Abstract
The Gram-positive anaerobic bacterium Clostridium perfringens is pathogenic to humans and animals, and the production of its toxins is strictly regulated during the exponential phase. We recently found that the 5′ leader sequence of the colA transcript encoding collagenase, which is a major toxin of this organism, is processed and stabilized in the presence of the small RNA VR-RNA. The primary colA 5′-untranslated region (5′UTR) forms a long stem-loop structure containing an internal bulge and masks its own ribosomal binding site. Here we found that VR-RNA directly regulates colA expression through base pairing with colA mRNA in vivo. However, when the internal bulge structure was closed by point mutations in colA mRNA, translation ceased despite the presence of VR-RNA. In addition, a mutation disrupting the colA stem-loop structure induced mRNA processing and ColA-FLAG translational activation in the absence of VR-RNA, indicating that the stem-loop and internal bulge structure of the colA 5′ leader sequence is important for regulation by VR-RNA. On the other hand, processing was required for maximal ColA expression but was not essential for VR-RNA-dependent colA regulation. Finally, colA processing and translational activation were induced at a high temperature without VR-RNA. These results suggest that inhibition of the colA 5′ leader structure through base pairing is the primary role of VR-RNA in colA regulation and that the colA 5′ leader structure is a possible thermosensor.
INTRODUCTION
Clostridium perfringens is a Gram-positive, anaerobic, spore-forming bacterium that causes diseases in humans and other animals (1, 2). Type A is the most common toxinotype of this bacterium and causes gas gangrene (clostridial myonecrosis) and food poisoning in humans. Whole-genome sequencing has revealed that C. perfringens has only a few genes that encode enzymes for amino acid biosynthesis, and thus its nutritional sources need to be generated through host cell degradation (3). Most genes that encode C. perfringens extracellular toxins and enzymes are expressed at the mid- and late exponential phases (2, 4, 5, 6). Therefore, the expression of genes encoding extracellular enzymes and toxins that are required to gain nutrients from host cells should be controlled tightly for efficient host cell infection and proliferation.
The VirR-VirS two-component system (TCS) is a key global regulator of C. perfringens gene expression (7, 8, 9). Phosphorylated VirR binds directly to a target promoter with two direct repeated sequences (VirR box) and then activates the transcription of target genes, including the θ-toxin gene (pfoA), the alpha-clostripain gene (ccp), and the vrr gene, encoding the small RNA (sRNA), i.e., VR-RNA (8, 10). The 386-nucleotide (nt) VR-RNA regulates the expression of plc (alpha-toxin), colA (κ-toxin), cpd2, and several housekeeping genes, suggesting that the sRNA plays a critical role in the virulence and metabolism of C. perfringens (9, 11, 12, 13, 14). Although the vrr gene has a potential open reading frame (ORF) (CPE0957; 73 amino acids), studies of truncated VR-RNA mutants have revealed that this ORF is not required for target gene regulation, indicating that VR-RNA functions as a bona fide sRNA (11, 15). Collagenase, encoded by colA, is a primary toxin of C. perfringens that degrades collagen, which is a main component of the connective tissues of host cells. Collagenase could play a role in clostridial virulence in terms of spreading toxins and cells to host tissue, but it is not essential for gas gangrene in the mouse myonecrosis model (16).
We recently found that VR-RNA induces cleavage of the colA mRNA 5′-untranslated region (5′UTR) and stabilizes colA mRNA (15). We predicted that the colA mRNA 5′UTR would form a stem-loop structure containing an internal bulge, which would mask the colA ribosome binding site and inhibit translation. Because this structure is broken in processed colA mRNA, processing enhanced the translational efficiency of colA and stabilized the transcript. The 3′ regions of VR-RNA and the colA mRNA 5′UTR have long complementary sequences, suggesting that VR-RNA regulates colA through base pairing. However, this has not been confirmed experimentally in vivo. In addition, the precise roles of the 5′ leader structure and the processing remain unclear.
Here we confirmed the importance of complementarity between VR-RNA and colA in C. perfringens cells and determined which complementary region is required for regulation. The VR-RNA-dependent activation of colA translation depended on the bulge structure in the colA mRNA 5′UTR, suggesting that VR-RNA and colA mRNA initially interact in a single-stranded region. On the other hand, only disruption of the secondary structure of colA mRNA induced the mRNA processing and translational activation of colA, even in the absence of VR-RNA. Thus, the primary function of VR-RNA on colA regulation could be to prevent the stem-loop structure of colA mRNA by base pairing. Our findings also indicated that a high temperature could disrupt the colA mRNA structure, regardless of whether VR-RNA was present. Processing is dispensable for regulation by VR-RNA at low temperatures but essential for high-temperature-dependent regulation. The present findings also suggest that disruption or melting of the colA mRNA conformation is essential for translational regulation and that the colA 5′ leader sequence is a possible RNA thermosensor.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Clostridium perfringens strain 13 (17) and its derivative strains harboring plasmids were cultured on brain heart infusion (BHI)-sheep blood agar plates (BD Difco, Franklin Lakes, NJ, and Nippon Biotest Laboratories Inc., Tokyo, Japan) at 37°C under anaerobic conditions, using an Anaeropack system (Mitsubishi Gas Chemical Co. Inc., Tokyo, Japan), or grown in Gifu anaerobic medium (GAM) broth (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) supplemented with 25 μg ml−1 chloramphenicol. Escherichia coli JM109 was cultured in LB medium supplemented with either 50 μg ml−1 ampicillin or 25 μg ml−1 chloramphenicol.
Plasmid construction.
Tables S1 and S2 in the supplemental material list the respective oligonucleotides and plasmids that were used in this study. VR-RNA and colA-gst coexpression vectors were constructed as follows. A DNA fragment containing the vrr promoter region, the vrr gene, and an intrinsic terminator was amplified from the genomic DNA of C. perfringens strain 13 by a PCR using primers NOB-0390 and NOB-0393. The PCR products were digested with SpeI and NaeI and ligated to SpeI/NaeI-digested pJIR418 and pCPE33 to generate pVrr and pCPE111, respectively.
The colA-FLAG expression vector, pCPE94, was constructed by amplifying colA-FLAG fragments by PCR with NOB-0068 and NOB-0084 primers and then cloning them into the BamHI and SalI sites of pCP (15).
A point mutation was introduced into vrr by two independent PCR amplifications with the NOB-0390–NOB-0401 and NOB-0400–NOB-0393 primer pairs, using pVrr as the template DNA. The NOB-0400 and NOB-401 primers are complementary. The two PCR products were then mixed to serve as the template for a second PCR using the NOB-0390 and NOB-0393 primers. The PCR product was ligated at the SpeI and NaeI sites of pJIR418 and pCPE111 to generate pVrrmut1 and pCPE112, respectively. Point mutations were introduced into the vrr, colA-gst, and colA-FLAG genes as described above, using the primers listed in Table S1 in the supplemental material.
The coding sequences of VR-RNA were amplified from the VR-RNA expression vectors by a PCR using the NOB-0383 and NOB-0386 primers to create the DNA template for transcription in vitro. The PCR products were ligated into the EcoRI-HindIII sites of pGEM3zf (+) (Promega, Madison, WI) to generate pNOE40. Each mutation was confirmed by DNA sequencing.
RNA synthesis in vitro.
