Abstract
Multiple sequencing of genomes belonging to a bacterial species allows one to analyze and compare statistics and dynamics of the gene complements of species, their pan-genomes. Here, we analyzed multiple genomes of Escherichia coli, Shigella spp., and Salmonella enterica. We demonstrate that the distribution of the number of genomes harboring a gene is well approximated by a sum of two power functions, describing frequent genes (present in many strains) and rare genes (present in few strains). The virtual absence of Shigella-specific genes not present in E. coli genomes confirms previous observations that Shigella is not an independent genus. While the pan-genome size is increasing with each new strain, the number of genes present in a fixed fraction of strains stabilizes quickly. For instance, slightly fewer than 4,000 genes are present in at least half of any group of E. coli genomes. Comparison of S. enterica and E. coli pan-genomes revealed the existence of a common periphery, that is, genes present in some but not all strains of both species. Analysis of phylogenetic trees demonstrates that rare genes from the periphery likely evolve under horizontal transfer, whereas frequent periphery genes may have been inherited from the periphery genome of the common ancestor.
INTRODUCTION
Analysis of sequenced bacterial genomes shows broad variability of their size and content, even for closely related strains. For instance, only 39% of the combined, nonredundant set of proteins were common for three initially sequenced Escherichia coli strains (1). To characterize a bacterial species genome based on genomic sequences of different strains, a pan-genome approach was suggested (2). The pan-genome is the total complement of genes from all sequenced strains of the same species (2), genus (3), or a larger group (4). The pan-genome consists of three parts: the universal genome with genes common for all strains, the unique genome with strain-specific genes (known as ORFans), and the periphery (genes that are present in a subset of strains) (4).
In most studied bacterial species, the gene content of strains varies widely, and each additional sequenced strain adds new genes to the pan-genome. The E. coli pan-genome is open, i.e., far from saturation (5). An example of a closed pan-genome is that of Bacillus anthracis. It does not increase after taking into account the first four genomes (2). This agrees with the observation that B. anthracis is a recently emerged species that has a low genome variability primarily limited to virulence plasmids (6, 7). The pan-genome of Salmonella enterica can be tentatively defined as closed, because the number of new genes brought by each particular strain genome is not large relative to the average genome size (8, 9).
E. coli and S. enterica are close relatives. The last common ancestor of E. coli and S. enterica existed about 100 million years ago, as estimated by using the protein clock model (10). Multilocus sequence typing data suggested that distinct Shigella spp. (S. boydii, S. dysenteriae, S. flexneri, and S. sonnei) are pathogenic strains of E. coli that originated independently in at least seven waves (35,000 to 270,000 years ago) and evolved convergently to acquire the “Shigella phenotype” (11). For comparison, the last common ancestor of E. coli K-12 and E. coli O157:H7 lived about 4.5 million years ago (12). All Shigella spp. have similar phenotypic features (they are nonmotile, obligate pathogens and have similar metabolisms) that have been the basis for initially describing them as a separate genus (13), and for pragmatic reasons these organisms remain classified in separate genera (14).
The use of extrapolation to estimate the pan-genome size for bacterial groups (5, 15) should be taken with a fair degree of caution. Strain sampling is uneven because pathogenic strains provoke stronger interest of the scientific community, and their genomes are overrepresented for some bacterial groups (16). The other reason is a large fraction of uncultivated strains in many bacterial groups (17, 18), as under modern sequencing technologies, the genomes of uncultivated strains are still underrepresented in the genome databases. The same applies to the estimates of the core genome size, but in this case the problem is less drastic as the core genome is almost saturated for many bacterial groups. Here, we suggest an approach that provides a robust estimate of the pan-genome composition.
Although pan-genome analysis was performed for several bacterial species, only few very recent studies consider more than a single prokaryote group (see, for example, reference 19). Here, we compare the pan-genomes of two groups, E. coli plus Shigella spp. and S. enterica, demonstrate the existence of the common periphery in these pan-genomes, and discuss its possible evolutionary history.
MATERIALS AND METHODS
Strain genome sequences.
Available (as of 1 September 2009) complete genome sequences of 48 strains of E. coli, Shigella spp., and S. enterica were used for the construction of ortholog groups (OGs). The genomes were taken from the NCBI Genome database (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) and are listed in Table S1 in the supplemental material.
Construction of OGs.
OGs were constructed as follows. Bidirectional best hits (BBHs) were constructed for each pair of strains using BLASTP (20). BLASTP results with identities of <50% or coverage of shorter sequence <67% were ignored. At the next step, if two paralogs were more similar to each other than to either BBH partner, both paralogs were added to the OG. Then, maximal connected components were constructed. This was done using ad hoc software based on the Relational Database Management System (RDBMS) ORACLE Express Edition.
