Abstract
Formation of nonnative disulfide bonds in the cytoplasm, so-called disulfide stress, is an integral component of oxidative stress. Quantification of the extent of disulfide bond formation in the cytoplasm of Escherichia coli revealed that disulfide stress is associated with oxidative stress caused by hydrogen peroxide, paraquat, and cadmium. To separate the impact of disulfide bond formation from unrelated effects of these oxidative stressors in subsequent experiments, we worked with two complementary approaches. We triggered disulfide stress either chemically by diamide treatment of cells or genetically in a mutant strain lacking the major disulfide-reducing systems TrxB and Gor. Studying the proteomic response of E. coli exposed to disulfide stress, we found that intracellular disulfide bond formation is a particularly strong inducer of the heat shock response. Real-time quantitative PCR experiments showed that disulfide stress induces the heat shock response in E. coli σ32 dependently. However, unlike heat shock treatment, which induces these genes transiently, transcripts of σ32-dependent genes accumulated over time in disulfide stress-treated cells. Analyzing the stability of σ32, we found that this constant induction can be attributed to an increase of the half-life of σ32 upon disulfide stress. This is concomitant with aggregation of E. coli proteins treated with diamide. We conclude that oxidative stress triggers the heat shock response in E. coli σ32 dependently. The component of oxidative stress responsible for the induction of heat shock genes is disulfide stress. Nonnative disulfide bond formation in the cytoplasm causes protein unfolding. This stabilizes σ32 by preventing its DnaK- and FtsH-dependent degradation.
INTRODUCTION
Oxidative stress is defined as an imbalance between the generation of reactive oxygen species (ROS), such as superoxide anion, hydrogen peroxide, and hydroxyl radicals, and their detoxification by cellular antioxidant systems. ROS have the potential to attack virtually any cellular macromolecule, including DNA, lipids, and proteins. To counteract the damaging effects, microorganisms respond to oxidative stress in various ways. The classical oxidative stress response is regulated by transcription factors, such as OxyR from Escherichia coli, OhrR from Bacillus subtilis, and Yap1 from Saccharomyces cerevisiae, which all act as reversible redox switches (1–3). Activated by the oxidation of highly conserved cysteines, these regulators induce the expression of antioxidant enzymes, such as peroxiredoxins, glutaredoxins, and thioredoxins.
A separate class of transcription factors which specifically sense disulfide stress has been reported in a number of organisms. These regulators are activated when the thiol-disulfide balance is perturbed, resulting in the formation of inter- and intramolecular disulfides within proteins or thiolation of proteinogenic amino acids with small-molecule thiols such as glutathione. This condition may be caused by an accumulation of naturally occurring electrophiles, such as quinones, and does not necessarily involve radical formation or the presence of ROS (4). In Streptomyces coelicolor, a distinct regulatory system consisting of RsrA and σR responds to disulfide stress, while in B. subtilis, the redox-sensing proteins Spx and YodB control parts of the disulfide stress response (5–8). As many as 275 genes have recently been shown to be under the control of Spx (9). In Pseudomonas aeruginosa, the LysR-type transcriptional regulator MexT confers resistance to disulfide stress by upregulating an efflux system for electrophilic compounds (10).
In many organisms, the response to ROS not only triggers an antioxidant response but also typically involves the upregulation of heat shock genes. In the Gram-positive model organism B. subtilis, it has been shown that the induction of class III heat shock genes is mediated by the disulfide stress-sensing transcription factor Spx (9).
In the Gram-negative model organism E. coli, all major chaperone systems, including DnaK, GroEL, and HtpG, along with other classical heat shock proteins, such as Clp proteases and the small heat shock proteins IbpA and IbpB, are upregulated upon hydrogen peroxide-induced oxidative stress (11). However, no specific disulfide stress-sensing mechanism responsible for the upregulation of these heat shock genes has been identified so far, and it is not well understood how the heat shock response is prompted by oxidative stress in this organism. In E. coli the heat shock response is controlled by the heat shock sigma factor σ32 (for a review, see reference 12). Under nonstress conditions, the concentration of σ32 is very low, and the sigma factor has a very short half-life in vivo (13, 14). Upon exposure of cells to heat shock, the concentration of σ32 increases; this increase involves numerous transcriptional, translational, and posttranslational regulatory mechanisms (15–26).
