Abstract
Hypotaurine (HT; 2-aminoethane-sulfinate) is known to be utilized by bacteria as a sole source of carbon, nitrogen, and energy for growth, as is taurine (2-aminoethane-sulfonate); however, the corresponding HT degradation pathway has remained undefined. Genome-sequenced Paracoccus denitrificans PD1222 utilized HT (and taurine) quantitatively for heterotrophic growth and released the HT sulfur as sulfite (and sulfate) and HT nitrogen as ammonium. Enzyme assays with cell extracts suggested that an HT-inducible HT:pyruvate aminotransferase (Hpa) catalyzes the deamination of HT in an initial reaction step. Partial purification of the Hpa activity and peptide fingerprinting-mass spectrometry (PF-MS) identified the Hpa candidate gene; it encoded an archetypal taurine:pyruvate aminotransferase (Tpa). The same gene product was identified via differential PAGE and PF-MS, as was the gene of a strongly HT-inducible aldehyde dehydrogenase (Adh). Both genes were overexpressed in Escherichia coli. The overexpressed, purified Hpa/Tpa showed HT:pyruvate-aminotransferase activity. Alanine, acetaldehyde, and sulfite were identified as the reaction products but not sulfinoacetaldehyde; the reaction of Hpa/Tpa with taurine yielded sulfoacetaldehyde, which is stable. The overexpressed, purified Adh oxidized the acetaldehyde generated during the Hpa reaction to acetate in an NAD+-dependent reaction. Based on these results, the following degradation pathway for HT in strain PD1222 can be depicted. The identified aminotransferase converts HT to sulfinoacetaldehyde, which desulfinates spontaneously to acetaldehyde and sulfite; the inducible aldehyde dehydrogenase oxidizes acetaldehyde to yield acetate, which is metabolized, and sulfite, which is excreted.
INTRODUCTION
The organosulfinate hypotaurine (HT; 2-aminoethane-sulfinate) (Fig. 1) is an osmolyte at high concentration (0.25 M), e.g., in deep-sea invertebrates, as is an analogue organosulfonate, taurine (2-aminoethane-sulfonate) (Fig. 1) (1–3). HT is the precursor of taurine in the taurine biosynthetic pathway, e.g., in mammalian tissue (4, 5). Many mammals synthesize taurine, but they cannot degrade it, and taurine is excreted. Taurine is utilized by environmental bacteria under various metabolic situations (reviewed in reference 6), e.g., as a carbon, nitrogen, and energy source for aerobic heterotrophic growth via the taurine degradation pathway, which has been studied in great detail, including the desulfonation reaction involved (7–9). At least one bacterium, the marine strain Ruegeria pomeroyi DSS-3, is known to utilize also HT as a source of carbon, nitrogen, and energy (10). However, the HT degradation pathway, and particularly the desulfination reaction involved, remained undefined. In this study, another genome-sequenced bacterium, Paracoccus denitrificans PD1222, was found to utilize HT as a carbon, nitrogen, and energy source, and it was used to explore the HT degradation pathway. P. denitrificans PD1222 also utilized taurine, via the pathway described biochemically and genetically by Brüggemann et al. (9).
Fig 1.
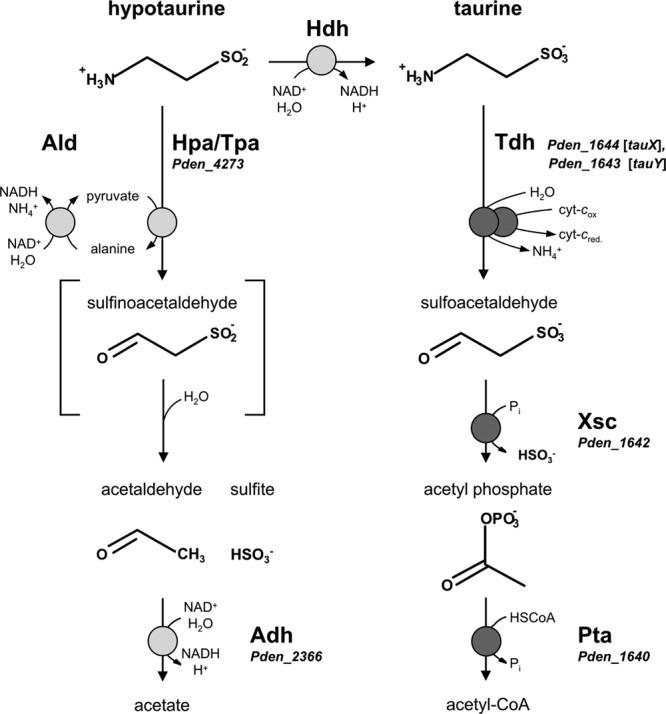
The known pathway for the utilization of taurine and candidate reactions for the utilization of hypotaurine in Paracoccus denitrificans PD1222. A hypothetical pathway is oxidation of HT to taurine and utilization of the taurine via the established taurine pathway (right). An alternative HT pathway (left) is via deamination of HT (see text). The known genes for the utilization of taurine are denoted by their locus tags (Pden_xxxx), as are candidate genes, if validated (see text), for possible reactions with HT. Tdh, deaminating, cytochrome c-dependent taurine dehydrogenase (membrane associated, two component; TauXY) (EC 1.4.99.2); Xsc, desulfonative sulfoacetaldehyde acetyltransferase (EC 2.3.3.15); Pta, phosphate acetyltransferase (EC 2.3.1.8); Hdh, NAD+-dependent HT dehydrogenase (EC 1.8.1.3); Hpa/Tpa, HT:pyruvate aminotransferase/archetypal taurine:pyruvate aminotransferase (EC 2.6.1.77); Ald, NAD+-dependent alanine dehydrogenase (EC 1.4.1.1); Adh, acetaldehyde dehydrogenase (EC 1.2.1.10).
Taurine is synthesized from, e.g., cysteine, which is oxygenated by cysteine dioxygenase (EC 1.13.11.20) to cysteinesulfinate and decarboxylated to HT by cysteinesulfinate decarboxylase (EC 4.1.1.29). The conversion of HT to taurine (Fig. 1) was described 50 years ago (11) as an NAD+-dependent HT dehydrogenase in rat liver and designated EC 1.8.1.3, but this enzyme activity has never been confirmed in vitro (12).
One option for the HT degradation pathway in P. denitrificans PD1222 is thus a primary attack on the sulfino moiety of HT and its oxidation to taurine by hypothetical HT dehydrogenase (Hdh in Fig. 1). The reaction product, taurine, could then be catabolized via the well-known taurine degradation pathway (Fig. 1; Table 1). First, membrane-bound, cytochrome c-coupled taurine dehydrogenase (Tdh, TauXY) (EC 1.4.99.2) yields sulfoacetaldehyde and ammonium (9, 13, 14). Second, desulfonation of sulfoacetaldehyde by sulfoacetaldehyde acetyltransferase (Xsc) (EC 2.3.3.15) (6, 8, 9) yields acetyl phosphate, which enters the amphibolic pathways via acetyl coenzyme A (acetyl-CoA) (Fig. 1). Alternatively, deamination can take place via taurine:pyruvate aminotransferase (Tpa) (EC 2.6.1.77), which catalyzes a transfer of the amino group to pyruvate, yielding alanine (15), while the regeneration of pyruvate is catalyzed by an NAD+-dependent alanine dehydrogenase (Ald) (EC 1.4.1.1) and ammonium is released (Fig. 1); these enzymes are active in taurine pathways of other bacteria (10, 16–19). For all pathways known, the next step in taurine degradation, desulfonation of sulfoacetaldehyde (Fig. 1), is catalyzed by Xsc, and the conversion of acetyl phosphate to acetyl-CoA is performed by phosphate acetyltransferase (Pta) (EC 2.3.1.8) (Fig. 1) (8, 20); the latter reaction can be bypassed via acetate kinase and acetate-CoA ligase (18).
Table 1.