Modification of OGs.
Ortholog tables were modified to remove genes of viral, insertion sequence, transposon origin, and hypothetical genes. If an OG contained a gene from a phage- or mobile element-related GO category, or annotated with terms associated with phage or mobile elements, the group was filtered out. In addition, we removed groups where all genes had the standard annotation “hypothetical protein” and none was assigned a GO function.
To account for possible misannotations (21), we used TBLASTN. Sequences with ≥50% identity (amino acids) with coverage ≥67% were annotated as genes encoding hypothetical protein. Start/stop codon existence was not checked. Hence, recent pseudogenes were accepted as genes.
GOstat analysis.
Each OG was assigned with GO terms linked to all constituent genes. Functional analysis was performed using the GOstat web server (22). Both overrepresented functional categories (GOterms) with an E value >0.1 and underrepresented ones were ignored. Manual curation was applied to eliminate nonunique terms and select the most illustrative categories (for instance, if a “cobalamin biosynthetic process” term was found, we removed the more general “cobalamin metabolic process” one).
Trees.
Phylogenetic trees were constructed for all modified OGs. Sequences found by tblastn were ignored. Amino acid sequences were aligned by CLUSTAL W (23). Trees were constructed using the PhyML software (tree topology, branch length, and substitution rate parameters were optimized; the tree topology search operation option was the best of NNI and the SPR search) (24) and rooted at midpoint using the BADASP software (25). Trees and unrooted trees were analyzed by using an ad hoc BioPerl script.
RESULTS AND DISCUSSION
Descriptive statistics of OGs.
Using the procedures described above, genes of E. coli, Shigella spp., and S. enterica (all 48 strains are listed in Table S1 in the supplemental material) were clustered in 9,916 OGs. For 6,448 genes (singletons), no orthologs were found.
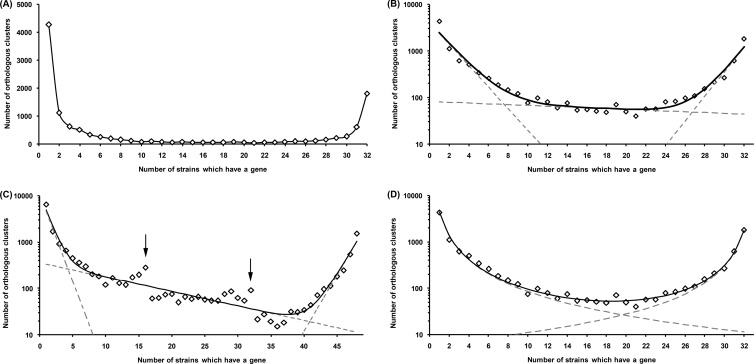
The distribution of the OGs by the number of strains in which they are present has a well-known U-shape form (Fig. 1). The periphery genes tend to be rare (i.e., present in two to three strains) or almost universal (i.e., absent in few strains). This holds true for the E. coli-plus-Shigella distribution (Fig. 1A) and for separate distributions of E. coli, Shigella spp., or S. enterica (see Fig. S1 in the supplemental material). As described previously (26), one can approximate this distribution by calculating the sum of three exponents corresponding to the unique genome, the periphery, and the universal genome: y(x) = e−0.53x+8.3 + e−0.02x+4.41 + e0.6x−12.27 (Fig. 1B).
Fig 1.
Distribution of OGs by the number of strains in which they are present. (A) 32 E. coli sensu lato strains; (B) 32 E. coli sensu lato strains, approximated by the sum of three exponents. Dashed exponential lines (for unique genes, the periphery, and the universal genome, respectively) provide the decomposition of the general trend line for the distribution [solid line; y(x) = e−0.53x+8.3 + e−0.02x+4.41 + e0.6x−12.27]. The total squared error is 1.48. (C) 48 strains, with notation as in panel B [y(x) = e−0.86x+9.28 + e−0.07x+5.88 + e0.55x−19.81]. Peaks marked with arrows contain OGs comprising genes specific for S. enterica and E. coli sensu lato. (D) 25 E. coli and 7 Shigella strains, showing decomposition to the sum of two power law functions [solid line; y(x) = 4,400x−1.7 + 1,746 × (33 − x)−1.62]. The total squared error is 0.59.
When the same procedure is applied to all 48 genomes, additional peaks appear in the middle (Fig. 1C). These peaks are generated by genes restricted to S. enterica (the peak at 16 genomes) or E. coli plus Shigella spp. (the peak at 32 genomes). This visually demonstrates the existence of gene pools restricted to specific groups and shows the inhomogeneity of the sample (see, for example, reference 19). In contrast, the absence of peaks for 7 or 25 genomes indicates that E. coli and Shigella spp. have a common pan-genome with virtually no genes restricted to either E. coli or Shigella spp. (also see below). Hence, we will hereafter use “E. coli sensu lato” to refer to Escherichia coli and Shigella clones of E. coli (see, for example, reference 11).