In the present study, we demonstrate that environmental stressors associated with oxidative stress all cause intracellular disulfide bond formation. Using chemical and genetic tools to separate disulfide stress from other effects of oxidative stress, we discovered that it is this disulfide stress component of oxidative stress that induces the heat shock response in E. coli. Upregulation of heat shock genes in E. coli upon oxidative stress directly depends on the heat shock sigma factor σ32, which is stabilized under disulfide stress conditions. The half-life of σ32 increased from below 1.6 min under control conditions to well over 20 min in disulfide-stressed cells. This stabilization coincides with the disulfide stress-induced aggregation of proteins. This mechanism provides E. coli with a strategy to upregulate the heat shock genes in response to oxidative stress without the need for a specialized disulfide stress-sensing regulator.
MATERIALS AND METHODS
Strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| DHB4 | F′ lac-pro lacIq Δ(ara-leu)7697 araD139 ΔlacX74 galE galK rpsL phoR Δ(phoA)PvuII ΔmalF3 thi | Lab strain collection |
| LL014 | DHB4/pAID135 | This work |
| WP778 | DHB4 gor-522. . . .mini-Tn10 Tc trxB::Km | 43 |
| BB7222 | MC4100 ara+ | 58 |
| BB7224 | MC4100 ara+ rpoH::Km zhf::Tn10 suhX401 | 58 |
| Plasmids | ||
| pAID135 | PhoAΔ2-22 under the control of the tac promoter | 28 |
| pEC5217 | pUC18 carrying E. coli rpoH Ap | 59 |
| pBO110 | pBAD24 carrying E. coli lpxC Ap | 37 |
Alkaline phosphatase assays.
E. coli LL014 (harboring pAID135) was grown aerobically in glucose MOPS (morpholinepropanesulfonic acid) minimal medium (27) containing 40 μg/ml l-leucine and 100 μg/ml thiamine at 37°C until an optical density at 600 nm (OD600) of 0.2 was reached. To induce the expression of leaderless alkaline phosphatase from pAID135, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 5 mM. At an OD600 of 0.4, disulfide stress was induced by the addition of the indicated concentrations of various oxidative stress-inducing agents. At specific time points, 900 μl cells was harvested directly onto 100 μl 1 M iodoacetamide solution and the cells were incubated for 20 min on ice to stop further disulfide exchange reactions (28). Cells were pelleted by centrifugation (13,000 × g, 2 min, 4°C) and washed twice in 1 ml ice-cold washing buffer (50 mM NaCl, 10 mM NH4Cl, 10 mM MgCl2, 10 mM iodoacetamide, 40 mM MOPS–KOH, pH 7.3). The pellet was resuspended in 100 μl lysis buffer (10 mM EDTA, 2 mg/ml lysozyme, 10 mM iodoacetamide, 20 mM Tris-HCl, pH 8.0) and incubated for 30 min on ice. Three freeze-thaw cycles (dry ice-ethanol, 28°C water bath) were used to completely lyse the cells. The lysate was resuspended in 900 μl resuspension buffer (10 mM MgCl2, 10 mM ZnCl2, 1 M Tris-HCl, pH 8.0) and incubated in a water bath at 28°C. One hundred microliters 0.4% para-nitrophenylphosphate (pNPP) solution in 1 M Tris-HCl, pH 8.0, was added, and as soon as the sample turned visibly yellow, the incubation time with pNPP was noted and 100 μl stop solution (1 M K2HPO4) was added. Then, the sample was stored for 10 min on ice. After centrifugation (13,000 × g, 2 min, 4°C) the absorption of the supernatant at 420 nm and at 550 nm was determined. One hundred microliters lysis buffer in 900 μl resuspension buffer, 100 μl pNPP solution, and 100 μl stop solution was used as a blank. Alkaline phosphatase activity (in arbitrary units) was calculated according to the following equation (29): 1,000 {[A420 − (1.75 × OD550)]/(time × OD600)} × dilution factor, where time is in seconds.
Disulfide stress treatment.
WP778 was grown in LB medium supplemented with 8 mM dithiothreitol (DTT) at 37°C until an OD600 of 0.5 was reached. To induce disulfide stress in WP778, the medium was exchanged by spinning down the cells (5,000 × g, 5 min, 37°C) and resuspension of the cells in the same amount of DTT-free LB medium. Strain DHB4 was grown in LB medium at 37°C. At an OD600 of about 0.5, diamide was added to a final concentration of 1 mM. A culture volume corresponding to 2 ml of a culture of an OD600 of 1 was harvested by centrifugation (10,000 × g, 10 min, 4°C) immediately before and at several time points after removal of DTT in the case of WP778 or after addition of diamide in the case of DHB4. Cells were washed twice with double-distilled H2O, resuspended in 550 μl two-dimensional (2D) rehydration buffer (7 M urea, 2 M thiourea, 1% [wt/vol] Serdolit MB-1, 1% [wt/vol] dithiothreitol, 4% [wt/vol] CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 0.5% [vol/vol] Pharmalyte 3-10), and then lysed by sonication on ice (power setting, 3.75; 10 pulses of 1 s on and 1 s off performed 2 times interrupted by 30 s; 550 Sonic Dismembrator; Fisher Scientific, Pittsburgh, PA).