Gene loci in the genome of Paracoccus denitrificans PD1222 attributed to desulfonation and desulfination pathways (see the text)
| Pathway | Locus tag | Orientationa | Annotation |
|---|---|---|---|
| Taurine | Pden_1640 | ← | Phosphate acetyltransferase (Pta1) |
| Pden_1641 | ← | Sulfite exporter (TauZ) | |
| Pden_1642 | ← | Sulfoacetaldehyde acetyltransferase (Xsc1) | |
| Pden_1643 | ← | Taurine dehydrogenase (Tdh), large subunit (TauX) | |
| Pden_1644 | ← | Taurine dehydrogenase (Tdh), small subunit (TauY) | |
| Pden_1645 | ← | Taurine transporter (TRAP type), large permease component (TauM) | |
| Pden_1646 | ← | Taurine transporter (TRAP type), small permease component (TauL) | |
| Pden_1647 | ← | Taurine transporter (TRAP type), periplasmic binding protein (TauK) | |
| Pden_1648 | → | Transcriptional regulator (GntR family) (TauR) | |
| Sulfoacetate | Pden_1008 | ← | Phosphate acetyltransferase (Pta3) |
| Pden_1009 | ← | Sulfite exporter (SauZ) | |
| Pden_1010 | ← | Sulfoacetaldehyde acetyltransferase (Xsc3) | |
| Pden_1011 | ← | Sulfoacetate transporter (major facilitator superfamily 1) (SauU) | |
| Pden_1012 | ← | Sulfoacetate-CoA ligase (SauT) | |
| Pden_1013 | ← | Sulfoacetaldehyde dehydrogenase (acylating) (SauS) | |
| Pden_1014 | → | Transcriptional regulator (LacI family) | |
| Isethionate | Pden_4275 | ← | Phosphate acetyltransferase (Pta2) |
| Pden_4276 | ← | Sulfite exporter (IseZ) | |
| Pden_4277 | ← | Sulfoacetaldehyde acetyltransferase (Xsc2) | |
| Pden_4278 | ← | Isethionate dehydrogenase (IseJ) | |
| Pden_4279 | ← | Isethionate transporter (major facilitator superfamily 1) (IseT) | |
| Pden_4280 | → | Transcriptional regulator (IcIR family) | |
| HT (deamination) | Pden_4273 | → | HT:pyruvate/taurine:pyruvate aminotransferase (Hpa/Tpa) |
| Pden_4274 | → | Transcriptional regulator (GntR family) | |
| HT (pyruvate regeneration) | Pden_1729 | ← | Alanine dehydrogenase |
| Unknown | Transcriptional regulator | ||
| HT (acetaldehyde oxidation) | Pden_2365 | → | Transcriptional regulator (Fis family) |
| Pden_2366 | → | Acetaldehyde dehydrogenase | |
| Pden_2367 | → | Alcohol dehydrogenase |
Carried on the forward (→) or reverse (←) strand of chromosome 1 (locus tags Pden_0001 to Pden_2832) or chromosome 2 (locus tags Pden_2833 to Pden_4514) of P. denitrificans PD1222.
Another option for HT degradation is a primary attack on the amino group, hence deamination of HT, in analogy to the first step of taurine degradation (Fig. 1). Indeed, this reaction has been shown to be catalyzed by archetypal taurine:pyruvate aminotransferase (Tpa) (see above) purified from taurine-induced Bilophila wadsworthia (15), as well as for a “taurine:oxoglutarate aminotransferase” (EC 2.6.1.55) purified from a beta-alanine-induced Achromobacter superficialis strain (21). Importantly, for the latter, acetaldehyde and sulfite were identified as the reaction products (22), which implies a spontaneous desulfination of the anticipated HT deamination product sulfinoacetaldehyde (Fig. 1). Interestingly, a candidate gene for taurine:pyruvate aminotransferase (Tpa) is included in the genome of P. denitrificans PD1222 but is not part of the taurine pathway (Table 1).
The aim of this study was to define the HT degradation pathway in P. denitrificans PD1222 in comparison to its well-known taurine pathway (Table 1). We used analytical chemistry, physiology, biochemistry, genetics, and genomics and confirmed the pathway that is shown on the left in Fig. 1.
MATERIALS AND METHODS
Chemicals.
HT (>98%) was purchased from Sigma-Aldrich. Sulfoacetaldehyde, as the bisulfite addition complex, was synthesized previously (23). Standard chemicals were of the highest purity available from Sigma-Aldrich, Fluka, Roth, or Merck. Biochemicals (NADH, NADPH, NAD+, and NADP+) were purchased from Biomol (Hamburg, Germany).
Growth conditions, harvesting of cells, and preparation of cell extracts.
Paracoccus denitrificans PD1222 was kindly provided by R. J. M. van Spanning (Vrije Universiteit Amsterdam, Amsterdam, The Netherlands). A phosphate-buffered, mineral salts medium, pH 7.2 (24), supplemented with acetate (10 mM), taurine (10 mM), or HT (10 mM) as the sole carbon source, was used. The HT growth medium was filter sterilized. Cultures in the 5-ml scale were incubated in glass tubes (Corning) in a roller at 30°C in the dark or in the 7- to 500-ml scale in Erlenmeyer flasks on a shaker. Cultures were inoculated (1%) with cultures pregrown with the same substrate and harvested in the late exponential growth phase by centrifugation (15,000 × g, 15 min, 4°C); the cell pellets were stored frozen (−20°C). Cells were resuspended in 50 mM Tris-HCl buffer (pH 9.0) containing 0.03 mg ml−1 DNase I (Sigma) and 5 mM MgCl2 and disrupted by three to four passages through a chilled French pressure cell (140 MPa, 4°C) (Aminco, Silver Spring, MD, USA). Whole cells and debris were removed by centrifugation (15,000 × g, 15 min, 4°C) to obtain crude extract, and the membrane fragments were removed by ultracentrifugation (70,000 × g, 60 min, 4°C) to obtain the soluble protein fraction and the membrane pellet. The membranes were resuspended in 50 mM Tris-HCl buffer (pH 9.0) containing 5 mM MgCl2 to obtain the membrane fraction.
Growth experiments were done in the 50-ml scale in 300-ml Erlenmeyer flasks when using phosphate-buffered mineral salts medium (24) with 0.5 mM instead of 20 mM ammonium chloride, in order to facilitate quantification of ammonium excretion during growth (see below). Samples were taken at intervals to measure optical density at 580 nm (OD580), to assay protein, and to determine the concentrations of HT, sulfate, sulfite, and ammonium.
Analytical methods.
HT, taurine, and alanine were analyzed after derivatization with 2,4-dinitrofluorobenzene (DNFB) by reversed-phase high-pressure liquid chromatography (HPLC) with UV detection (DNFB/HPLC-UV) (25); briefly, a Nucleosil C18 column (125 by 3 mm; particle size, 5 μm; Macherey-Nagel, Germany) and a gradient system (mobile phase A, 20 mM potassium phosphate puffer, pH 2.2; B, 100% methanol; flow rate, 0.5 ml min−1) were used, and the gradient was set from 0% B to 80% B in 17 min and the detection was set to 360 nm. Sulfoacetaldehyde, acetaldehyde, and pyruvate were analyzed after derivatization with 2-(diphenylacetyl)-indan-1,3-dian-1-hydrazone (DIH) by reversed-phase HPLC (DIH/HPLC-UV) (26); briefly, a Nucleosil C18 column (above) and a gradient system (mobile phase A, 0.11 M NaClO4; B, 100% acetonitrile; flow rate, 0.5 ml min−1) were used, and the gradient was set from 0% B to 100% B in 17 min and the detection was set to 400 nm.
For the analysis of HT and taurine in nonderivatized samples, an HPLC method was developed using a hydrophilic-interaction liquid chromatography (HILIC) column (SeQuant ZIC-HILIC; 150 by 4.6 mm; particle size, 5 μm; Merck, Germany) coupled to an evaporative light-scattering detector (ELSD); the mobile phases used were 0.1 M ammonium acetate in water containing 10% acetonitrile (A) and 100% acetonitrile (B). For the analysis of HT and taurine (and of sulfite/sulfate) in samples from growth experiments, an isocratic method (isocratic-HILIC-ELSD) was set to 40% buffer A and 60% eluent B for 12 min; the flow rate was 0.75 ml min−1. Taurine and HT eluted at 8.3 min and 9.0 min, respectively, and sulfite and sulfate coeluted at a 10.5-min retention time. Furthermore, alanine coeluted with HT. A gradient method was used (gradient-HILIC-ELSD) for the separation of HT and alanine in samples taken from enzyme assays (same column, mobile phases, and flow rate); the gradient was started after 0.1 min at 75% B to 60% B in 18 min. HT and alanine eluted at 10.1 and 9.6 min, respectively. For codetection of NAD+ and NADH in samples taken from enzyme reactions, the gradient-HILIC-ELSD method was further modified (after 0.1 min at 90% B gradient to 60% B in 18 min), and a UV detector was used (254 nm) in addition to the ELSD (gradient-HILIC-UV-ELSD). Alanine, HT, NADH, and NAD+ eluted at 15.6, 16.3, 16.9, and 18.8 min, respectively, but HT coeluted with pyrophosphate (see Results). The HPLC system LC-20 AT with diode array detector (DAD) SPD-M20A (Shimadzu) was used, and as ELSD, ZAM 3000 (Schambeck, Bad Honnef, Germany) was used.
Total protein was determined according to a protocol based on the work of Lowry (27) with bovine serum albumin (BSA) as the standard. Soluble protein was assayed by protein dye binding (28). Sulfate release during growth was quantified turbidimetrically as a suspension of BaSO4 (29), and sulfite was quantified colorimetrically as the fuchsin adduct (23). The ammonium ion was assayed colorimetrically by the Berthelot reaction (30). Acetate was quantified by an acetic acid quantification kit (NZYTech-Genes and Enzymes, Lisbon, Portugal).
Protein electrophoresis and PF-MS.