An alternative approximation approach is to describe the U-shape distribution as a sum of two power law functions: y(x) = 4,400x−1.7 + 1,746(33 − x)−1.62 (Fig. 1D). Here, the first term describes the genes present in a few strains (almost unique), and the second term reflects the distribution of genes present in most genes (almost universal).
To compare the two approximations, we applied a chi-squared statistics. For the sum of two power law functions, the χ2 value equals 128, while for the sum of three exponents, it equals 1,909. Since at the same time the number of free parameters in the power law approximation is lower, the sum of two power functions seems to be a better way to describe the distribution of OGs, although the fit is still not perfect.
Since we were mainly interested in functional differences between strains, we excluded from further analysis hypothetical and virus-related genes and added (probably) misannotated genes or recent pseudogenes, found using tblastn (20). The details are given in Materials and Methods. All subsequent analyses were performed with this modified variant of OGs.
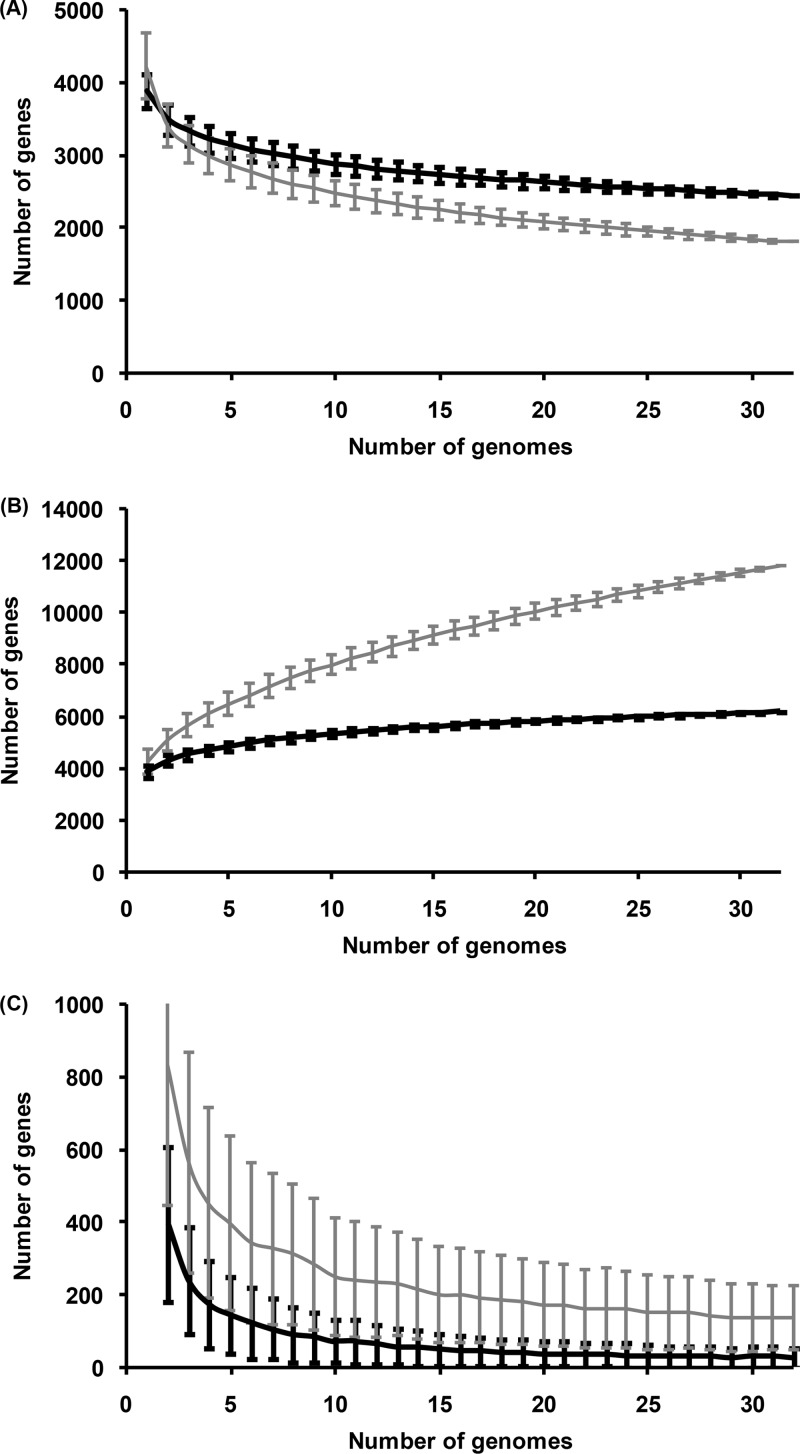
Figure 2 summarizes the basic OG statistics and highlights differences between the initial and modified OGs (for E. coli sensu lato). The core genome (Fig. 2A) of modified OGs contains significantly more genes (a total of 660 genes were added to the core genome of all 48 strains).