2D gel electrophoresis.
2D gel electrophoresis was performed using immobilized pH gradient strips, pH 4 to 7 (Amersham Biosciences, Piscataway, NJ), as previously described (30). Gels were stained as described previously (31) and scanned using an Expression 1680 scanner with a transparency unit (Epson America Inc., Long Beach, CA) at a resolution of 200 dots per inch with a 16-bit grayscale. Gel images were analyzed using Delta2D software (Decodon GmbH, Greifswald, Germany).
Determination of diamide concentration in medium.
Strain DHB4 was grown aerobically in glucose MOPS minimal medium (27) containing 40 μg/ml l-leucine and 100 μg/ml thiamine at 37°C. At an OD600 of 0.4, diamide was added to a final concentration of 1 mM. To monitor the diamide concentration in the medium, 1.5-ml samples were harvested, cells were spun down (13,000 × g, 2 min, 4°C), and the absorption of the filtered (0.2-μm-pore-size syringe filters) supernatant was determined at a maximum λ (diamide) of 296 nm (ε = 3,000 M−1 cm−1 [32]).
Quantitative real-time PCR (RT-PCR).
Strains BB7222 and BB7224 were grown aerobically at 30°C in LB medium. When an OD600 of 0.5 was reached, the cultures were shifted to 43°C to induce heat shock conditions. Disulfide stress was induced by addition of 1 mM diamide to the medium. For RNA extraction, 1.2 ml of culture was harvested on 600 μl ice-cold killing buffer (20 mM NaN3, 5 mM MgCl2, 20 mM Tris-HCl, pH 7.5) and the RNA was extracted using an RNeasy minikit (Qiagen, Valencia, CA) according to the protocol provided by the manufacturer.
RNA was subjected to a second DNase I treatment using an Ambion DNA-free kit (Ambion Inc., Austin, TX). The RNA concentration and A260/A280 as well as A260/A230 ratios were measured with a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE) (see Table S1 in the supplemental material). For cDNA synthesis, a total of 400 ng RNA was incubated for 5 min at 65°C with 1 μl of 10 mM deoxynucleoside triphosphates and 1 μl of 50 μM random hexamers (Invitrogen, Grand Island, NY) in a total volume of 13 μl. A mixture containing 4 μl 5× first-strand buffer, 1 μl 0.1 M DTT, 1 μl RNaseOUT recombinant RNase inhibitor (Invitrogen, Grand Island, NY), and 1 μl SuperScript III reverse transcriptase (Invitrogen, Grand Island, NY) was added, and cDNA synthesis was performed for 60 min at 50°C, followed by enzyme inactivation at 70°C for 15 min. Primer design was performed using Primer3 software (v.0.4.0) (33) with the following parameters: product size range, 180 to 230 bp; primer size, 20 to 27 bp; primer melting temperature, 60 to 64°C.
Real-time PCR was performed in a total volume of 10 μl containing 200 ng of cDNA, 5 μl SYBR GreenER (Invitrogen, Grand Island, NY), and 0.5 μl of each gene-specific primer (100 pmol/μl). The primers used are listed in Table 2. After initial denaturation for 4 min at 95°C, amplification was performed for 40 cycles (30 s at 95°C, 30 s at 62°C, 30 s at 72°C) prior to melting curve analysis. Expression of each gene was analyzed in triplicate using two biological replicates. No-template controls were included for each primer pair. Since amplification efficiencies were between 96% and 102%, data analysis was performed using the 2−ΔΔCT method, where CT is the threshold cycle (34). Expression of test genes upon stress treatment was normalized against accD expression (first normalization) and expression before stress treatment (second normalization). accD is a gene that has been used as a suitable control in experiments with E. coli exposed to various stress conditions (35). To test the suitability of accD as a gene for normalization, phoP was used as an alternative normalization gene. An assessment of the expression values of hslR under diamide stress yielded comparable results for both normalization genes (see Fig. S2 in the supplemental material).
Table 2.