Probe RNAs were transcribed using T7 RNA polymerase (Epicentre Biotechnologies, Madison, WI). HindIII-digested pNOE40-46 and PCR products generated from the colA-gst expression vectors by use of primers NOB-0370 and NOB-0011 served as templates for transcription of the VR-RNA and colA mRNA 5′UTR variants, respectively, in vitro. The synthesized RNAs were resolved by 6% urea-denaturing polyacrylamide gel electrophoresis (PAGE), extracted, and dissolved in sterile double-deionized water.
RNA extraction and Northern blot analysis.
Total RNAs extracted from C. perfringens derivatives grown in GAM broth were Northern blotted as described previously (6). Digoxigenin-labeled DNA probes were generated using DIG-High Prime kits according to the supplier's instructions (DIG application manual; Roche, Basel, Switzerland). Template DNAs for generating colA and VR-RNA probes were amplified from strain 13 genomic DNA by PCRs using the described primers.
Gel mobility shift assay.
Interaction between VR-RNA and colA mRNA was analyzed as described previously (15). We end labeled in vitro-synthesized colA mRNA (0.5 pmol) by using 1 μl of T4 polynucleotide kinase (PNK), 2 μl of 10× PNK buffer, and 5 μl of [γ-32P]ATP in 20 μl of reaction buffer at 37°C for 30 min. T4 PNK was then inactivated by heating at 95°C for 2 min. Unlabeled VR-RNA (1, 2, or 4 nM) and 5′-end-labeled colA mRNAs (1 nM) were incubated at 37°C for 30 min in reaction mixtures (10 μl) comprising 20 mM HEPES, pH 7.9, containing 100 mM KCl, 1 mM MgCl2, 1 mM dithiothreitol, and 1 μg of tRNA. Loading dye (5 μl) containing 50% glycerol, 0.1% bromophenol blue, and 0.1% xylene cyanol was added, and then the mixtures were resolved in 4% nondenaturing acrylamide gels in 1× Tris-borate-EDTA (TBE) buffer at 4°C.
Western blot analysis.
Extracellular proteins in culture supernatants were precipitated with 10% (wt/vol) trichloroacetic acid, washed with cold acetone, and resuspended in LETS buffer (100 mM LiCl, 10 mM EDTA, 10 mM Tris-HCl, pH 7.5, and 1% [wt/vol] sodium dodecyl sulfate). A volume equivalent to a culture optical density at 600 nm (OD600) of 0.02 was separated by SDS-PAGE and then electroblotted onto polyvinylidene difluoride membranes. Nonspecific binding on the membranes was blocked with 2.5% skim milk in Tris-buffered saline containing 0.2% Tween 80, and then the membranes were probed with anti-glutathione S-transferase (anti-GST) or anti-DYKDDDDK (Wako Pure Chemical Industries, Ltd., Osaka, Japan) diluted 1:5,000. Horseradish peroxidase-conjugated anti-mouse secondary antibodies (GE Healthcare UK Ltd., Buckinghamshire, England) were used at a dilution of 1:50,000, and then bound antibodies were detected using an Immunostar LD system (Wako Pure Chemical Industries, Ltd.).
RNA structure mapping.
The colA mRNA 5′UTR synthesized in vitro was dephosphorylated by incubation with calf intestinal alkaline phosphatase (CIAP; TaKaRa Bio Co. Ltd., Shiga, Japan) according to the supplier's instructions and then end labeled with T4 PNK as described above. Labeled RNAs were resolved by electrophoresis on 6% urea-denaturing polyacrylamide gels, extracted, purified, and dissolved in sterile double-deionized water. The RNAs were digested with 0.2, 0.02, or 0.002 U of T1 nuclease, 5, 0.5, or 0.05 U of S1 nuclease, or 0.2, 0.02, or 0.002 U of V1 nuclease according to the manufacturer's instructions (Life Technologies Inc., Carlsbad, CA). The T1, S1, and V1 RNases specifically cleave RNA after unpaired guanine residues, single-stranded RNA, and double-stranded RNA, respectively. The digested RNAs were extracted with phenol-chloroform, precipitated with ethanol, and resolved by electrophoresis on 8 M urea-6% denaturing polyacrylamide gels.
RESULTS
The colA mRNA 5′UTR forms a stem-loop structure in vitro.
The predicted long stem-loop structure of the colA mRNA 5′UTR masks the colA Shine-Dalgarno (SD) sequence and inhibits translation (15). We structurally analyzed the colA mRNA 5′UTR by using RNA structure mapping. Figure 1 shows that the colA mRNA 5′UTR transcribed in vitro formed a long stem-loop. The SD sequence was located in a single-stranded region of the bulge structure, suggesting that the upstream or downstream stem structure inhibits ribosome binding to the SD sequence, as we reported previously (15). Thus, we confirmed that the actual secondary structure of the colA 5′UTR corresponds to the predicted structure.
Fig 1.
Secondary structure prediction of colA mRNA 5′UTR. (A) RNA structure mapping of colA mRNA 5′UTR synthesized in vitro. Digested RNAs were resolved by electrophoresis on 6% urea-acrylamide gels. T1, S1, and V1, RNAs digested by RNase T1, RNase S1, and RNase V1, respectively. RNAs hydrolyzed in alkali (OH) and denatured RNAs digested by RNase T1 (G) are also shown (see Materials and Methods). Areas corresponding to the bulge in the stem-loop structure and the colA SD sequence are indicated. (B) Probed colA mRNA 5′UTR structure predicted using the RNAfold WebServer site (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Guanidine residues cleaved with RNase T1 are circled. Arrows and lines connected to filled circles represent RNase S1 and RNase V1 cleavage sites, respectively. Processing sites induced by VR-RNA have been identified (15) and are indicated by filled triangles.
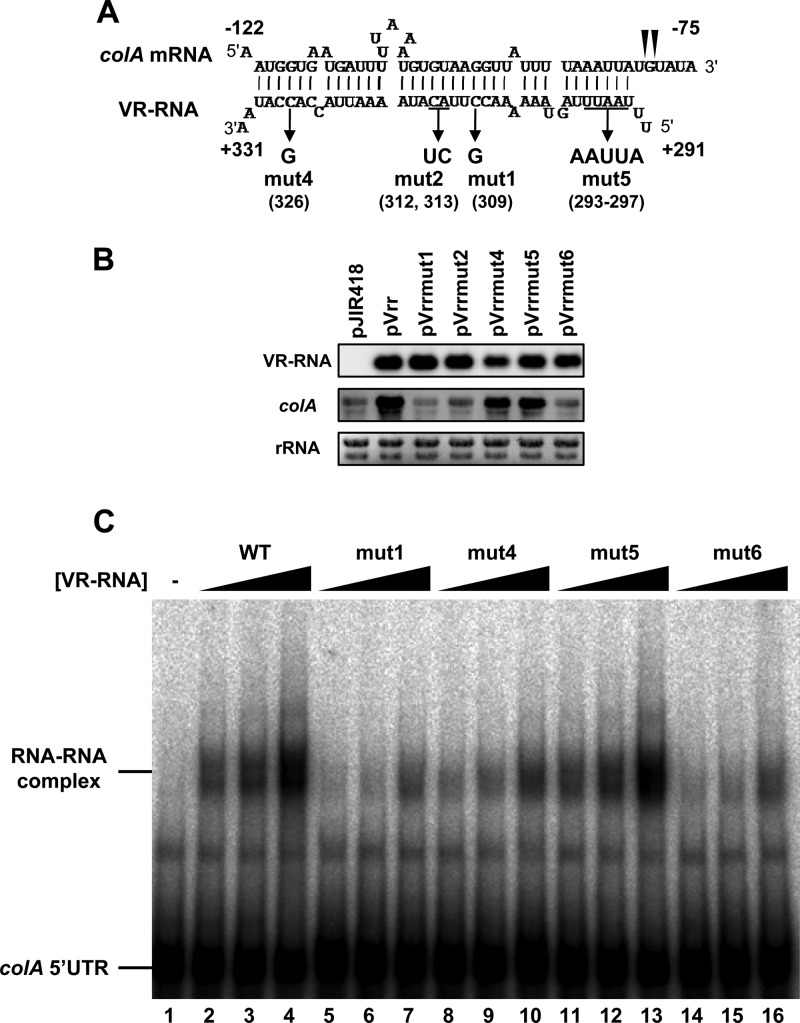
Implication of the region in the VR-RNA sequence that is essential for VR-RNA–colA base pairing and colA regulation.