Fig 2.
(A to C) Sizes of the core genome (A), pan-genome (B), and new genes observed upon adding a new genome (C) for E. coli sensu lato strains. Gray lines, initial OGs; black lines, modified OGs; error bars, standard deviations.
The pan-genome is not saturating for both E. coli sensu lato and S. enterica groups (Fig. 2B and Fig. S2B in the supplemental material). The number of new genes observed upon the addition of new genomes (Fig. 2C and Fig. S2C in the supplemental material) is lower for modified OGs; the last genome adds, on average, 18 genes for modified OGs and 132 for initial ones. There are two reasons for this difference. First, most strain-specific genes are phage or transposon related or hypothetical and not present in modified OGs. Second, some orphans formed OGs after the tblastn search.
Percent pan-genome of E. coli.
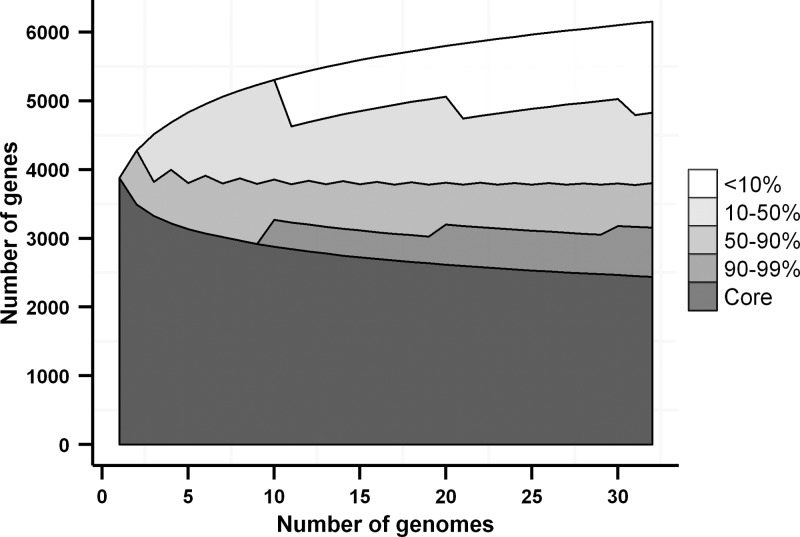
The narrow definition of the pan-genome size is the number of genes that exist in at least one strain of a given species (see the introduction for details). As shown above, the pan-genome is far from saturation for E. coli and still increasing as new strain sequences become available. However, to characterize the structure of the E. coli pan-genome, we may modify the condition, requiring that a gene is present in at least a fraction of strains. As shown in Fig. 3 (for E. coli sensu lato), such percentile pan-genomes are saturating fast. The jagged pattern is a consequence of the rounding procedure. We see that about 4,000 genes are present in at least 50% of strains, and this number shows very little change when new strains are added. After the first few strains are processed, only two plots are not saturated: the pan-genome and the core genome sensu stricto. This means that only two percentile intervals are expanding: very rare (mainly unique) genes and almost universal genes present in all but a few strains. Similarly, “soft core” is defined as genes present in 85% of strains was shown to be close to saturation in 186 genomes (27). Other categories are stable and do not change dramatically.
Fig 3.
Number of genes present in a given fraction of E. coli sensu lato genomes as dependent on the number of considered genomes. The topmost boundary shows the pan-genome size, the lowest boundary shows the core genome size, and the remaining boundaries show the percentile pan-genome sizes.
Comparison of pan-genomes and the pan-genome structure.
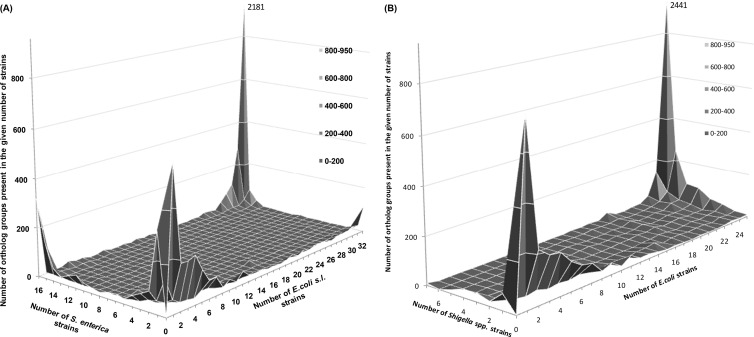
To characterize the gene distribution in the E. coli sensu lato and S. enterica pan-genomes, we considered the joint distribution of OGs by the number of strains in which they are present (Fig. 4A). To account for multiple sequencing of very similar strains and mistakes of automated OG prediction, we used relaxed conditions to define pan-genome parts. For example, the highest peak on the chart consists of universal genes that can be found in all strains. However, we considered as universal not only OGs from this peak but also genes from the nine adjacent cells (see Table S2 in the supplemental material for the definition of double pan-genome parts).