Denatured proteins were separated on 13% SDS-PAGE gels and stained with Coomassie brilliant blue R-250 (31). For analysis of the soluble protein expression pattern in acetate-, taurine-, or HT-grown cells, proteins were separated by two-dimensional (2D) isoelectric focusing (IEF)–SDS-PAGE (2D-PAGE) using the Bio-Rad ReadyStrip immobilized pH gradient (IPG) system for the first-dimension separation (17-cm length; pI range, pH 4 to 7) and 12% SDS-PAGE gels (17 cm by 20 cm; no stacking gel) for the second-dimension separation. Sample preparation and IEF separation conditions were essentially as described in the manufacturer's instructions (Bio-Rad ReadyStrip IPG strip instruction manual) with the modifications described previously (32). Stained protein bands or spots were cut from the gel and subjected to peptide fingerprinting-mass spectrometry (PF-MS) at the Proteomics Facility of the University of Konstanz (www.proteomics-facility.uni-konstanz.de). The parameters for mass spectral analysis and Mascot database searching and scoring were set as previously described (32).
Separation and enrichment of native enzymes.
Fast protein liquid chromatography (FPLC) of soluble protein extract (up to 5 ml) was done using an anion-exchange column (Mono Q HR 10/10; GE Healthcare) equilibrated with Tris-H2SO4 buffer (pH 9.0) at a flow rate of 1 ml min−1. Bound proteins were eluted by a gradient of Na2SO4 from 0 M to 0.5 M, and all collected fractions (2 ml) were tested for aminotransferase activity; the aminotransferase eluted at about 0.1 M Na2SO4.
Enzyme assays.
The assay for Tpa and Hpa was performed discontinuously at 30°C. The formation of alanine as well as the disappearance of taurine or HT was quantified in samples (50 μl) taken at intervals during the reactions, when using the DNFB/HPLC-UV method (see above). The standard reaction mixture (0.5 ml) contained 50 mM Tris-HCl (pH 9.0), 5 mM MgCl2, 5 mM substrate, 0.1 mM pyridoxal-5-phosphate, 10 mM pyruvate, and 250 to 400 μg protein. Ald was assayed spectrophotometrically as formation of the coproduct NADH at 365 nm (16); the standard reaction mixture contained 0.1 M 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) (pH 10.0), 1 mM NAD+, 10 mM l-alanine, and 50 to 100 μg total protein. Tdh (TauXY) was assayed spectrophotometrically by the reduction of cytochrome c at 550 nm (9); the reaction mixture (1 ml) contained 50 mM Tris-HCl (pH 9.0), 5 mM MgCl2, 50 μM cytochrome c, 5 mM substrate, and 50 to 100 μg protein. Hdh was assayed spectrophotometrically as formation of NADH (11); the reaction mixture (1 ml) contained 66 mM Tris-HCl buffer (pH 7.5), 1.5 mM HT, 13 mM NAD+, and 50 to 500 μg protein. Adh was assayed spectrophotometrically as formation of NADH (33); the reaction mixture (1 ml) contained 50 mM sodium phosphate and 50 mM sodium pyrophosphate (pH 9.0), 1 mM dithiothreitol (DTT), 25 to 100 μM acetaldehyde, 1 mM NAD+, and 20 to 30 μg protein. Xsc was assayed by the colorimetric determination of acetyl phosphate from sulfoacetaldehyde (8), and SorA-type sulfite dehydrogenase (Sdh) was assayed photometrically with K3Fe(CN)6 as electron acceptor (34).
Recombinant, purified Hpa and Adh in combination were assayed first by quantification of the conversion of NAD+ to NADH and by formation of alanine using HILIC (gradient-HILIC-UV-ELSD, above) and second by formation of acetate using the acetic acid quantification kit (see above). The reaction mixture (2 ml) contained 50 mM sodium phosphate and 50 mM sodium pyrophosphate (pH 9.0), 1 mM HT, 2 mM pyruvate, 0.1 mM pyridoxal-5-phosphate, 2 mM NAD+, 2 mM dithiothreitol, 30 μg recombinant Hpa, and 60 μg recombinant Adh.
Heterologous expression of the candidate Hpa and Adh genes in Escherichia coli and purification of the recombinant proteins.
Chromosomal DNA of P. denitrificans PD1222 was isolated (Illustra bacterial genomicPrep Mini Spin kit; GE Healthcare), and the candidate genes for Hpa (locus tag Pden_4273) and Adh (locus tag Pden_2366) were amplified by PCR (Phusion HF DNA polymerase; Finnzymes) using the following primer pairs (Microsynth, Balgach, Switzerland): Pden4273f and Pden4273r, CACCATGACGCTCGATCTGAACCCCA and GCCGCGGATCAAGGGTCA; Pden2366f and Pden2366r, CACCATGAAGATGAGACTGAGGAGTCTT and GGCAAGGCGGGGAAGG (the directional overhang is underlined). Pden_4273 was amplified by 30 cycles of 15 s at 98°C, 20 s at 58°C, and 60 s at 72°C, and Pden_2366 was amplified by 30 cycles of 15 s at 98°C, 20 s at 66.3°C, and 70 s at 72°C. The PCR products were purified (QIAquick gel extraction kit; Qiagen) and ligated into an N-terminal His6-tagged expression vector (Champion pET 100 directional TOPO expression kit; Invitrogen). OneShot TOP10 E. coli cells (Invitrogen) were transformed with the construct, and the correct integration of the insert was confirmed by sequencing (GATC-Biotech, Konstanz, Germany). BL21 Star (DE3) OneShot E. coli cells (Invitrogen) were transformed with the constructs and grown at 37°C in LB medium (100 mg liter−1 ampicillin). The cultures were induced (0.5 mM isopropyl-β-d-thiogalactopyranoside [IPTG]) at an optical density at 580 nm (OD580) of 0.7 and grown for an additional 5 h at 20°C, and the cells were harvested by centrifugation (15,000 × g, 20 min, 4°C) and stored frozen (−20°C). For purification, cells were resuspended in 20 mM Tris-HCl, pH 8.0, containing 0.03 mg ml−1 DNase I (Sigma) and disrupted by four passages through a chilled French pressure cell (140 MPa) (Aminco, Silver Spring, MD, USA). Whole cells and debris were removed by centrifugation (15,000 × g, 15 min, 4°C), and the membrane fragments were removed by ultracentrifugation (70,000 × g, 60 min, 4°C). The soluble protein fractions were loaded on Ni+-chelating agarose affinity columns (1-ml column volume; Macherey-Nagel, Germany) equilibrated with buffer A (20 mM Tris-HCl, pH 8.0, 100 mM KCl). The columns were washed (30 mM imidazole in buffer A), and His6-tagged proteins were eluted (200 mM imidazole in buffer A) and stored at −20°C in glycerol (30% [vol/vol] final concentration).
RESULTS
Physiology of growth of P. denitrificans PD1222 with HT.
HT (10 mM) was stable in aerobic mineral salts medium at 30°C, as confirmed when neither HT disappearance nor formation of taurine, sulfite, or sulfate was detectable in sterile controls during a 9-day incubation (not shown). After inoculation with P. denitrificans PD1222, growth was exponential till about 30 h, when it slowed, reaching stationary phase after 40 h (Fig. 2A). HT utilization initially accelerated till about 27 h, when it slowed, and HT was exhausted by 46 h (Fig. 2B), in agreement with growth. Release of ammonium ion reached a stable final concentration at about 40 h, in agreement with growth; this concentration (5.5 mM), with the nitrogen in biomass, represented mass balance. The sulfinate moiety was released as sulfite, from which sulfate continued to be formed after growth was complete; recovery of sulfinate sulfur was about 95%. The formation of sulfite peaked at about 4 mM between 27 and 30 h, when growth slowed. The molar growth yield of 4.4 g protein (mol carbon)−1 determined after 65 h suggested quantitative utilization of the HT carbon (normal, 4 to 6 g mol−1 [35]). Replicate growth experiments (not shown) showed the same pattern, but the time at which HT had been completely utilized was delayed by up to 80 h while the sulfite concentration at which the growth slowed never exceeded 4 mM (Fig. 2). These observations were interpreted to represent a retardation of growth if strain PD1222 was exposed to increased sulfite stress (here, at >4 mM sulfite) (see Discussion). The maximal growth rate (μ) of 0.1 h−1 (Fig. 2) and the growth yield allowed a maximal specific HT degradation rate of 3.1 mkat (kg protein)−1 to be calculated.
Fig 2.