We previously showed that colA mRNA processing and stabilization require the 3′ portion of VR-RNA (15). We found significant complementarity between VR-RNA and the colA 5′UTR (Fig. 2A) and observed that these regions are necessary for forming VR-RNA–colA mRNA complexes in vitro (15). We introduced various point mutations into the vrr gene encoding VR-RNA to define which region is important for base pairing to colA mRNA and for colA regulation in the VR-RNA 3′ region (Fig. 2A). The mutated vrr genes were cloned into the pJIR418 vector, an E. coli-C. perfringens shuttle vector, and then transformed into TS140, a VR-RNA-deficient strain. Northern blots of colA mRNAs from these transformants revealed that wild-type (WT) VR-RNA and the VR-RNA mut4 and mut5 mutants had restored chromosomally encoded colA expression, whereas VR-RNA mut1 and mut2 did not (Fig. 2B). These results indicated that nucleotides 306 to 316 of VR-RNA, which form an 11-bp RNA duplex with colA mRNA, are important and that C326 or U293-U297 in the VR-RNA sequence is dispensable for colA regulation. Indeed, VR-RNA–colA complexes were clearly detectable when equimolar amounts of VR-RNA and the colA 5′UTR were mixed in vitro (Fig. 2C, lane 2), and the mutation of nt 293 to 297 in VR-RNA (VR-RNA mut5) did not affect the formation of VR-RNA–colA complexes (Fig. 2C, lanes 11 to 13). However, the amount of VR-RNA mut4–colA complexes was decreased compared with the amount of wild-type VR-RNA-containing complexes (Fig. 2C, lanes 8 to 10), suggesting that G326 affects the interaction between VR-RNA and the colA mRNA 5′UTR in vitro but not in vivo. On the other hand, VR-RNA mut1–colA complexes were detectable only by gel mobility shift assays using a 4-fold molar excess of VR-RNA mut1 (Fig. 2C, lane 7). Therefore, pairing of the central 11 bp (Fig. 2A, nt 306 to 316 in VR-RNA and nt −102 to −92 in colA) could be important for interaction between VR-RNA and colA mRNA and for colA regulation. We then constructed VR-RNA mut6 by simultaneously introducing the mut4 and mut5 point mutations into the VR-RNA sequence from U293 to U297 and at G326 to determine whether only the central 11-bp RNA duplex regulates colA expression (Fig. 2A). The expression of colA was not restored by VR-RNA mut6, indicating that the central 11-bp pairing is not sufficient to regulate colA (Fig. 2B). In addition, colA and an equimolar amount of VR-RNA mut6 could not form an RNA-RNA complex (Fig. 2C, lane 14). This finding also supported the notion that the central 11-bp pairing is important but not sufficient for efficient VR-RNA–colA base pairing in vitro as well as in vivo.
Fig 2.
Determination of essential region within VR-RNA–colA mRNA base-paired duplex for colA regulation. (A) Base pairing between VR-RNA and colA mRNA 5′UTR. Filled triangles indicate processing sites within the colA mRNA sequence. Point mutations introduced into the VR-RNA sequence are also shown. (B) Complementation of colA expression by plasmid-borne mutated VR-RNA. Total RNAs (1 μg) isolated from VR-RNA-deficient strains harboring mutated VR-RNA expression vectors and grown to mid-exponential phase at 37°C were resolved in 1.2% denaturing agarose and then blotted on nylon membranes. VR-RNA and colA mRNA were probed with these gene-specific probes. Methylene blue-stained 23S and 16S rRNA blots are indicated at the bottom, as loading controls. (C) Interaction between VR-RNA and colA mRNA in vitro. The colA mRNA 5′UTR (1 nM) synthesized in vitro was separated by 4% PAGE, with or without wild-type or mutated VR-RNA (1, 2, and 4 nM).
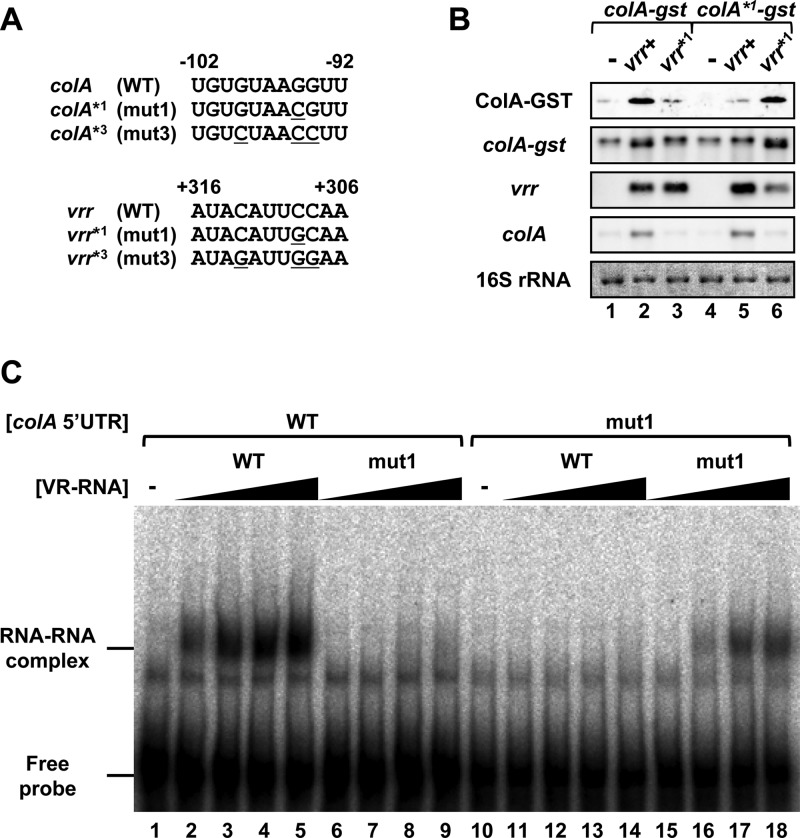
VR-RNA directly regulates colA expression though base pairing with the 5′UTR of colA mRNA.