Fig 4.
(A and B) Numbers of OGs present in a given number of strains in two groups: S. enterica versus E. coli sensu lato (A) or Shigella clones versus other E. coli strains (B). Horizontal axes show the numbers of strains in the specified group. Vertical axes show the numbers of OGs present in the given number of strains. The maximal value at the vertical axis is restricted to 950 to show small peaks in more detail, and the height of the universal-genome peak is indicated.
We then used GOstat to detect overrepresented functional categories in each pan-genome part (Table 1). The overrepresented categories in the universal genome of all 48 strains are linked to housekeeping functions (translation, RNA processing, etc.). Other overrepresented categories are characteristic features of the Enterobacteriaceae, such as “aerobic respiration” or metabolic pathways, e.g., “vitamin synthesis.”
Table 1.
Overrepresented functional categories (GOstat) for fractions of the E. coli sensu lato and S. enterica pan-genomesa
| Group (no. of genes) | No. of genesb | GOterm | GO category | E-value |
|---|---|---|---|---|
| Unique genome (1,614) | 30/42 | GO:0040029 | Regulation of gene expression, epigenetic | 7.73E–17 |
| 27/41 | GO:0000746 | Conjugation | 2.97E–13 | |
| 11/13 | GO:0006278 | RNA-dependent DNA replication | 6.27E–06 | |
| 35/92 | GO:0019861 | Flagellum | 1.62E–05 | |
| 42/135 | GO:0009289 | Fimbrium | 0.00102 | |
| 9/16 | GO:0009307 | DNA restriction-modification system | 0.00514 | |
| 5/7 | GO:0046689 | Response to mercury ion | 0.01727 | |
| S. enterica periphery (436) | 3/3 | GO:0019310 | Inositol catabolic process | 0.01968 |
| 3/6 | GO:0046685 | Response to arsenic | 0.05093 | |
| S. enterica-specific genome (479) | 32/142 | GO:0009405 | Pathogenesis | 2.09E–14 |
| 16/36 | GO:0009236 | Cobalamin biosynthetic process | 5.35E–09 | |
| 56/588 | GO:0045449 | Regulation of transcription | 0.00381 | |
| 3/3 | GO:0006101 | Citrate metabolic process | 0.00472 | |
| E. coli sensu lato periphery (1,278) | 42/103 | GO:0009306 | Protein secretion | 3.73E–09 |
| 46/142 | GO:0009405 | Pathogenesis | 1.19E–05 | |
| 12/18 | GO:0019439 | Aromatic compound catabolic process | 8.70E–05 | |
| 32/96 | GO:0007155 | Cell adhesion | 0.00031 | |
| 41/135 | GO:0009289 | Fimbrium | 0.00036 | |
| 7/10 | GO:0043101 | Purine salvage | 0.00347 | |
| 51/201 | GO:0008643 | Carbohydrate transport | 0.00745 | |
| 3/3 | GO:0006578 | Betaine biosynthetic process | 0.03384 | |
| E. coli sensu lato specific genome (178) | 48/1,123 | GO:0006810 | Transport | 0.00096 |
| 19/293 | GO:0022804 | Active transmembrane transporter activity | 0.00096 | |
| 2/6 | GO:0015925 | Galactosidase activity | 0.07839 | |
| S. enterica universal and E. coli sensu lato periphery (308) | 16/103 | GO:0009306 | Protein secretion | 0.00069 |
| 14/86 | GO:0015031 | Protein transport | 0.00104 | |
| 14/92 | GO:0019861 | Flagellum | 0.00207 | |
| 3/3 | GO:0019543 | Propionate catabolic process | 0.00313 | |
| 3/6 | GO:0006805 | Xenobiotic metabolic process | 0.03491 | |
| Near-unique common periphery (202) | 7/41 | GO:0000746 | Conjugation | 0.00151 |
| 3/16 | GO:0009307 | DNA restriction-modification system | 0.06693 | |
| 2/5 | GO:0006276 | Plasmid maintenance | 0.07658 | |
| Near-universal common periphery (122) | 4/10 | GO:0046487 | Glyoxylate metabolic process | 0.00177 |
| 12/214 | GO:0044248 | Cellular catabolic process | 0.00884 | |
| Universal genome (2,804) | 546/710 | GO:0005737 | Cytoplasm | 3.25E–75 |
| 426/522 | GO:0044249 | Cellular biosynthetic process | 1.01E–69 | |
| 556/826 | GO:0005886 | Plasma membrane | 1.08E–44 | |
| 113/117 | GO:0006412 | Translation | 5.04E–28 | |
| 78/84 | GO:0006396 | RNA processing | 3.22E–17 | |
| 276/477 | GO:0005975 | Carbohydrate metabolic process | 1.57E–08 | |
| 61/92 | GO:0009110 | Vitamin biosynthetic process | 0.00041 | |
| 16/17 | GO:0009082 | Branched chain family amino acid synthesis | 0.00117 | |
| 21/25 | GO:0009060 | Aerobic respiration | 0.00155 | |
| 4/4 | GO:0030244 | Cellulose biosynthetic process | 0.16172 |
As defined in Table S2 in the supplemental material.