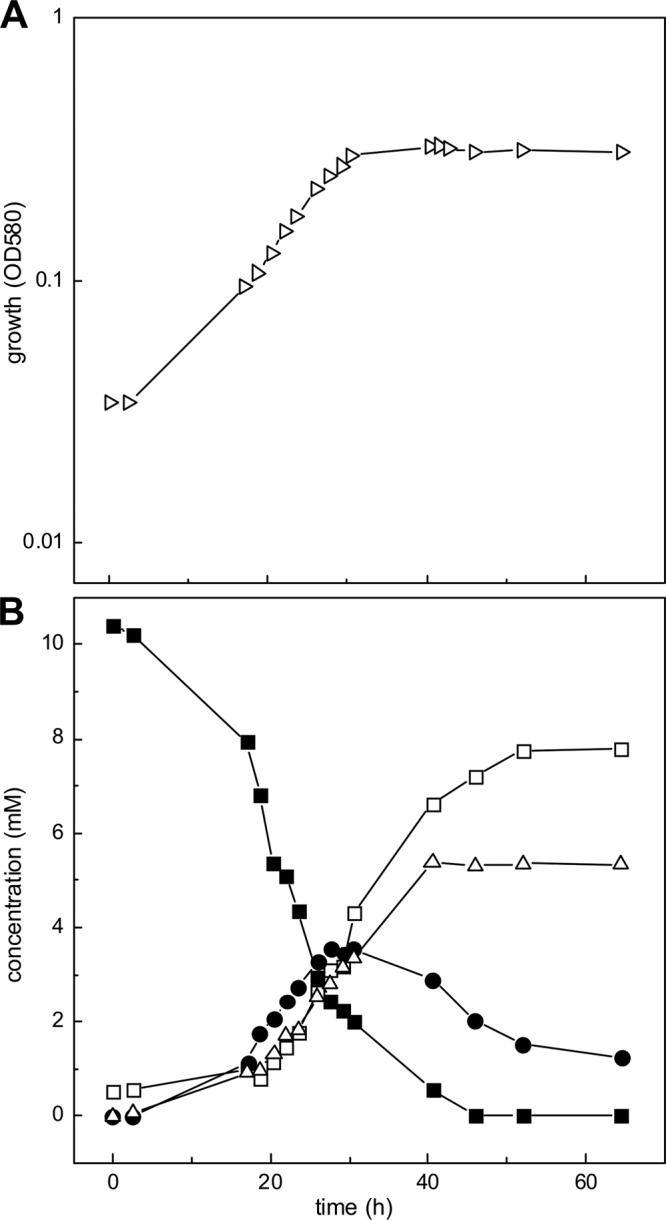
Growth of P. denitrificans PD1222 with HT. (A) Growth was monitored as turbidity (OD580). (B) The culture supernatant fluid was assayed for HT (solid squares), sulfite (solid circles), sulfate (open squares), and ammonium ion (open triangles) (see Materials and Methods).
Analysis of protein expression by differential PAGE and PF-MS.
The protein expression pattern in P. denitrificans PD1222 during growth with acetate, taurine, or HT was compared by one-dimensional (1D)- and 2D-PAGE when crude extracts (1D-PAGE; Fig. 3) and soluble protein fractions (2D-PAGE; see Fig. S1 in the supplemental material) were examined. On the 1D gel (Fig. 3), two additional, strong protein bands at about 50 and 55 kDa were visible only for HT-grown cells, suggesting two HT-inducible proteins. These bands were excised and submitted to PF-MS. The 50-kDa band (band b in Fig. 3) was identified as the gene product of (locus tag) Pden_4273 on chromosome 2 of strain PD1222, annotated to encode an aminotransferase class III protein (see Table S1 in the supplemental material), i.e., an archetypal taurine:pyruvate aminotransferase (Tpa) (Fig. 1). The 55-kDa band (band a in Fig. 3) was identified as Pden_2366 (on chromosome 1), annotated to encode an NAD+-dependent aldehyde dehydrogenase (Adh) (see Table S1). These identifications were confirmed, and expanded upon, when the 2D gels were analyzed (see Fig. S1 and Table S1 in the supplemental material). First, the Tpa (Pden_4273) and Adh (Pden_2366) genes were reidentified by PF-MS of prominent protein spots visible specifically on the gel of HT-grown cells but not on the gels of taurine- and acetate-grown cells (see Fig. S1, spots A to D at about 50 to 55 kDa in molecular mass and different pI). Another protein spot visible uniquely for HT-grown cells (spot E in Fig. S1) identified an alcohol dehydrogenase gene, Pden_2367, which is carried directly downstream of the identified Adh gene (Pden_2366) (see Table S1). Furthermore, two spots visible for both taurine- and HT-grown cells, but not for acetate-grown cells, identified two genes of the taurine pathway operon in P. denitrificans, for TauX (Pden_1644), the small subunit of membrane-bound, deaminating taurine dehydrogenase (TauXY) (spots F and H; see Fig. S1), and for Xsc (Pden_1642) (spots G and I in Fig. S1; also see the bands at around 64 kDa in Fig. 3); the latter identification, however, is ambiguous, due to the high sequence identity of Pden_1642 (taurine pathway) with the other two Xsc candidate genes in strain PD1222 (Pden_4277 [isethionate pathway] and Pden_1010 [sulfoacetate pathway]; see the Discussion).
Fig 3.
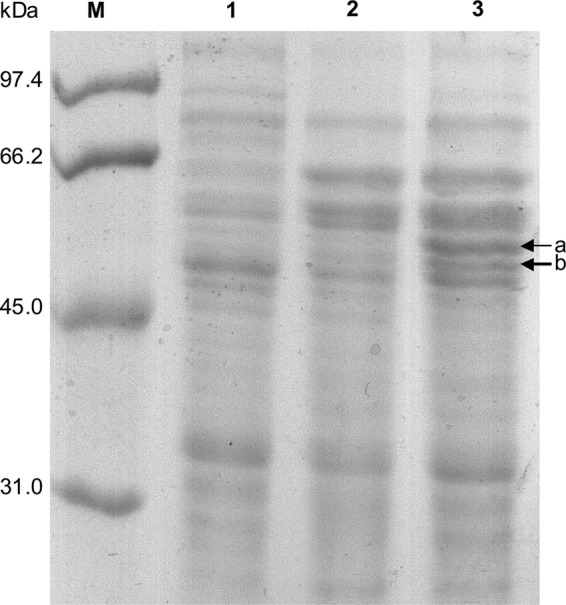
Comparison of the protein expression patterns in acetate-, taurine-, and HT-grown cells of P. denitrificans PD1222 by 1D SDS-PAGE. Crude extracts of P. denitrificans PD1222 were used. Two prominent protein bands (a and b) that suggested proteins specifically induced during growth with HT were excised and identified by PF-MS (see text). Lane M, molecular mass markers; lane 1, acetate; lane 2, taurine; lane 3, HT.
Enzyme activities detectable in cell extracts and enrichment of an HT:pyruvate aminotransferase activity.
The results of the proteomic work suggested that the taurine-desulfonation gene cluster is expressed at a high level during growth with taurine, as well as during growth with HT, which might reflect a coinduction of the taurine genes during growth with HT due to the structural analogy of HT and taurine as inducer (see the Discussion). Two proteins appeared to be expressed at very high levels specifically in HT-grown cells, predicted taurine:pyruvate aminotransferase (Tpa) and NAD+-dependent aldehyde dehydrogenase (Adh), and they were therefore considered the best candidates for an involvement specifically in HT degradation. In order to confirm and expand upon the proteomic results, enzyme assays were performed with cell extracts from acetate-, taurine-, or HT-grown cells (Table 2).
Table 2.
Enzyme activities detectable in crude extract, membrane fraction, or soluble protein fraction as a function of the growth substrate used
| Enzyme assay | Substrate | Sp act (mkat [kg protein])−1 (SD) in extracts of cells grown with substrate: |
||
|---|---|---|---|---|
| Acetate | Taurine | HT | ||
| Hpa/Tpaa | HT | —d | 1.2 (0.2) | 2.8 (0.5) |
| Taurine | — | 0.5 (0.3) | 1.6 (0.5) | |
| Tdhb | HT | — | 0.1 (0.1) | 0.6 (0.1) |
| Taurine | — | 4.2 (0.1) | 1.0 (0.1) | |
| Alda | Alanine | — | — | 1.1 (0.2) |
| Xscc | Sulfoacetaldehyde | — | 0.7 (0.1) | 0.4 (0.1) |
Crude extracts were used for the enzyme assays.
Membrane fractions were used for the enzyme assays.
Soluble fractions were used for the enzyme assays.
—, below detection limit (<0.1 mkat [kg protein]−1).