No experimental evidence has yet supported the notion that VR-RNA directly regulates colA expression through base pairing in vivo. We introduced complementary mutations into the 3′ region of VR-RNA (vrr*1) and the 5′UTR of the colA mRNA (colA*1) to confirm that VR-RNA regulates colA through base pairing in vivo (Fig. 3A). Wild-type and mutant vrr genes were inserted into the colA-gst expression vector (15). The resulting plasmid vector, which coexpressed colA-gst and VR-RNA, was transformed into TS140, and then the amount of extracellular ColA-GST fusion protein isolated from the transformants was measured by Western blotting. Large and very small amounts of ColA-GST protein accumulated when coexpressed with wild-type and mutated VR-RNA, respectively (Fig. 3B, lanes 1 to 3). A compensatory point mutation was then introduced into the colA-gst 5′UTR (designated colA*1-gst) (Fig. 3A). The fusion protein from the colA*1-gst gene accumulated only when the compensated mutated VR-RNA was coexpressed (Fig. 3B, lanes 4 to 6). In addition, the processed colA-gst mRNA was detected when complementary VR-RNA was expressed from the plasmid (Fig. 3B, lanes 2 and 6). These results show that complementarity between C309 of VR-RNA and G−95 of colA is necessary to activate colA mRNA processing and translation in vivo. Moreover, ColA-GST protein expression depends on the presence of VR-RNA, although similar amounts of colA-gst mRNA were detected with and without VR-RNA, suggesting that the primary regulatory effect of VR-RNA on colA-gst is translational. We also constructed and tested an additional mutant set: vrr*2 and colA*2-gst. The results shown in Fig. S1 in the supplemental material suggested that base pairing between C319/A320 of VR-RNA and G−99/U−98 of colA mRNA is also important for the regulation. The effects of the point mutations on interaction between VR-RNA and the colA mRNA 5′UTR were analyzed using gel mobility shift assays (Fig. 3C). The wild-type colA mRNA 5′UTR no longer interacted with VR-RNA mut1, whereas colA mRNA 5′UTR mut1 interacted only with the mutated complementary VR-RNA (Fig. 3C), suggesting that mutations in the 11-bp VR-RNA–colA RNA duplex inhibited their interaction. Therefore, VR-RNA directly activates processing and protein expression through base pairing with the colA mRNA 5′UTR in vivo.
Fig 3.
Complementarity between VR-RNA and colA mRNA 5′UTR is important for colA regulation. (A) Mutation sites within the VR-RNA–colA RNA duplex. Mutation sites in the colA mRNA 5′UTR or VR-RNA are underlined. (B) Western and Northern blots of VR-RNA-deficient strains carrying colA-gst or mutated colA-gst and vrr, a gene encoding VR-RNA, or a mutated vrr coexpression vector. Each lane was loaded with 0.02 A280 unit of extracellular protein or with 0.5 μg of total RNA from cells grown to mid-exponential growth phase at 37°C. The GST fusion protein was probed with an anti-GST antibody (top panel). Chromosomally encoded colA mRNA, colA-gst, and plasmid-encoded VR-RNA were detected using specific probes. Methylene blue-stained 16S rRNAs are indicated at the bottom, as loading controls. (C) Gel mobility shift assay to analyze the interaction between mutated colA and VR-RNA. Lanes contained 1 nM radiolabeled wild-type (lanes 1 to 9) or mutated (lanes 10 to 18) colA mRNA 5′UTR. Before electrophoresis, colA was incubated without or with 1, 2, 4, and 8 nM wild-type (lanes 2 to 5 and 11 to 14) and mutated (lanes 6 to 9 and 15 to 18) VR-RNA. The free colA mRNA 5′UTR and the colA mRNA 5′UTR–VR-RNA complex are indicated.
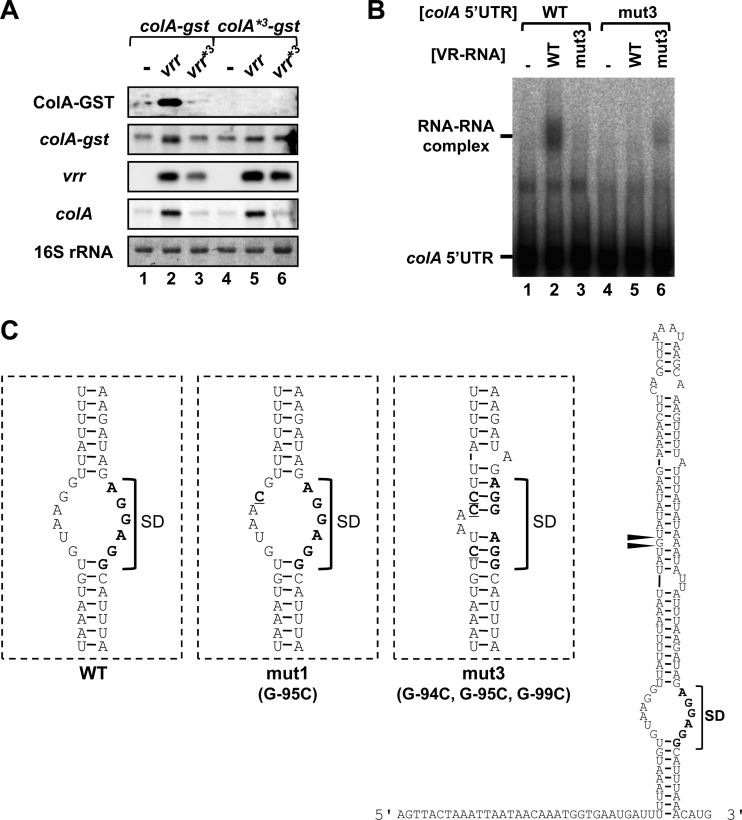
The single-stranded region of the bulge structure in colA mRNA is necessary for VR-RNA-dependent translational activation.
We constructed an additional complementary mutant set, vrr*3 and colA*3 (mut3), in which three nucleotides (C308G, C309G, and C313G in VR-RNA and G−99C, G−95C, and G−94C in colA) in the central 11-bp pairing region were replaced (Fig. 3A). The coexpression of VR-RNA mut3 did not activate ColA-GST expression (Fig. 4A, lane 3). Although the mutated VR-RNA mut3 construct could not form RNA complexes with the wild-type colA 5′UTR, it interacted with the mutated complementary colA*3 mut3 construct in vitro (Fig. 4B). However, colA*3-gst translation was no longer activated, even in the presence of mutated complementary VR-RNA (Fig. 4A, lanes 4 to 6, vrr*3). The predicted secondary structure of the mutated colA 5′UTR suggested that the bulge structure in the stem was closed in colA*3-gst mRNA, whereas the structure of the colA*1-gst mRNA remained unaltered (Fig. 4C). These findings suggested that interaction between VR-RNA and the single-stranded region in the bulge structure of the colA mRNA 5′UTR is necessary for colA translational activation in vivo.
Fig 4.
A stem-loop structure of colA mRNA that is strengthened by mutation inhibits translation and interaction with VR-RNA. (A) Western and Northern blots of VR-RNA-deficient strains harboring colA-gst*3 and vrr*3 coexpression vectors. Lanes were loaded with 0.02 A280 unit of extracellular protein or with 0.5 μg of total RNA from cells grown to mid-exponential phase at 37°C. The GST fusion protein was probed with an anti-GST antibody (top panel). Chromosomally encoded colA mRNA, colA-gst, and plasmid-encoded VR-RNA were detected using specific probes. Methylene blue-stained 16S rRNAs are indicated at the bottom, as loading controls. (B) Gel mobility shift assay with 1 nM radiolabeled colA mRNA 5′UTR and 2 nM wild-type or mutant VR-RNA. (C) Secondary structure predictions of mutated colA mRNAs. Bold type and underlining represent the SD sequence and mutated bases, respectively. Filled triangles indicate processing sites.