The first value indicates the number of genes in the indicated group within the pan-genome; the second value indicates the number of genes of the indicated category within the entire pan-genome.
The opposite corner of the chart in Fig. 4A (with small coordinates at horizontal axes) contains unique genes (the unique genome). Overrepresented functions in the unique genome tend to be plasmid related, e.g., “DNA restriction-modification system” or “response to mercury ion.”
Some genes are universal in one genus but absent in the other. For E. coli, the number of such genes is 178 (the smaller peak at the right corner of the chart in Fig. 4A and the adjacent area). The S. enterica-specific set comprises 479 genes (the left peak and the adjacent area). Although similar genetic signatures have been described before (28), we suggest that a weaker definition is biologically more reasonable, since rare gene loss in one of many strains may be an unstable evolutionary accident.
The S. enterica-specific universal genome is larger than the E. coli-specific one, likely because E. coli strains are phenotypically diverse and may live in different environments (see Table S1 in the supplemental material). The second possible explanation is the larger number of E. coli strains in the data set, because each new strain could decrease, but not increase, the peak's height. However, this technical explanation does not hold, since when E. coli and S. enterica subsamples of same size are compared, the S. enterica-specific genome is still larger (data not shown).
As shown in Table 1, these genus-specific groups have different overrepresented GO categories. For instance, the common ancestor of E. coli and S. enterica had lost the ability to synthesize cobalamin and, approximately 71 million years ago, the common ancestor of S. arizonae and S. enterica reestablished this ability by horizontal gene transfer (HGT) (29, 30). As a result, the “cobalamin biosynthetic process” (GO:0009236) overrepresented category can be observed in the S. enterica-specific pan-genome. The most significant GO term for the E. coli-specific universal genome is “transport” (48 of 178 genes). Again, a possible explanation is a more diverse lifestyle of the E. coli strains.
The plateau at the center contains the shared periphery. Here, the number of genes in each cell is small, but overall the shared periphery comprises 324 genes. Few functional categories (in particular “glyoxylate catabolism” and “conjugation”) are overrepresented in this group (data not shown). However, when we split this group into almost unique (rare) and almost universal (frequent) shared peripheries, different overrepresented functional categories emerge. The almost-unique common periphery contains functional categories that are overrepresented here and also in unique genomes. We suggest that a significant fraction of such genes are spread by HGT (see below). In contrast, the frequent periphery contains numerous metabolism genes, likely independently lost by a minority of strains of E. coli and S. enterica.
Some functional categories, e.g., “flagellum,” are overrepresented in more that one pan-genome part. It is well known that Shigella strains are nonmotile and have lost flagellar function due to mutations in many different genes (31). Some flagellar genes are lost in individual Shigella genomes. For example, S. sonnei Ss046 has a flagellin gene (SSON_1193) that has orthologs in all other considered strains except S. dysenteriae Sd197 and three E. coli strains. This results in overrepresentation of the flagellum category among the S. enterica universal/E. coli periphery group. The occurrence of flagellum-linked genes in the unique genome is caused by the Flag-2 gene cluster present in three E. coli strains: ATCC 8739, SMS-3-5, and UMN026. This locus was shown to be present in 15 of 72 strains of the ECOR collection and is believed to be an ancestral one, subsequently lost by most E. coli strains (32, 33).
Another category overrepresented in several pan-genome parts is “pathogenesis.” Many of the analyzed strains are pathogens of humans or animals (see Table S1 in the supplemental material). Genes that are linked to the pathogenesis are overrepresented in the S. enterica-specific genome (“pathogenesis” is the most overrepresented GO term in this pan-genome part that contains >20% of all pathogenesis genes) and in the E. coli periphery. This is caused by the presence of commensal strains of E. coli in the sample and demonstrates the existence of district mechanisms of pathogenicity in E. coli and S. enterica. All S. enterica strains are strong pathogens, and this set of 32 genes likely defines pathogenesis mechanisms that are common for all S. enterica strains. For instance, orthologs of Salmonella pathogenicity island 1 are absent in E. coli strains. This island has been acquired by the common ancestor of S. enterica and S. bongori (close to 100 to 140 million years ago) and carries a type III secretion system that allows these strains to invade epithelial cells (30).
