuous spectrophotometric assays for methionine aminopeptidase. Anal. Biochem. 280:159–165 [DOI] [PubMed] [Google Scholar]
- 29. Bekker M, Kramer G, Hartog AF, Wagner MJ, de Koster CG, Hellingwerf KJ, de Mattos MJ. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153:1974–1980 [DOI] [PubMed] [Google Scholar]
- 30. Zhuang Z, Song F, Martin BM, Dunaway-Mariano D. 2002. The YbgC protein encoded by the ybgC gene of the tol-pal gene cluster of Haemophilus influenzae catalyzes acyl-coenzyme A thioester hydrolysis. FEBS Lett. 516:161–163 [DOI] [PubMed] [Google Scholar]
- 31. Dubuisson JF, Vianney A, Hugouvieux-Cotte-Pattat N, Lazzaroni JC. 2005. Tol-Pal proteins are critical cell envelope components of Erwinia chrysanthemi affecting cell morphology and virulence. Microbiology 151:3337–3347 [DOI] [PubMed] [Google Scholar]
- 32. Dillon SC, Bateman A. 2004. The hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics 5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leduc D, Battesti A, Bouveret E. 2007. The hotdog thioesterase EntH (YbdB) plays a role in vivo in optimal enterobactin biosynthesis by interacting with the ArCP domain of EntB. J. Bacteriol. 189:7112–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heath RJ, Rock CO. 1996. Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein: dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271:27795–27801 [DOI] [PubMed] [Google Scholar]
- 35. Park SJ, Lee SY. 2003. Identification and characterization of a new enoyl coenzyme A hydratase involved in biosynthesis of medium-chain-length polyhydroxyalkanoates in recombinant Escherichia coli. J. Bacteriol. 185:5391–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naggert J, Narasimhang ML, DeVeauxg L, Chog H, Randhawa ZI, Cronan JE, Jr, Greenll BN, Smith S. 1991. Cloning, sequencing, and characterization of Escherichia coli thioesterase II. J. Biol. Chem. 266:11044–11050 [PubMed] [Google Scholar]
- 37. Narasimhan ML, Lampi JL, Cronan JE., Jr 1986. Genetic and biochemical characterization of an Escherichia coli K-12 mutant deficient in acyl-coenzyme A thioesterase II. J. Bacteriol. 165:911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D. 2008. Divergence of function in the hot dog fold enzyme superfamily: the bacterial thioesterase YciA. Biochemistry 47:2789–2796 [DOI] [PubMed] [Google Scholar]
- 39. Kunishima N, Asada Y, Sugahara M, Ishijima J, Nodake Y, Sugahara M, Miyano M, Kuramitsu S, Yokoyama S, Sugahara M. 2005. A novel induced-fit reaction mechanism of asymmetric hot dog thioesterase PaaI. J. Mol. Biol. 352:212–228 [DOI] [PubMed] [Google Scholar]
- 40. Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD. 2006. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J. Biol. Chem. 281:11028–11038 [DOI] [PubMed] [Google Scholar]
- 41. Nie L, Ren Y, Schulz H. 2008. Identification and characterization of Escherichia coli thioesterase III that functions in fatty acid beta-oxidation. Biochemistry 47:7744–7751 [DOI] [PubMed] [Google Scholar]
- 42. Crosa JH, Walsh CT. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wissenbach U, Kroger A, Unden G. 1990. The specific functions of menaquinone and demethylmenaquinone in anaerobic respiration with fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate by Escherichia coli. Arch. Microbiol. 154:60–66 [DOI] [PubMed] [Google Scholar]
- 44. Wissenbach U, Ternes D, Unden G. 1992. An Escherichia coli mutant containing only demethylmenaquinone, but no menaquinone: effects on fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate respiration. Arch. Microbiol. 158:68–73 [DOI] [PubMed] [Google Scholar]
- 45. Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJ. 2005. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60:787–796 [DOI] [PubMed] [Google Scholar]
- 46. Thoden JB, Zhuang Z, Dunaway-Mariano D, Holden HM. 2003. The structure of 4-hydroxybenzoyl-CoA thioesterase from Arthrobacter sp. strain SU. J. Biol. Chem. 278:43709–43716 [DOI] [PubMed] [Google Scholar]