The aminotransferase reaction with taurine or HT as the substrate in the presence of pyruvate (Fig. 1) was routinely followed discontinuously by HPLC-UV after derivatization of the amino groups of HT, taurine, and alanine (DNFB/HPLC-UV). The activities determined (Table 2) indicated an about 2-fold-higher aminotransferase activity with HT as the substrate (2.8 mkat [kg protein]−1) than with taurine as the substrate (1.6 mkat [kg protein]−1) in crude extracts of HT-grown cells. Furthermore, this HT:pyruvate/taurine:pyruvate aminotransferase (“Hpa/Tpa”) activity was about 2-fold lower in extracts of taurine-grown cells than in those of HT-grown cells and not detectable for acetate-grown cells (Table 2). Hpa/Tpa showed negligible activity with 2-oxoglutarate instead of pyruvate as amino group acceptor. Furthermore, an NAD+-dependent alanine dehydrogenase (Ald) activity, catalyzing the regeneration of pyruvate for Hpa/Tpa (Fig. 1), was found specifically in crude extract of HT-grown cells but not in that of taurine- and acetate-grown cells (Table 2). In contrast, high activity of a deaminating taurine dehydrogenase (Tdh) corresponding to membrane-associated TauXY of the taurine degradation pathway in P. denitrificans (Fig. 1) was found in the membrane fraction of taurine-grown cells, with taurine as the substrate. This Tdh activity was about 25% in the membrane fraction of HT-grown cells, with taurine as the substrate (Table 2). The activity with HT as the substrate was <10% of that with taurine as the substrate (Table 2); hence, the Tdh in P. denitrificans is not good at deaminating HT, in contrast to Hpa/Tpa (above). Finally, high activity of desulfonative Xsc, corresponding to the taurine degradation pathway (Fig. 1), was found in the soluble fraction of both taurine- and HT-grown cells, although the specific activity determined for taurine-grown cells was about 2-fold higher than that for HT-grown cells, and no activity was detectable for acetate-grown cells (Table 2). It is clear that the taurine degradative pathway is partially induced in HT-grown cells.
No activity of an NAD+-dependent HT dehydrogenase (Hdh) was detectable when either crude extract or the membrane or soluble fraction of HT-grown cells was tested. Also, no significant activity was detectable for the predicted HT-inducible, NAD+-dependent acetaldehyde dehydrogenase (Adh) (see above) unless pyrophosphate was present in the reaction. Finally, no activity of a SorA-type sulfite dehydrogenase (Sdh) for HT-, taurine-, or acetate-grown cells was measurable, corresponding to the observation that, initially, sulfite is excreted during growth with HT (Fig. 2B) and that no candidate gene for Sdh is found in the genome of P. denitrificans PD1222.
The Hpa/Tpa activity in the soluble fraction of HT-grown cells could be enriched through one anion-exchange chromatography step (see Materials and Methods), and analysis of the obtained active fraction in comparison to nonactive fractions by SDS-PAGE indicated an enrichment of a protein of about 50 to 55 kDa (see Fig. S2 in the supplemental material). PF-MS of this band confirmed gene locus Pden_4273 for Hpa/Tpa and also gene locus Pden_2366 for Adh (see Table S1).
Heterologous expression of the Hpa/Tpa candidate gene and identification of the HT-deamination products.
Pden_4273 was overexpressed in E. coli and purified as a His6-tagged protein via affinity chromatography (Fig. 4A). As followed by DNFB/HPLC-UV, DIH/HPLC-UV, and/or HILIC-UV-ELSD (see Materials and Methods), the purified protein with taurine as a substrate catalyzed a pyruvate-dependent deamination of taurine to alanine and sulfoacetaldehyde (Fig. 5A), as expected for archetypal Tpa. With HT as the substrate, the enzyme catalyzed a pyruvate-dependent HT disappearance concomitant with alanine formation (Fig. 5B), but in contrast, no formation of sulfoacetaldehyde, or of sulfinoacetaldehyde, could be detected by HPLC (or by matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]; data not shown). However, we observed a new peak on the DIH/HPLC-UV chromatograms, i.e., after derivatization of carbonylic compounds with DIH, indicating a reaction product that increased concomitantly with both HT disappearance and alanine formation. This peak cochromatographed with DIH-derivatized, authentic acetaldehyde (data not shown) and could be quantified in stoichiometric amounts with an authentic acetaldehyde standard (Fig. 5B). Furthermore, a formation of sulfite as the second reaction product could be confirmed and be quantified during the reaction (Fig. 5B) by a colorimetric assay (see Materials and Methods).
Fig 4.
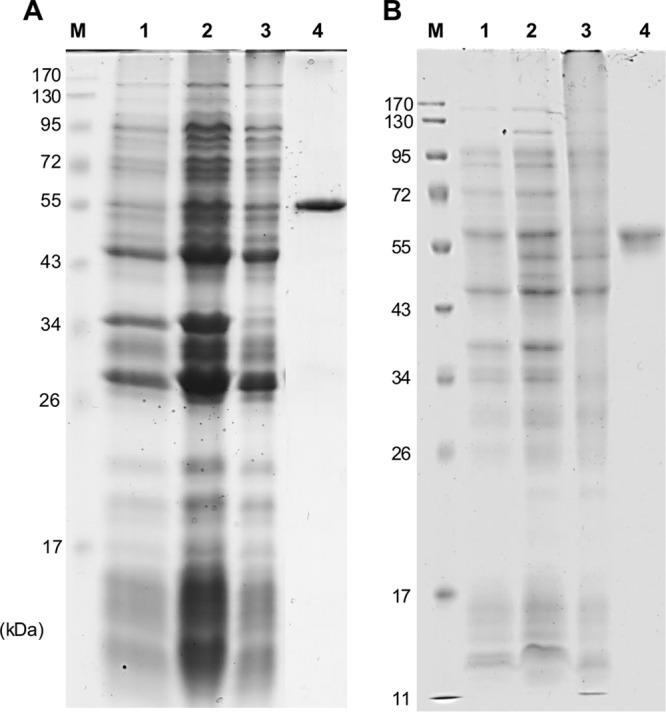
Analysis by SDS-PAGE of the overexpression of the Hpa (A) and Adh (B) candidate genes in E. coli and of the purification of the His-tagged proteins. Lane M, molecular mass markers; lane 1, whole cells prior to IPTG induction; lane 2, whole cells 5 h after induction; lane 3, soluble protein fraction after ultracentrifugation (100 μg protein); lane 4, protein fraction obtained from affinity chromatography (30 μg protein).
Fig 5.
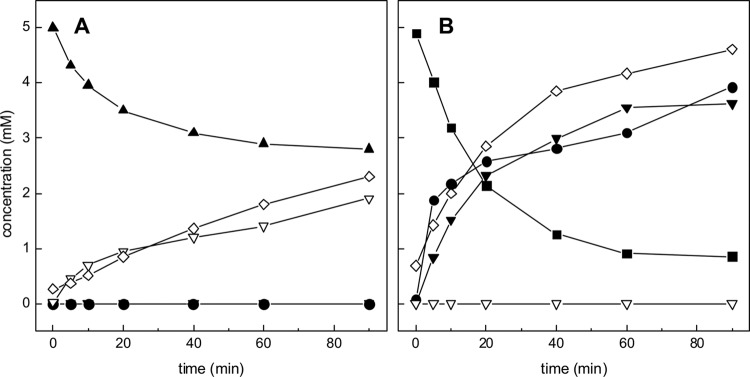
Reaction of purified recombinant candidate Hpa with taurine (A) and HT (B). Samples were taken at intervals for subsequent analyses by different HPLC methods (see Materials and Methods). (A) The reaction with taurine as the substrate in the presence of pyruvate led to disappearance of taurine (solid triangles) and pyruvate (not shown) concomitantly with formation of alanine (open diamonds) and sulfoacetaldehyde (open triangles). (B) The reaction with HT as the substrate led to disappearance of HT (solid squares) and pyruvate (not shown) concomitantly with formation of alanine (open diamonds), acetaldehyde (solid triangles), and sulfite (solid circles) but not to formation of sulfoacetaldehyde (open triangles).
Heterologous expression of the Adh candidate gene and confirmation of its role in conversion of the HT-desulfination product acetaldehyde to acetate.
Pden_2366 was overexpressed in E. coli and purified as a His6-tagged protein via affinity chromatography (Fig. 4B). The protein catalyzed an NAD+-dependent oxidation of acetaldehyde to acetate, as confirmed when either authentic acetaldehyde (not shown) or the acetaldehyde that was formed during the Hpa reaction, i.e., in combination with purified Hpa/Tpa, HT, and pyruvate, was used for the reaction, and when followed discontinuously by DIH/HPLC-UV, HILIC-UV-ELSD, and an acetate-concentration assay (Fig. 6): acetate was formed in stoichiometric amounts relative to alanine formation and NAD+-to-NADH conversion. The purified enzyme tested negative for a reaction with HT in the presence of NAD+, and no taurine was formed; hence, Pden_2366 did not catalyze an HT-dehydrogenase reaction.
Fig 6.