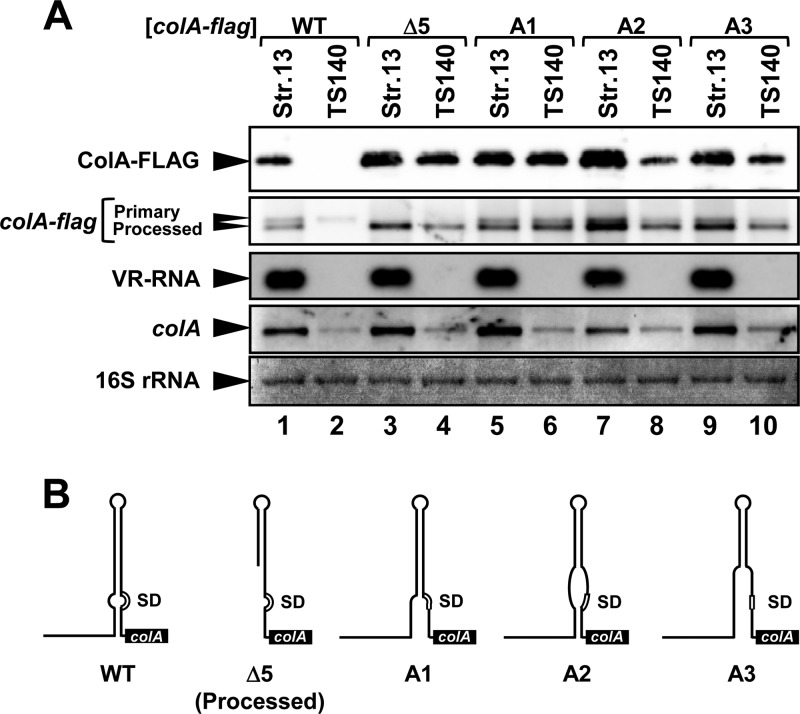
Disruption of the stem structure in the colA mRNA 5′UTR leads to processing and translational activation.
VR-RNA directly binds to the colA mRNA 5′UTR, which would disrupt the stem-loop structure in the colA mRNA 5′UTR that inhibits colA translation. We examined whether destruction of the inhibitory structure without VR-RNA was sufficient to release the colA SD sequence and enhance translation. A new reporter gene was needed to analyze mRNA stability, processing, and protein expression, because the transcript of colA-gst, namely, the reporter gene colA used above, is very stable with or without VR-RNA, which differs from genome-encoded colA mRNA expression (see Fig. S2 in the supplemental material). We inserted the colA-FLAG gene (in which the promoter, 5′UTR, and 285 codons of colA are fused to the N terminus of the FLAG tag) into pJIR418 to generate pCPE94. Expression of the colA-FLAG mRNA and ColA-FLAG protein in strain 13, an isogenic wild-type strain, and TS140, a VR-RNA-deficient mutant, was analyzed by Northern and Western blotting, respectively. The primary and processed colA-FLAG mRNAs were distinguishable on Northern blots, and only a small amount of the primary type was detected in TS140, suggesting that the colA-FLAG and genome-carried colA genes behaved similarly (Fig. 5A, lanes 1 and 2). Removal of the processing region in pCPE94 to generate pCPE94Δ5 caused the fusion protein to accumulate in strain 13 and in TS140, consistent with our published findings (Fig. 5A, lanes 3 and 4) (15). The accumulation of colA-FLAG transcripts also depended on ribosome binding to the SD sequence (see Fig. S3). We then introduced mutations to disrupt the stem-loop structure of colA mRNA into pCPE94 (Fig. 5B, mutants A1 to A3; see Fig. S4). The colA SD sequence and vicinity would be located on a single-stranded region in these mutated colA 5′UTRs (Fig. 5B). All mutations caused obvious ColA-FLAG protein accumulation, suggesting that the disruption of base pairing around the colA SD sequence was sufficient to enhance translation (Fig. 5A, lanes 5 to 10). Meanwhile, these mutations induced processing in the 5′UTR regardless of whether VR-RNA was present, although A2 and A3 mutant expression was still regulated by VR-RNA (Fig. 5A). This result was not anticipated, because processing is thought to depend on VR-RNA, and it indicated that the colA mRNA 5′UTR is processed without VR-RNA when the intramolecular secondary structure is destroyed, which would promote translation. Thus, mutations disrupting the stem in the colA mRNA 5′UTR are sufficient to activate colA processing and translation. Furthermore, the stem structures upstream and downstream of the bulge are essential for basal colA repression and important for VR-RNA-dependent regulation.
Fig 5.
Disruption of secondary structure in the colA mRNA 5′UTR leads to translational activation and processing. (A) Western and Northern blots of wild-type (strain 13) and VR-RNA-deficient (TS140) strains carrying pCPE94 derivatives. Lanes were loaded with 0.02 OD600 unit of extracellular protein or with 1 μg of total RNA from cells at mid-exponential growth phase. The FLAG-tagged protein was probed with an anti-FLAG antibody (top panel). Chromosomally encoded colA or colA-FLAG mRNA and VR-RNA were detected using colA- and VR-RNA-specific probes, respectively. Methylene blue-stained 16S rRNAs are indicated at the bottom, as loading controls. (B) Schematic drawings of secondary structures of colA mRNA 5′UTRs expressed by pCPE94 derivatives. Structures were predicted using the RNAfold WebServer site.
Processing further enhances colA translational activation.
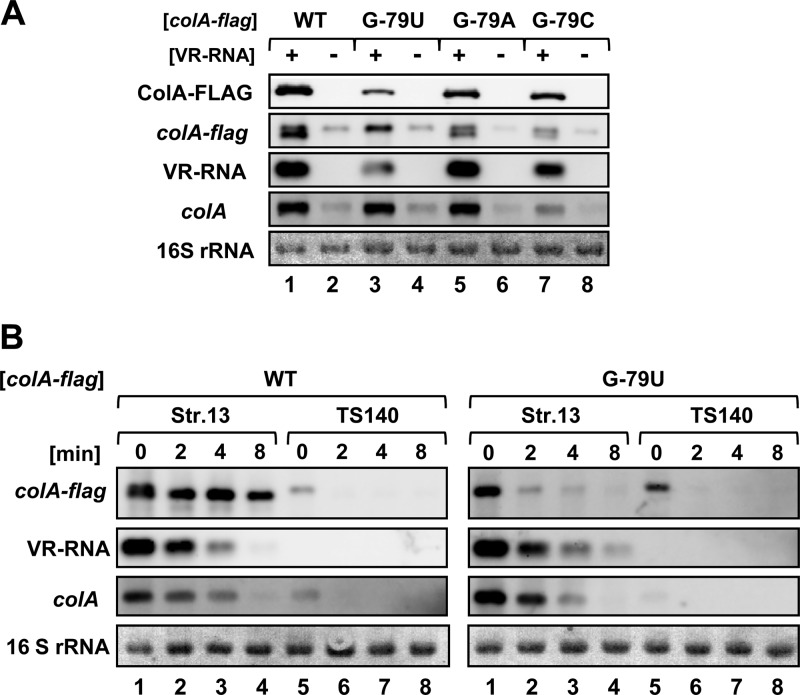
Disruption of the secondary structure in the colA mRNA 5′UTR was sufficient to induce processing and translational activation. However, the precise role of processing in colA regulation remained unclear. We therefore constructed a processing-deficient mutant and introduced a point mutation into the G−79 processing site of the colA-FLAG reporter gene. Northern blots showed that the mRNAs of colA-FLAG G−79A and the G−79C mutant were processed in the presence of VR-RNA (Fig. 6A, lanes 5 and 7). On the other hand, processed colA-FLAG mRNA was undetectable when G−79 was replaced with U (Fig. 6A, lane 3). This result suggests that the G−79U mutation in the colA-FLAG 5′UTR inhibits processing. Although processing did not occur, the colA-FLAG mutant was translationally activated in a VR-RNA-dependent manner (Fig. 6A, lanes 3 and 4). However, WT colA-FLAG produced more ColA-FLAG protein than G−79U mutant colA-FLAG in the presence of VR-RNA. The primary WT colA-FLAG mRNA disappeared within 2 min of adding rifampin, and the processed transcripts persisted for 8 min (half-life of >8 min) (Fig. 6B). The primary transcripts of G−79U mutant colA-FLAG were also detectable at 8 min but were less stable than the processed WT colA-FLAG mRNA (Fig. 6B). Therefore, although unnecessary for VR-RNA-dependent translational colA activation, processing enhances the efficiency of translational activation and/or the stability of the transcripts.