Primers used for RT-PCR
| Primer | Sequence | Use |
|---|---|---|
| hslO_F | TGCCGCAACATGACCAATTACATC | Test gene |
| hslO_R | TCACCATCAAACTTCAGCGTAGCGGT | Test gene |
| hslR_F | CTGTTGAGGTTCGACTGGATAAATG | Test gene |
| hslR_R | GCTGTTCAGTAATCGCCTTTACAAT | Test gene |
| dnaJ_2_FW | GAAGCTTATGAAGTTCTGACCGACT | Test gene |
| dnaJ_2_RV | GCTCCATGTTATAGCGTAAATCAGC | Test gene |
| dnaK_F | ACAGCACCCGTAAGCAGGTTGAAGAA | Test gene |
| dnaK_R | TGGGCGATTTCCATCAGTTTCTGGGA | Test gene |
| accD_F | CTGGATTGAACGAATTAAAAGCAAC | Reference gene |
| accD_F | AGGCTTCCTTCATCTAACAGGCTAT | Reference gene |
Crude extract aggregation assay.
Thirty milliliters of LB medium was inoculated with E. coli strain BB7222 or BB7224 and incubated overnight at 37°C with shaking. Overnight cultures were subcultured to 2 liters LB medium and incubated at 37°C with shaking until an OD600 of 0.6 to 0.8 was reached. Cells were harvested (10 min, 8,000 rpm, 4°C), washed in 25 ml of ice-cold lysis buffer (50 mM HEPES–KOH, pH 8.0, 175 mM NaCl, 1 mM MgCl2), and resuspended in 10 ml lysis buffer. Cells were lysed by two passages through a French pressure cell at 1,200 lb/in2. Cell debris was removed by centrifugation at 3,000 × g for 5 min. The supernatant was aliquoted and stored at −80°C. Cell-free lysates were thawed on ice, and aggregates were removed by centrifugation for 20 min at 16,100 × g at 4°C. Supernatants were treated with diamide (0 to 100 mM) at 30°C (disulfide stress) or incubated at 43°C for 20 min (heat stress). After centrifugation for 20 min at 16,100 × g at 4°C, 10 μl 5× reducing SDS buffer was added to the supernatant. Pellets were rinsed with 100 μl lysis buffer prior to addition of 50 μl 1× reducing SDS buffer. Proteins were analyzed on Mini-PROTEAN TGX precast gels (Bio-Rad).
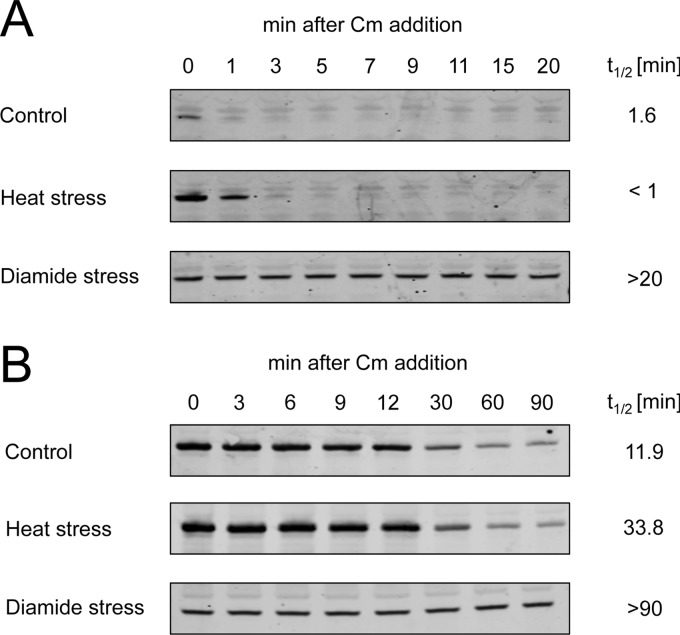
In vivo stability of σ32.
The in vivo stability of σ32 and LpxC was analyzed as previously described (36, 37). E. coli MC4100 carrying the rpoH (encoding σ32) expression plasmid pEC5217 or the lpxC expression plasmid pBO110 was grown in LB medium until the OD600 reached 0.5. The culture was split into three subcultures, and expression of σ32 or LpxC was induced by the addition of 1 mM IPTG or 0.025% arabinose, respectively. While one culture was cultivated at 30°C (control), 1 mM diamide (disulfide stress) was added to the second culture after 5 min of IPTG or arabinose induction. The third culture was transferred to 43°C (heat stress) after 20 min of IPTG or arabinose induction. Protein synthesis was stopped in all cultures by adding chloramphenicol to a final concentration of 200 μg/ml after a total of 25 min of IPTG or arabinose addition. Samples of 1 ml were collected 0, 1, 3, 5, 7, 9, 11, 15, and 20 min (σ32) or 0, 3, 6, 9, 12, 30, 60, and 90 min (LpxC) after chloramphenicol addition. Samples were immediately frozen in liquid nitrogen and thawed on ice. Cells were harvested by centrifugation at 4°C for 5 min at 13,200 rpm. Pellets were resuspended in 40 μl TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). After addition of 10 μl 5× SDS sample buffer, samples were vigorously vortexed and incubated for 10 min at 95°C. Ten microliters was loaded on denaturing SDS gels. Western blotting was performed using an iBlot system according to the manufacturer's instructions (Life Technologies, Darmstadt, Germany). Immunodetection of RpoH and LpxC was performed using polyclonal rabbit anti-RpoH and anti-LpxC antibodies (Stressgene) and a secondary IRDye 680LT goat anti-rabbit IgG (H+L; Li-cor Biosciences, Bad Homburg, Germany). Visualization of RpoH and LpxC was carried out using an Odyssey infrared imaging system (Li-cor Biosciences, Bad Homburg, Germany). Band intensities were quantified using the Adobe Photoshop CS5 analysis/record measurements function.