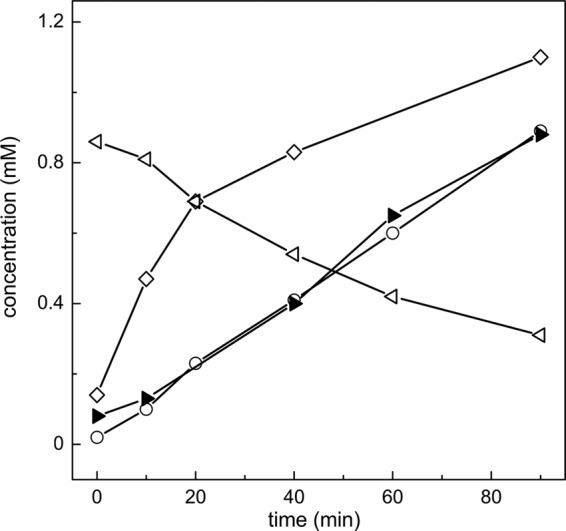
Reaction of purified recombinant Hpa in combination with purified recombinant Adh. Samples were taken at intervals for subsequent analyses by HPLC and determination of acetate concentration (see Materials and Methods). During a reaction of Hpa with HT and pyruvate, and in the presence of Adh and NAD+, a formation of acetate (solid triangles), but not of acetaldehyde (not shown; Fig. 5B), was observed. The two reactions were monitored as formation of alanine (open diamonds) and as conversion of NAD+ (open triangles) to NADH (open circles), respectively. Notably, HT could not be analyzed by HPLC in this assay, since the essential Adh reaction buffer component pyrophosphate coeluted with HT during HILIC-UV-ELSD.
DISCUSSION
The carbon-sulfur bond in most organosulfonates is very stable (36, 37), and the removal of sulfonate groups by bacteria has been recognized to involve many desulfonative pathways, enzymes, and reaction mechanisms. For dissimilation of substituted C2-sulfonates (e.g., taurine, isethionate, and sulfoacetate), the known pathways converge at the level of sulfoacetaldehyde for desulfonation through Xsc (6). Alternatively, taurine as a sulfur source is desulfonated via a specific inducible monooxygenase (TauD) (38). For the dissimilation of substituted C3-sulfonates (3-sulfolactate and l-cysteate), two desulfonative enzymes have recently been described (see review in reference 39), 3-sulfolactate sulfolyase (SuyAB) (EC 4.4.1.24) (40) and l-cysteate sulfolyase (CuyA) (EC 4.4.1.25) (41). Finally, for organosulfonates that lack substituents that aid in the destabilization of the carbon-sulfur bond, e.g., propanesulfonate or benzenesulfonate, a less subtle mechanism is employed to remove the sulfonate group, oxygenation (e.g., see review in reference 42).
The carbon-sulfur bond in most organosulfinates at physiological pH is also stable; however, their sulfinic acids can easily auto-oxidize/disproportionate to give the corresponding sulfonic acids, S-esters, or thiosulfonic acids (5, 43, 44). A higher reactivity of the HT-sulfinate group than of the taurine-sulfonate group under physiological conditions is reflected in the functions of HT to detoxify sulfide in deep-sea invertebrates (45–48) and to act as an antioxidant and free-radical-trapping agent in mammalian cells (49, 50). However, HT is stable under the conditions that we used, i.e., in oxic culture medium.
P. denitrificans is able to utilize not only taurine and isethionate (Table 1) but also HT for growth. The growth experiments (Fig. 2A and B) confirmed a mass balance for the utilization of HT carbon and biomass formation and for the release of HT sulfur and nitrogen. Strain PD1222 apparently suffered from sulfite stress due to the increased amount of sulfite released into the growth medium (51), a phenomenon that is observed also for other organosulfonate-degrading, sulfite-excreting bacteria that do not express, or contain, a SorA-type sulfite dehydrogenase for effective detoxification of sulfite, e.g., for growth of Ruegeria pomeroyi DSS-3 with >4 mM cysteate (52). However, there is sulfite-to-sulfate conversion in cultures (Fig. 2B), significantly faster than auto-oxidation of sulfite, and thus some indication for an enzymatically catalyzed (though slow) conversion of sulfite to sulfate in P. denitrificans PD1222. Strain PD1222 contains a sulfite exporter, Pden_1641 (TauZ, Table 1), a gene that is part of the taurine utilization operon (Pden_1648–1640), in order to decrease the cytoplasmic toxicity of sulfite (discussed further below).
The results obtained from the proteomic and enzymatic work strongly support the option (Fig. 1) that the initial reaction on HT is catalyzed by Hpa Pden_4273, which is an archetypal Tpa in P. denitrificans PD1222. No indication for a conversion of HT to taurine, e.g., through Hdh, was obtained (Fig. 1), and hence, it still remains unclear whether an HT-to-taurine conversion is also enzymatically catalyzed (12) or proceeds solely in radical-scavenging reactions (50, 53). The HT pathway is inducible, as is the taurine pathway (Table 1), and two highly inducible proteins (Fig. 3) were attributed by PF-MS to two candidate genes that, when cloned and overexpressed as pure proteins (Fig. 4), represented the two enzymes necessary to convert HT to a central metabolite, acetate (Fig. 5 and 6). There was no sulfinoacetaldehyde formation visible with HT as the substrate for deamination, but instead acetaldehyde and sulfite formation (Fig. 5B). This indicates that the Hpa reaction is followed by three nonenzymatic reactions: (i) the HT-deamination product sulfinoacetaldehyde spontaneously desulfinates to ethenol and sulfur dioxide, which is mechanistically analogous to the spontaneous decarboxylation of beta-keto acids, and furthermore, (ii) ethenol tautomerizes to acetaldehyde and (iii) sulfur dioxide hydrates to sulfite (Fig. 7), as was observed (Fig. 5B) previously with an oxoglutarate-coupled aminotransferase that represented most likely a beta-alanine:pyruvate aminotransferase in Achromobacter superficialis (6, 22). Hence, the observed spontaneous desulfination of sulfinoacetaldehyde (Fig. 7) stands in contrast to the enzymatically catalyzed desulfonation of the analogue in taurine degradation, sulfoacetaldehyde, and to the enzymatically catalyzed desulfinations known so far, of three other organosulfinates: (i) the sulfinate analogue of aspartate, cysteine sulfinate, is desulfinated by aspartate beta-decarboxylase (EC 4.1.1.12) of Alcaligenes faecalis (54) and (engineered) aspartate aminotransferase (EC 2.6.1.1) of E. coli (55), as well as by a cysteine sulfinate desulfinase/selenocysteine lyase (EC 4.4.1.16) of E. coli (56). Moreover, (ii) 2′-hydroxybiphenyl-2-sulfinate is desulfinated by a hydrolase (EC 3.13.1.3) involved in dibenzothiophene sulfur acquisition in Rhodococcus erythropolis (57), and (iii) 3-sulfinopropionyl-CoA is desulfinated by a new desulfinase, an acyl-CoA dehydrogenase-superfamily enzyme (EC 1.3.8.x) involved in dissimilation of 3,3′-dithiodipropionate in Advenella mimigardefordensis (58).
Fig 7.
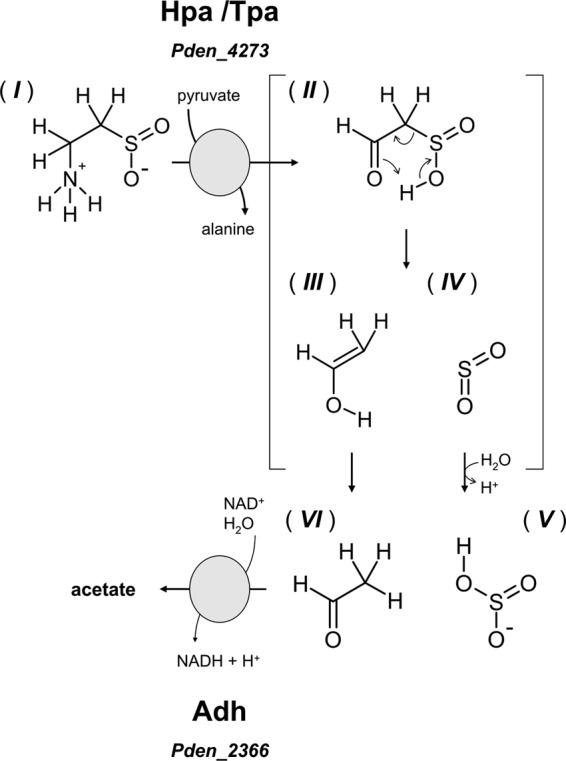
Illustration of the two enzymatically catalyzed reactions for HT utilization in Paracoccus denitrificans PD1222 and of the proposed mechanism of spontaneous sulfinoacetaldehyde desulfination to give acetaldehyde and sulfite. Compounds: I, hypotaurine; II, sulfinoacetaldehyde; III, ethenol; IV, sulfur dioxide; VI, acetaldehyde; V, sulfite.