Fig 6.
Processing is dispensable for colA regulation by VR-RNA but required to further activate translation. (A) Western and Northern blots of strains 13 and TS140 harboring plasmids expressing the colA-FLAG gene with a mutated G−79 processing site. Lanes were loaded with 0.02 OD600 unit of extracellular protein or with 1 μg of total RNA from cells at mid-exponential growth phase. The FLAG-tagged protein was probed with an anti-FLAG antibody (top panel). Chromosomally encoded colA or colA-FLAG mRNA and VR-RNA were detected using colA- and VR-RNA-specific probes, respectively. (B) Stability of colA-FLAG mRNA. C. perfringens strains 13 and TS140 harboring colA-FLAG expression vectors with the G−79 processing site replaced with U were grown to mid-exponential phase. Total RNAs (1 μg) isolated from cultures before or at the indicated times after adding rifampin (200 mg ml−1) were Northern blotted. Methylene blue-stained 16S rRNAs are indicated at the bottom, as loading controls.
High temperature also induces colA mRNA processing and translational activation.
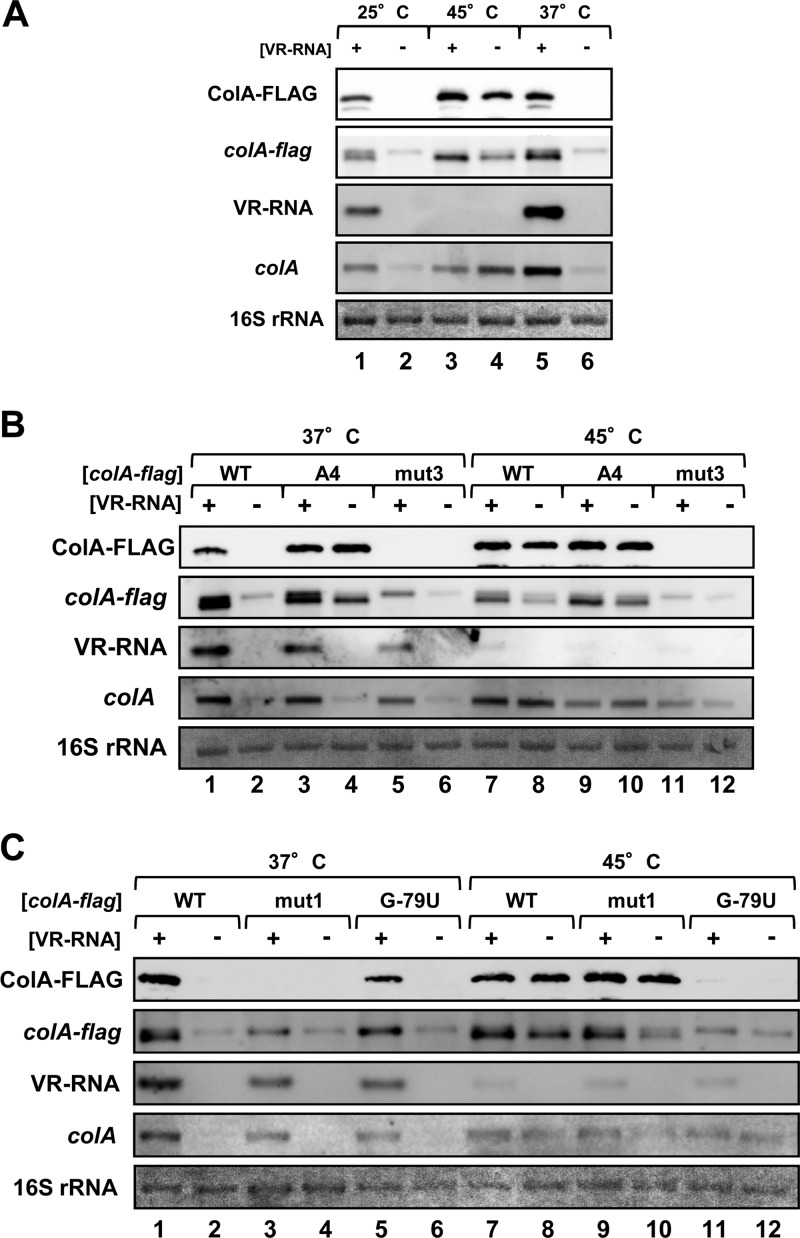
The above results showed that disrupting the colA mRNA structure leads to ColA processing and translational activation. Since changes in temperature usually alter the RNA structure, we speculated that the stem-loop structure of colA mRNA could be stabilized or disrupted by changes in temperature. We analyzed ColA-FLAG protein and colA-FLAG mRNA expression in cells cultured at 25°C, 37°C, or 45°C (Fig. 7A). ColA-FLAG protein and processed transcripts accumulated in the presence of VR-RNA at 25°C and 37°C, whereas protein expression was activated at 45°C without VR-RNA, suggesting that this temperature melted the stem-loop structure of the colA mRNA 5′UTR and induced processing and translational activation (Fig. 7A). In addition, VR-RNA expression was hardly detectable at 45°C, suggesting that VR-RNA is very unstable or poorly transcribed at high temperatures. To determine whether or not the RNA secondary structure was involved in colA activation at a high temperature, we analyzed a mutant colA-FLAG mRNA with an altered mRNA secondary structure in the 5′UTR. The A4 mutation, which disrupts the stem structure of colA mRNA and activates translation without VR-RNA (see Fig. S5 in the supplemental material), caused almost identical amounts of protein accumulation under all tested conditions (Fig. 7B, lanes 3, 4, 9, and 10). ColA-FLAG protein expression from the mut3 mutant, in which we predicted the bulge structure in the mRNA 5′UTR would close and further inhibit translation (Fig. 3 and 4; see Fig. S5), was also not activated at 45°C (Fig. 7B, lanes 5, 6, 11, and 12). These results support the notion that inhibiting colA mRNA structure formation at 45°C also activates ColA protein expression. In addition, such regulation was independent of VR-RNA, because the expression of mut1 mutant colA-FLAG, in which the essential residue base pairing to VR-RNA was mutated (Fig. 2 and 3), was also modulated at 45°C (Fig. 7C, lanes 3, 4, 9, and 10). On the other hand, G−79U mutant colA-FLAG was not activated at 45°C. The G−79U residue base paired with the opposite A residue, thus removing a mismatch in the stem structure. Therefore, thermoregulation of colA-FLAG requires processing and/or strengthening the stem inhibits alterations in the colA mRNA structure.
Fig 7.