As described above (Fig. 1C), we did not observe additional peaks for 7 and 25 genomes of Shigella and non-Shigella strains, respectively. To further demonstrate the homogeneity of the E. coli strains, we compared the pan-genomes of 7 Shigella clones to those of 25 E. coli strains (Fig. 4B). In this case, group-specific genes are virtually absent, similar to the comparison of commensal and pathogenic E. coli (see Fig. S3A in the supplemental material). It means that there are no gene pool barriers between Shigella clones and other E. coli strains or between commensal and pathogenic E. coli strains.
HGT of the common periphery.
Clearly, most universal genes have been vertically inherited (in one genus or in a larger group), and a significant fraction of unique genes have been gained by HGT from other species or are unrecognized prophage or integrated plasmid genes (1, 34, 35). However, the common periphery of E. coli and S. enterica can consist of both classes of genes. Indeed, the pan-genome of the last common ancestor of E. coli and S. enterica also had a periphery. After separation of the genera, a fraction of the ancestral periphery genes may have remained in the common periphery. Alternatively, a gene may have been gained by one strain of both genera independently and subsequently spread to some related strains by lateral transfer. These two situations (vertical or horizontal origin of common periphery genes) can be resolved based on the gene tree. If a gene had been vertically inherited, one would observe monophyletic E. coli and S. enterica subtrees. Contrariwise, if a gene is spreading horizontally, species-specific nodes of the tree may be intermixed. At that, one has to keep in mind that independent gains from related species could still yield a resolved tree.
For each OG, a rooted maximum-likelihood tree was constructed. Sequences found by tblastn were ignored, because these may be pseudogenes with abnormally high evolution rates. We performed this analysis for genes from the rare periphery, the frequent periphery, and the universal genome (see Table S2 in the supplemental material).
If a tree cannot be divided into two monophyletic subtrees, i.e., E. coli sensu lato and S. enterica, this indicates HGT. The statistics of these trees are presented in Table 2. The fraction of trees with possible HGT ranges from 12% (universal) to 55% (rare periphery). If trees containing paralogs are excluded, the fraction of suspected HGT events decreases, but the trend persists: fewer HGTs for universal genes, more HGTs for frequent periphery, and even more HGTs for rare periphery (Table 2). Considering unrooted trees, we obtain the same result (Table 2).
Table 2.
Fraction of trees with possible HGT events in three regions of the pan-genome matrixa
| Group | % OGs |
||
|---|---|---|---|
| Overall | Paralogs excluded | Paralogs excluded, trees unrooted | |
| Near-unique shared periphery | 55 (102/187) | 42 (55/130) | 20 (26/130) |
| Near-universal shared periphery | 26 (59/227) | 15 (25/172) | 4 (7/172) |
| Universal genes | 12 (284/2,392) | 7 (161/2,168) | 2 (48/2,168) |
As defined in Table S2 in the supplemental material. In columns 3 and 4, OGs containing paralogs were excluded. In column 4, the trees were unrooted. See the text for details and further discussion.
Conclusions.
The gene complements of E. coli and Shigella spp. are very similar, and these species cannot be separated into two groups, since they have a common gene pool. This is consistent with the analysis of gene phylogenetic trees (11). At the same time, despite intensive HGT between Enterobacteriaceae species (36) and specifically between E. coli and S. enterica (35), the latter maintain stable species-restricted gene pools. Hence, comparative pan-genome analysis complements current sequence- and phenotype-based methods of classifying prokaryotes and provides a new perspective to the understanding of bacterial evolution.
The existence of species-specific, universal genes, reflected in the peaks in Fig. 1C and Fig. 4A, may be used to define species. An analysis of the Vibrionaceae demonstrated that similar considerations may be used to delineate fine taxonomic relationships within a larger taxonomic group (19).
Genes that are shared between some, but not all, strains naturally form two categories, dependent on the fraction of strains carrying a gene, as shown in Fig. 1D. The genes of the shared periphery may have different evolutionary histories, being inherited vertically from the ancestral pan-genome (mainly genes from the frequent periphery) or transferred horizontally (mainly rare periphery). These groups differ also in functional characteristics, being enriched in different functional classes, both metabolic and structural.