Tpa was initially discovered in Pseudomonas aeruginosa TAU5 (59), purified from Bilophila wadsworthia (15), and detected in several other bacteria that degrade taurine under different metabolic situations (e.g., references 8, 17–19, and 23). Tpa is a homomultimer with a subunit of about 51 kDa (Fig. 3 and 4), and the cofactor is pyridoxal-5′-phosphate (15). A wider substrate range of Tpa has previously been recognized (15), when Tpa deaminated taurine (rate, 100%) and HT (rate, 218%), as found with the Hpa/Tpa of P. denitrificans (Table 2; Fig. 5A and B). Hence, in P. denitrificans PD1222, we found a physiological function of this wider substrate range of Hpa/Tpa, as the first inducible enzyme to access HT as a carbon, nitrogen, and energy source. Pyruvate as the specific acceptor of the amino group for Hpa/Tpa is regenerated by an HT-inducible alanine dehydrogenase (Ald) (16) in P. denitrificans PD1222 (Table 2). Adh Pden_2366, the third inducible enzyme involved in HT metabolism, oxidizes and detoxifies acetaldehyde to acetate. The in vitro activity of Adh was strictly dependent on the presence of pyrophosphate in the reaction buffer (33). The HT-inducible alcohol dehydrogenase, Pden_2367, which we found (see Fig. S1 and Table S1 in the supplemental material) and which is encoded directly downstream of the identified Adh gene, Pden_2366, appears to be gratuitously expressed during growth with HT, presumably through a coinduction of Pden_2366-2367.
The genome of P. denitrificans PD1222 (two chromosomes) harbors three candidate gene clusters for organosulfonate degradation (Table 1), first the taurine operon (transport, TauXY, Xsc, Pta, and sulfite export) (chromosome 1) and then a gene cluster each for isethionate (chromosome 2) and sulfoacetate (chromosome 1) degradation, based on our previous work with orthologous gene clusters (9, 60, 61). The three desulfonation pathways would converge at the level of sulfoacetaldehyde, but each of these clusters (presumed operons) contains its own set of Xsc and Pta genes (Table 1), which share up to 99% sequence identity. The proteomic data (see Fig. S1 and Table S1 in the supplemental material) confirmed that at least TauX, probably Xsc, and presumably the whole of the taurine operon are expressed in strain PD1222 during growth with taurine, as well as during growth with HT (Table 2). This coinduction most likely reflects the structural analogy of HT and taurine as inducers.
However, the coinduction of the taurine pathway might also play a key role for the HT catabolism in strain PD1222. It might transport HT into the cell via the taurine transport system (TauKLM) (9) and extrude toxic sulfite via the sulfite exporter (TauZ; see above), both of which are coencoded in the taurine operon (9) (Table 1). The taurine operon is strictly regulated, as seen in comparison to growth with acetate (Table 2; see also Fig. S1 in the supplemental material), and also the HT pathway is independently regulated from the taurine operon (Table 2 and Fig. 3; see also Fig. S1), as is the isethionate pathway (9). There are candidate regulator genes cocarried with each of the gene clusters (presumed operons) that encode HT, taurine, isethionate, and sulfoacetate uptake and degradation in strain PD1222 (Table 1). A more detailed picture of the regulation of these many sulfinate and sulfonate utilization genes can readily be addressed in a future study, e.g., by transcriptional and proteomic analyses.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Klaus Hollemeyer for MALDI-TOF MS and to Thomas Huhn and Bernhard Schink for helpful discussion.
The work of A.-K.F. and M.W. is supported by the Konstanz Research School Chemical Biology (KoRS-CB), and the work of D.S. is supported by a Deutsche Forschungsgemeinschaft (DFG) grant (SCHL 1936/1-1) and by the University of Konstanz, the KoRS-CB, and the Konstanz Young Scholar Fund (YSF).
Footnotes
Published ahead of print 19 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00307-13.
REFERENCES
- 1. Yin M, Palmer HR, Fyfe-Johnson AL, Bedford JJ, Smith RA, Yancey PH. 2000. Hypotaurine, N-methyltaurine, taurine, and glycine betaine as dominant osmolytes of vestimentiferan tubeworms from hydrothermal vents and cold seeps. Physiol. Biochem. Zool. 73:629–637 [DOI] [PubMed] [Google Scholar]
- 2. Yancey PH, Blake WR, Conley J. 2002. Unusual organic osmolytes in deep-sea animals: adaptations to hydrostatic pressure and other perturbants. Comp. Biochem. Phys. A 133:667–676 [DOI] [PubMed] [Google Scholar]
- 3. Rosenberg NK, Lee RW, Yancey PH. 2006. High contents of hypotaurine and thiotaurine in hydrothermal-vent gastropods without thiotrophic endosymbionts. J. Exp. Zool. A 305:655–662 [DOI] [PubMed] [Google Scholar]
- 4. Wright CE, Tallan HH, Lin YY, Gaull GE. 1986. Taurine: biological update. Annu. Rev. Biochem. 55:427–453 [DOI] [PubMed] [Google Scholar]
- 5. Huxtable RJ. 1992. Physiological actions of taurine. Physiol. Rev. 72:101–163 [DOI] [PubMed] [Google Scholar]
- 6. Cook AM, Denger K. 2002. Dissimilation of the C2 sulfonates. Arch. Microbiol. 179:1–6 [DOI] [PubMed] [Google Scholar]
- 7. Kondo H, Anada H, Ohsawa K, Ishimoto M. 1971. Formation of sulfoacetaldehyde from taurine in bacterial extracts. J. Biochem. 69:621–623 [PubMed] [Google Scholar]
- 8. Ruff J, Denger K, Cook AM. 2003. Sulphoacetaldehyde acetyltransferase yields acetyl phosphate: purification from Alcaligenes defragrans and gene clusters in taurine degradation. Biochem. J. 369:275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brüggemann C, Denger K, Cook AM, Ruff J. 2004. Enzymes and genes of taurine and isethionate dissimilation in Paracoccus denitrificans. Microbiology 150:805–816 [DOI] [PubMed] [Google Scholar]
- 10. Gorzynska AK, Denger K, Cook AM, Smits TH. 2006. Inducible transcription of genes involved in taurine uptake and dissimilation by Silicibacter pomeroyi DSS-3T. Arch. Microbiol. 185:402–406 [DOI] [PubMed] [Google Scholar]
- 11. Sumizu K. 1962. Oxidation of hypotaurine in rat liver. Biochim. Biophys. Acta 63:210–212 [DOI] [PubMed] [Google Scholar]
- 12. Stipanuk MH. 2004. Role of the liver in regulation of body cysteine and taurine levels: a brief review. Neurochem. Res. 29:105–110 [DOI] [PubMed] [Google Scholar]
- 13. Kondo H, Ishimoto M. 1987. Taurine dehydrogenase. Methods Enzymol. 143:496–499 [DOI] [PubMed] [Google Scholar]
- 14. Denger K, Smits THM, Cook AM. 2006. Genome-enabled analysis of the utilization of taurine as sole source of carbon or nitrogen by Rhodobacter sphaeroides 2.4.1. Microbiology 152:3197–3206 [DOI] [PubMed] [Google Scholar]
- 15. Laue H, Cook AM. 2000. Biochemical and molecular characterization of taurine:pyruvate aminotransferase from the anaerobe Bilophila wadsworthia. Eur. J. Biochem. 267:6841–6848 [DOI] [PubMed] [Google Scholar]
- 16. Laue H, Cook AM. 2000. Purification, properties and primary structure of alanine dehydrogenase involved in taurine metabolism in the anaerobe Bilophila wadsworthia. Arch. Microbiol. 174:162–167 [DOI] [PubMed] [Google Scholar]
- 17. Denger K, Ruff J, Schleheck D, Cook AM. 2004. Rhodococcus opacus expresses the xsc gene to utilize taurine as a carbon source or as a nitrogen source but not as a sulfur source. Microbiology 150:1859–1867 [DOI] [PubMed] [Google Scholar]
- 18. Baldock MI, Denger K, Smits THM, Cook AM. 2007. Roseovarius sp. strain 217: aerobic taurine dissimilation via acetate kinase and acetate-CoA ligase. FEMS Microbiol. Lett. 271:202–206 [DOI] [PubMed] [Google Scholar]
- 19. Krejčík Z, Denger K, Weinitschke S, Hollemeyer K, Pačes V, Cook AM, Smits TH. 2008. Sulfoacetate released during the assimilation of taurine-nitrogen by Neptuniibacter caesariensis: purification of sulfoacetaldehyde dehydrogenase. Arch. Microbiol. 190:159–168 [DOI] [PubMed] [Google Scholar]
- 20. Stadtman ER, Novelli GD, Lipmann F. 1951. Coenzyme A function in and acetyl transfer by the phosphotransacetylase system. J. Biol. Chem. 191:365–376 [PubMed] [Google Scholar]