High temperature induces colA translational activation without VR-RNA. Western and Northern blotting proceeded with an anti-FLAG antibody and colA- or VR-RNA-specific probes. Lanes were loaded with 0.02 OD600 unit of extracellular protein or with 1 μg of total RNA from cells at mid-exponential growth phase. (A) Proteins and RNAs were isolated from C. perfringens strain 13 and from TS140 harboring the colA-FLAG gene, grown at 25°C, 37°C, or 45°C. (B) Cells harboring mutated colA-FLAG genes with disrupted (A4) or strengthened (mut3) stem-loop structures in the colA-FLAG mRNA 5′UTR. (C) Cells harboring mutated colA-FLAG genes with an mRNA 5′UTR that could not interact with VR-RNA (mut1) or could not be processed at G79. Methylene blue-stained 16S rRNAs are indicated at the bottom, as loading controls.
DISCUSSION
We confirmed that VR-RNA regulates colA expression by direct base pairing by using a complementary mutation in VR-RNA and colA (Fig. 3; see Fig. S1 in the supplemental material). We previously predicted that VR-RNA–colA interaction generates 6+6+11+3+7-bp RNA duplexes (16) (Fig. 2A). The point mutations introduced into VR-RNA in vivo and in vitro revealed that interaction with the colA bulge structure opposite the SD sequence within the core 11-bp RNA duplex (VR-RNA positions +306 to +316 and colA positions −102 to −92) could be essential for colA regulation and that base pairing outside the core region strengthens interactions between these RNAs. Therefore, base pairing in addition to the important core duplex is also necessary for VR-RNA to regulate colA expression in vivo. The predicted structure of the colA mRNA 5′UTR is a stem-loop with an internal bulge where the SD sequence is located (Fig. 1). Mutational analysis of the bulge sequence in colA mRNA indicated that the single-stranded region within the bulge is important for formation of the VR-RNA–colA mRNA complex and for colA regulation by VR-RNA (Fig. 4; see Fig. S5 in the supplemental material). The Staphylococcus aureus sRNA, RNAIII, regulates the expression of rot and coa mRNAs, which encode a transcriptional repressor of toxin and staphylocoagulase, respectively, through sRNA-mRNA base pairing (18, 19). The C-rich loop in the stem-loop structure of RNAIII is thought to initially base pair with a G-rich unpaired region of target mRNAs and form a kissing loop-like structure (20). Thus, the single-stranded region in each RNA sequence is important for sRNA-mRNA interaction. The predicted secondary structure of VR-RNA indicated that the region important for base pairing is located within the single-stranded region upstream of the transcriptional terminator (see Fig. S6 in the supplemental material). Therefore, we propose that VR-RNA base pairing with colA is initiated within the single-stranded region of each RNA, and then additional base pairing outside the core strengthens interactions between these RNAs, causing a disruption in the secondary structure of colA mRNA (Fig. 8).
Fig 8.
Possible kissing interaction between VR-RNA and colA mRNA. The single-stranded regions of VR-RNA and colA mRNA predicted by CentroidFold (30) and RNA structure probing, respectively, should initially base pair. Subsequent additional base pairing between them strengthens the interaction and disrupts the colA stem structure, which might trigger colA processing by a single-strand-specific RNase. SD sequences and start codons of colA are indicated with brackets and underlined, respectively.
In contrast, when a mutation disrupting the stem-loop structure was introduced into the colA-FLAG gene, colA processing and translational activation were induced without VR-RNA (Fig. 5). Both the stems located upstream and downstream of the bulge are important for basal colA repression and VR-RNA-dependent regulation, because all of the mutations disrupting the stems in colA mRNA derepressed ColA-FLAG expression (Fig. 5; see Fig. S4 and S5 in the supplemental material). However, VR-RNA still affected A2 and A3 mutant expression, because it base paired with colA-FLAG mRNA at positions −102 to −94. Thus, it might have enhanced further disruption of the stem and/or stabilized the transcript. On the other hand, the UGU sequence just upstream of the colA bulge (positions −102 to −100) was mutated in the A1 and A4 mutants, in which VR-RNA scarcely affected expression. VR-RNA appeared to interact with the A2 and A3 mutants but not the A1 and A4 mutants. In addition, a high temperature (e.g., 45°C) was sufficient to melt the RNA structures and to induce processing and translational activation in the absence of VR-RNA (Fig. 7). These findings suggest that only disruption of the colA mRNA structure can trigger processing, and thus the primary function of VR-RNA in colA regulation is to prevent formation of the colA 5′ leader structure through base pairing.
We found that a point mutation in which the processing site G−79 was replaced with U (numbering relative to the ATG start codon; +1 is A) inhibited colA mRNA processing. The ColA-FLAG protein was expressed from the processing-deficient mutant in the presence of VR-RNA, but the amount of protein was less than that from processed colA-FLAG mRNA (Fig. 6A, lanes 1, 3, and 4). Similarly, primary colA-FLAG mRNA was stabilized in the presence of VR-RNA without processing, but it was more labile than processed transcripts (Fig. 6B). These results suggest that colA mRNA processing is required for maximally efficient colA activation. Processing would enhance colA translational efficiency or contribute to the further stabilization of colA mRNA, as the SD sequence in the processed transcripts was not masked by its own sequences and a short stem-loop structure formed near the 5′ end of the mRNA (see Fig. S4B in the supplemental material). These features of the transcript would render ribosome binding to the SD sequence highly efficient and block RNase, respectively (21). Thus, processing is important for rapid ColA protein accumulation in response to environmental changes that induce the expression of VR-RNA that is activated by the VirR-VirS two-component system.
We showed here that a temperature of 45°C controls colA expression and that such regulation depends on the conformation of the mRNA 5′UTR (Fig. 7). Thus, we suggest that the colA 5′ leader region is a possible RNA thermosensor like those in the alpha- and gammaproteobacteria, Listeria monocytogenes, and Synechocystis, which contribute to posttranscriptional gene regulation (22–26). Translation of the small heat shock gene agsA is activated at 45°C by an RNA thermosensor containing a four-U element in which four uridine residues base pair with AGGA of the SD sequence in Salmonella enterica (23). We found four uridine residues in the stem structure of colA mRNA, but they did not base pair with the SD sequence. However, the colA mRNA 5′UTR partially resembles the prfA RNA thermosensor of L. monocytogenes in terms of the mode of regulation. The expression of prfA, encoding a modulator protein that regulates toxin genes, is regulated by sRNAs and by high temperature. The 115-nt prfA mRNA 5′UTR forms a long stem-loop structure that inhibits ribosome binding to the SD sequence, like the case in colA mRNA, causing translational repression at 30°C (22). On the other hand, the RNA structure is melted and translation is initiated at higher temperatures, such as 37°C. In addition, S-adenosylmethionine (SAM) riboswitches are also involved in prfA regulation through RNA-RNA interaction (27). The sreA and sreB genes are regulated by the intracellular concentration of S-adenosylmethionine, which leads to transcriptional termination in the sequence of the S-box riboswitch. The preterminated sreA and sreB riboswitches base pair with the 5′UTR of the prfA mRNA, causing translational repression (27). The prfA SD sequence is also located in the bulge of the long stem-loop structure of the prfA mRNA 5′UTR. A mutation that causes base pairing in the bulge structure inhibits thermoregulation of prfA (22). The colA translation activated by sRNA and high temperature was also inhibited by a mutation which strengthened the stem structure in C. perfringens, suggesting that the bulge is important for the regulation of both the colA and prfA genes that is mediated by a conformational change in mRNA. C. perfringens grows more rapidly at 45°C than at 37°C, and temperatures of 43°C to 47°C are optimal for the growth of this organism. However, it thrives at around 37°C during infection in humans and other animals, and the physiological relevance of colA activation without VR-RNA at 45°C remains unknown.