Hence, comparative analysis of genomes allowed for the identification of a new type of genes, shared by some strains in different species. They are to some extent analogous to persisting polymorphisms inherited from a common ancestor by eukaryotes. In the latter case, persistence may be explained by balancing selection or simply insufficient time for fixation of one allele. Methods of population genetics, already applied to the analysis of gene distribution in single pan-genomes (37), may be used to describe the dynamics of gene gain and loss in bacteria after speciation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. I. Wolf and E. V. Koonin for helpful discussions, A. V. Favorov for the advice on statistics, I. V. Glotova for the advice on programming, and O. O. Bochkaryova for valuable comments.
This study was partially supported by The Ministry of Education and Science of Russian Federation, projects 8049 (M.S.G.) and 8135 (M.D.K.), Russian Foundation of Basic Research grant 13-04-01105, and program 30 “Living Nature” of the Russian Academy of Sciences.
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02285-12.
REFERENCES
- 1. Welch RA, Burland V, Plunkett G, III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Snipen L, Ussery DW. 2010. Standard operating procedure for computing pangenome trees. Stand. Genomic Sci. 2:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lapierre P, Gogarten JP. 2009. Estimating the size of the bacterial pan-genome. Trends Genet. 25:107–110 [DOI] [PubMed] [Google Scholar]
- 5. Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, Okinaka R, Jackson PJ, Hugh-Jones ME. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sacchi CT, Whitney AM, Mayer LW, Morey R, Steigerwalt A, Boras A, Weyant RS, Popovic T. 2002. Sequencing of 16S rRNA gene: a rapid tool for identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C. 2011. The Salmonella enterica pan-genome. Microb. Ecol. 62:487–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11:472–477 [DOI] [PubMed] [Google Scholar]
- 10. Doolittle RF, Feng DF, Tsang S, Cho G, Little E. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271:470–477 [DOI] [PubMed] [Google Scholar]
- 11. Pupo GM, Lan R, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U. S. A. 97:10567–10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64–67 [DOI] [PubMed] [Google Scholar]
- 13. Castellani A, Chalmers AJ. 1919. Manual of tropical medicine, 3rd ed Baillière, Tindall, and Cox, London, United Kingdom [Google Scholar]
- 14. Brenner DN. 1984. Enterobacteriaceae, p 408–420 In Holt JG, et al. (ed), Bergey's manual of systematic bacteriology, vol 1 The Williams & Wilkins Co, Baltimore, MD [Google Scholar]
- 15. Deng W, Liou SR, Plunkett G, III, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Behr MA. 2008. Mycobacterium du jour: what's on tomorrow's menu? Microbes Infect. 10:968–972 [DOI] [PubMed] [Google Scholar]
- 17. Davis KE, Joseph SJ, Janssen PH. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA, Janssen PH. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kahlke T, Goesmann A, Hjerde E, Willassen NP, Haugen P. 2012. Unique core genomes of the bacterial family Vibrionaceae: insights into niche adaptation and speciation. BMC Genomics 13:179. 10.1186/1471-2164-13-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhaxybayeva O, Nesbo CL, Doolittle WF. 2007. Systematic overestimation of gene gain through false diagnosis of gene absence. Genome Biol. 8:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beissbarth T, Speed TP. 2004. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics 20:1464–1465 [DOI] [PubMed] [Google Scholar]
- 23. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 24. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 25. Edwards RJ, Shields DC. 2005. BADASP: predicting functional specificity in protein families using ancestral sequences. Bioinformatics 21:4190–4191 [DOI] [PubMed] [Google Scholar]
- 26. Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:2567–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawrence JG, Ochman H. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383–397 [DOI] [PubMed] [Google Scholar]
- 30. Vernikos GS, Thomson NR, Parkhill J. 2007. Genetic flux over time in the Salmonella lineage. Genome Biol. 8:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, Xue Y, Zhu Y, Xu X, Sun L, Chen S, Nie H, Peng J, Xu J, Wang Y, Yuan Z, Wen Y, Yao Z, Shen Y, Qiang B, Hou Y, Yu J, Jin Q. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fricke WF, Wright MS, Lindell AH, Harkins DM, Baker-Austin C, Ravel J, Stepanauskas R. 2008. Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J. Bacteriol. 190:6779–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ren CP, Beatson SA, Parkhill J, Pallen MJ. 2005. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 187:1430–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Olschlager T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103:12879–12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karberg KA, Olsen GJ, Davis JJ. 2011. Similarity of genes horizontally acquired by Escherichia coli and Salmonella enterica is evidence of a supraspecies pangenome. Proc. Natl. Acad. Sci. U. S. A. 108:20154–20159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt WD. 2012. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. U. S. A. 109:1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Collins RE, Higgs PG. 2012. Testing the infinitely many genes model for the evolution of the bacterial core genome and pangenome. Mol. Biol. Evol. 29:3413–3425 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.