- 21. Toyama S, Misono H, Soda K. 1972. Crystalline taurine: alpha-ketoglutarate aminotransferase from Achromobacter superficialis. Biochem. Biophys. Res. Comm. 46:1374–1379 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka H, Toyama S, Tsukahar H, Soda K. 1974. Transamination of hypotaurine by taurine:alpha-ketoglutarate aminotransferase. FEBS Lett. 45:111–113 [DOI] [PubMed] [Google Scholar]
- 23. Denger K, Ruff J, Rein U, Cook AM. 2001. Sulphoacetaldehyde sulpho-lyase (EC 4.4.1.12) from Desulfonispora thiosulfatigenes: purification, properties and primary sequence. Biochem. J. 357:581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thurnheer T, Kohler T, Cook AM, Leisinger T. 1986. Orthanilic acid and analogs as carbon-sources for bacteria—growth physiology and enzymatic desulfonation. J. Gen. Microbiol. 132:1215–1220 [Google Scholar]
- 25. Denger K, Laue H, Cook AM. 1997. Anaerobic taurine oxidation: a novel reaction by a nitrate-reducing Alcaligenes sp. Microbiology 143:1919–1924 [DOI] [PubMed] [Google Scholar]
- 26. Cunningham C, Tipton KF, Dixon HBF. 1998. Conversion of taurine into N-chlorotaurine (taurine chloramine) and sulphoacetaldehyde in response to oxidative stress. Biochem. J. 330:939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kennedy SIT, Fewson CA. 1968. Enzymes of the mandelate pathway in bacterium N.C.I.B. 8250. Biochem. J. 107:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 29. Sörbo B. 1987. Sulfate: turbidimetric and nephelometric methods. Methods Enzymol. 143:3–6 [DOI] [PubMed] [Google Scholar]
- 30. Fachgruppe Wasserchemie in der Gesellschaft Deutscher Chemiker NWiD 1996. Deutsche Einheitsverfahren zur Wasser-, Abwasser- und Schlammuntersuchung, Band II. VCH, Weinheim, Germany [Google Scholar]
- 31. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 32. Schmidt A, Müller N, Schink B, Schleheck D. 2013. A proteomic view at the biochemistry of syntrophic butyrate oxidation in Syntrophomonas wolfei. PLoS One 8:e56905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taniyama K, Itoh H, Takuwa A, Sasaki Y, Yajima S, Toyofuku M, Nomura N, Takaya N. 2012. Group X aldehyde dehydrogenases of Pseudomonas aeruginosa PAO1 degrade hydrazones. J. Bacteriol. 194:1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Denger K, Weinitschke S, Smits TH, Schleheck D, Cook AM. 2008. Bacterial sulfite dehydrogenases in organotrophic metabolism: separation and identification in Cupriavidus necator H16 and in Delftia acidovorans SPH-1. Microbiology 154:256–263 [DOI] [PubMed] [Google Scholar]
- 35. Cook AM. 1987. Biodegradation of s-triazine xenobiotics. FEMS Microbiol. Rev. 46:93–116 [Google Scholar]
- 36. Wagner FC, Reid EE. 1931. The stability of the carbon-sulfur bond in some aliphatic sulfonic acids. J. Am. Chem. Soc. 53:3407–3413 [Google Scholar]
- 37. Freedman LD, Doak GO. 1957. The preparation and properties of phosphonic acids. Chem. Rev. 57:479–523 [Google Scholar]
- 38. Eichhorn E, van der Ploeg JR, Kertesz MA, Leisinger T. 1997. Characterization of α-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J. Biol. Chem. 272:23031–23036 [DOI] [PubMed] [Google Scholar]
- 39. Cook AM, Denger K, Smits THM. 2006. Dissimilation of C3-sulfonates. Arch. Microbiol. 185:83–90 [DOI] [PubMed] [Google Scholar]
- 40. Rein U, Gueta R, Denger K, Ruff J, Hollemeyer K, Cook AM. 2005. Dissimilation of cysteate via 3-sulfolactate sulfo-lyase and a sulfate exporter in Paracoccus pantotrophus NKNCYSA. Microbiology 151:737–747 [DOI] [PubMed] [Google Scholar]
- 41. Denger K, Smits TH, Cook AM. 2006. L-cysteate sulpho-lyase, a widespread pyridoxal 5′-phosphate-coupled desulphonative enzyme purified from Silicibacter pomeroyi DSS-3T. Biochem. J. 394:657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kertesz MA. 2000. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24:135–175 [DOI] [PubMed] [Google Scholar]
- 43. Horner L, Basedow OH. 1958. Zum Mechanismus der Autoxydation der Benzolsulfinsäure. Chem. Ber. 612:108–131 [Google Scholar]
- 44. Schubart R. 2000. Sulfinic acids and derivatives. Ullmann's encyclopedia of industrial chemistry. Wiley-VCH Verlag, Weinheim, Germany [Google Scholar]
- 45. Fiess JC, Hom JR, Hudson HA, Kato C, Yancey PH. 2002. Phosphodiester amine, taurine and derivatives, and other osmolytes in vesicomyid bivalves: correlations with depth and symbiont metabolism. Cah. Biol. Mar. 43:337–340 [Google Scholar]
- 46. Ortega JA, Ortega JM, Julian D. 2008. Hypotaurine and sulfhydryl-containing antioxidants reduce H2S toxicity in erythrocytes from a marine invertebrate. J. Exp. Biol. 211:3816–3825 [DOI] [PubMed] [Google Scholar]
- 47. Pruski AM, Fiala-Médioni A, Prodon R, Colomindes JC. 2000b. Thiotaurine is a biomarker for sulfide-based symbiosis in deep-sea bivalves. Limnol. Oceanogr. 45:1960–1967 [Google Scholar]
- 48. Yancey PH, Ishikawa J, Meyer B, Girguis PR, Lee RW. 2009. Thiotaurine and hypotaurine contents in hydrothermal-vent polychaetes without thiotrophic endosymbionts: correlation with sulfide exposure. J. Exp. Zool. A 311:439–447 [DOI] [PubMed] [Google Scholar]
- 49. Aruoma OI, Halliwell B, Hoey BM, Butler J. 1988. The antioxidant action of taurine, hypotaurine, and their metabolic precursors. J. Biochem. 256:251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fontana M, Amendola D, Orsini E, Boffi A, Pecci L. 2005. Oxidation of hypotaurine and cysteine sulphinic acid by peroxynitrite. Biochem. J. 389:233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gunnison AF. 1981. Sulphite toxicity: a critical review of in vitro and in vivo data. Food Cosmet. Toxicol. 19:667–682 [DOI] [PubMed] [Google Scholar]
- 52. Lehmann S. 2013. Sulfite dehydrogenases in organotrophic bacteria: enzymes, genes and regulation. PhD thesis University of Konstanz, Konstanz, Germany [Google Scholar]
- 53. Pecci L, Montefoschi G, Fontana M, Duprè S, Costa M, Cavallini D. 2000. Hypotaurine and superoxide dismutase: protection of the enzyme against inactivation by hydrogen peroxide and peroxidation to taurine. Adv. Exp. Med. Biol. 483:163–168 [DOI] [PubMed] [Google Scholar]
- 54. Soda K, Novogrodsky A, Meister A. 1964. Enzymatic desulfination of cysteine sulfinic acid. Biochemistry 3:1450–1454 [DOI] [PubMed] [Google Scholar]
- 55. Fernandez FJ, de Vries D, Pena-Soler E, Coll M, Christen P, Gehring H, Vega MC. 2012. Structure and mechanism of a cysteine sulfinate desulfinase engineered on the aspartate aminotransferase scaffold. Biochim. Biophys. Acta 1824:339–349 [DOI] [PubMed] [Google Scholar]
- 56. Mihara H, Kurihara T, Yoshimura T, Soda K, Esaki N. 1997. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 272:22417–22424 [DOI] [PubMed] [Google Scholar]
- 57. Nakayama N, Matsubara T, Ohshiro T, Moroto Y, Kawata Y, Koizumi K, Hirakawa Y, Suzuki M, Maruhashi K, Izumi Y, Kurane R. 2002. A novel enzyme, 2′-hydroxybiphenyl-2-sulfinate desulfinase (DszB), from a dibenzothiophene-desulfurizing bacterium Rhodococcus erythropolis KA2-5-1: gene overexpression and enzyme characterization. Biochim. Biophys. Acta 1598:122–130 [DOI] [PubMed] [Google Scholar]
- 58. Schürmann M, Deters A, Wübbeler JH, Steinbüchel A. 2013. A novel 3-sulfinopropionyl coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis strain DPN7T acting as a key enzyme during catabolism of 3,3′-dithiodipropionic acid is a member of the acyl-CoA dehydrogenase superfamily. J. Bacteriol. 195:1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shimamoto G, Berk RS. 1979. Catabolism of taurine in Pseudomonas aeruginosa. Biochim. Biophys. Acta 569:287–292 [DOI] [PubMed] [Google Scholar]
- 60. Weinitschke S, Sharma PI, Stingl U, Cook AM, Smits TH. 2010. Gene clusters involved in isethionate degradation by terrestrial and marine bacteria. Appl. Environ. Microbiol. 76:618–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Weinitschke S, Hollemeyer K, Kusian B, Bowien B, Smits TH, Cook AM. 2010. Sulfoacetate is degraded via a novel pathway involving sulfoacetyl-CoA and sulfoacetaldehyde in Cupriavidus necator H16. J. Biol. Chem. 285:35249–35254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