The sRNA RNAIII regulates several virulence factors in S. aureus by base pairing with target mRNA, which leads to mRNA degradation by the double-strand-specific endoribonuclease RNase III (28). We examined whether RNase III is involved in colA mRNA processing in C. perfringens. The rnc gene, which encodes RNase III in C. perfringens, was disrupted by inserting the erythromycin resistance gene. The Northern blots shown in Fig. S7 in the supplemental material show that VR-RNA and colA expression by the rnc mutant strain did not change, indicating that RNase III is not involved in either colA regulation by VR-RNA or processing of the colA mRNA 5′UTR. Identification and further analysis of the RNase involved in processing are needed to deepen our understanding of colA regulation.
Thermoregulation did not occur in the processing-deficient colA-FLAG G−79U mutant (Fig. 7). The U residue could base pair with the opposite A residue, which would strengthen the colA stem structure. This could explain why the mRNA is not processed and not translated at high temperatures. Thus, the RNase responsible for processing might not recognize the processing site in the colA G−79U mutant because the residue is not in a single-stranded region, and strengthening the stem could prevent it from opening at high temperatures. Therefore, we considered the following model for colA regulation. At low temperatures (such as ≤37°C), the VR-RNA base pairs with colA, opens up the stem structure, and induces processing and translation. Processing, while not necessary for expression, would render the activation permanent and shift the equilibrium toward higher levels of expression. At high temperatures (such as 45°C), VR-RNA seems to be expressed poorly or highly unstable (Fig. 7). The fact that the level of colA expression is constitutively high suggests that VR-RNA-dependent colA regulation is not efficient at higher temperatures and that colA RNA acts as a VR-RNA-independent thermosensor. Mutating U−79 increases the stability of the stem structure so that it can no longer open at 45°C and might block processing by a single-strand-specific endoribonuclease.
We found that the role of the sRNA, i.e., VR-RNA, in colA regulation is to prevent the formation of the colA 5′ leader structure through base pairing and that the bulge structure as well as the upstream and downstream stems in the colA 5′ leader sequence is necessary for VR-RNA-dependent colA regulation. In addition, the structure could be a possible thermosensor. If so, to our knowledge, this is the first description of a gene activated by sRNA and high temperature. Collagenase is not essential for the development of gas gangrene in the mouse myonecrosis model (16), but how collagenase activity contributes to cell growth under natural conditions remains unknown. The complexity of colA gene regulation by the conformation of the 5′ leader sequence suggests that the appropriate control and rapid induction of collagenase production in response to environmental conditions are important for the proliferation of C. perfringens under natural conditions, such as in soil or in host cells in the human intestine.
Supplementary Material
ACKNOWLEDGMENTS
We thank Norma Foster for a critical reading of the manuscript.
This work was supported by the Advanced Low Carbon Technology Research and Development Program (ALCA) and Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology Agency (JST).
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00148-13.
REFERENCES
- 1. Rood JI, Cole ST. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Petit L, Gibert M, Popoff MR. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104–110 [DOI] [PubMed] [Google Scholar]
- 3. Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohtani K, Hayashi H, Shimizu T. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171–179 [DOI] [PubMed] [Google Scholar]
- 5. Abe K, Obana N, Nakamura K. 2010. Effects of depletion of RNA-binding protein Tex on the expression of toxin genes in Clostridium perfringens. Biosci. Biotechnol. Biochem. 74:1564–1571 [DOI] [PubMed] [Google Scholar]
- 6. Obana N, Nakamura K. 2011. A novel toxin regulator, the CPE1446-CPE1447 protein heteromeric complex, controls toxin genes in Clostridium perfringens. J. Bacteriol. 193:4417–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shimizu T, Ba-Thein W, Tamaki M, Hayashi H. 1994. The virR gene, a member of a class of two-component response regulators, regulates the production of perfringolysin O, collagenase, and hemagglutinin in Clostridium perfringens. J. Bacteriol. 176:1616–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okumura K, Ohtani K, Hayashi H, Shimizu T. 2008. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 190:7719–7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ohtani K, Hirakawa H, Tashiro K, Yoshizawa S, Kuhara S, Shimizu T. 2010. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16:258–264 [DOI] [PubMed] [Google Scholar]
- 10. Cheung JK, Rood JI. 2000. The VirR response regulator from Clostridium perfringens binds independently to two imperfect direct repeats located upstream of the pfoA promoter. J. Bacteriol. 182:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimizu T, Yaguchi H, Ohtani K, Banu S, Hayashi H. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 43:257–265 [DOI] [PubMed] [Google Scholar]
- 12. Kawsar HI, Ohtani K, Okumura K, Hayashi H, Shimizu T. 2004. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 235:289–295 [DOI] [PubMed] [Google Scholar]
- 13. André G, Haudecoeur E, Monot M, Ohtani K, Shimizu T, Dupuy B, Martin-Verstraete I. 2010. Global regulation of gene expression in response to cysteine availability in Clostridium perfringens. BMC Microbiol. 10:234. 10.1186/1471-2180-10-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan Y, Ohtani K, Yoshizawa S, Shimizu T. 2012. Complex transcriptional regulation of citrate metabolism in Clostridium perfringens. Anaerobe 18:48–54 [DOI] [PubMed] [Google Scholar]
- 15. Obana N, Shirahama Y, Abe K, Nakamura K. 2010. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Mol. Microbiol. 77:1416–1428 [DOI] [PubMed] [Google Scholar]
- 16. Awad MM, Ellemor DM, Bryant AE, Matsushita O, Boyd RL, Stevens DL, Emmins JJ, Rood JI. 2000. Construction and virulence testing of a collagenase mutant of Clostridium perfringens. Microb. Pathog. 28:107–117 [DOI] [PubMed] [Google Scholar]
- 17. Mahony DE, Moore TI. 1976. Stable L-forms of Clostridium perfringens and their growth on glass surfaces. Can. J. Microbiol. 22:953–959 [DOI] [PubMed] [Google Scholar]
- 18. Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61:1038–1048 [DOI] [PubMed] [Google Scholar]
- 19. Chevalier C, Boisset S, Romilly C, Masquida B, Fechter P, Geissmann T, Vandenesch F, Romby P. 2010. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 6:e1000809. 10.1371/journal.ppat.1000809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21:1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bechhofer DH. 2009. Messenger RNA decay and maturation in Bacillus subtilis. Prog. Mol. Biol. Transl. Sci. 85:231–273 [DOI] [PubMed] [Google Scholar]
- 22. Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551–561 [DOI] [PubMed] [Google Scholar]
- 23. Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F. 2005. RNA thermometers are common in alpha- and gamma-proteobacteria. Biol. Chem. 386:1279–1286 [DOI] [PubMed] [Google Scholar]
- 24. Giuliodori AM, Di Pietro F, Marzi S, Masquida B, Wagner R, Romby P, Gualerzi CO, Pon CL. 2010. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37:21–33 [DOI] [PubMed] [Google Scholar]
- 25. Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F. 2011. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res. 39:2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, Dersch P. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. 10.1371/journal.ppat.1002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770–779 [DOI] [PubMed] [Google Scholar]
- 28. Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, Jacquier A, Vandenesch F, Romby P. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sloan J, Warner TA, Scott PT, Bannam TL, Berryman DI, Rood JI. 1992. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid 27:207–219 [DOI] [PubMed] [Google Scholar]
- 30. Sato K, Hamada M, Asai K, Mituyama T. 2009. CentroidFold: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37:W277–W280 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.