RESULTS
Oxidative stress causes disulfide bond formation in the E. coli cytoplasm.
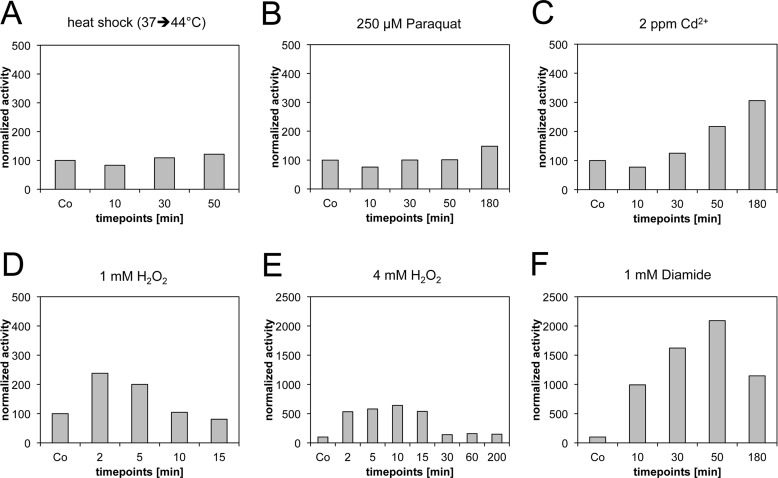
Several stressors, including cadmium, hydrogen peroxide, paraquat, diamide, and heat shock itself, have previously been described to induce the expression of heat shock proteins in E. coli (11, 38, 39). Given the known oxidizing effects of these chemicals, we hypothesized that disulfide stress might be the common denominator that induces the heat shock response. To monitor which of these stressors induces disulfide stress in vivo, we used a system based on leaderless alkaline phosphatase. This system was originally introduced by Derman and coworkers to identify mutations that allow disulfide bond formation in the cytoplasm (28). It uses a variant of alkaline phosphatase termed PhoAΔ2-22 as the reporter enzyme. Wild-type alkaline phosphatase is a periplasmic enzyme that needs two disulfide bonds to gain full catalytic activity (40). The PhoAΔ2-22 variant, however, lacks a signal sequence and is not exported into the periplasm. Therefore, PhoAΔ2-22 can gain activity only if disulfide bonds are formed in the cytoplasm, making it a versatile tool to test the disulfide stress-inducing abilities of oxidative stressors. We observed no significant activation of PhoAΔ2-22 under heat shock conditions (Fig. 1). Low millimolar concentrations of hydrogen peroxide (1 to 4 mM), however, transiently activated PhoAΔ2-22 (up to ∼7-fold induction after 10 min of exposure to 4 mM H2O2), indicating that peroxide does induce disulfide bond formation in E. coli. The relatively low level of PhoA activity confirms our previous observation that hydrogen peroxide does not lead to a large excess of disulfide bond formation in cytoplasmic proteins (41). Cadmium (2 ppm Cd2+) and paraquat (an inducer of superoxide [42]) stress also caused a low level of activation of PhoAΔ2-22 (∼3- and ∼1.5-fold, respectively). This activation was, however, observed only at later time points (50 to 180 min) (Fig. 1). The most potent activator of PhoAΔ2-22 was diamide. Within minutes of addition of 1 mM diamide, an ∼10-fold gain of alkaline phosphatase activity was observed (Fig. 1 and 2). The alkaline phosphatase activity remained high (up to ∼24-fold induction) for more than 180 min, until diamide was cleared from the medium (Fig. 2). The increase of alkaline phosphatase over time indicates the relatively slow kinetics of alkaline phosphatase oxidation by diamide. Western blots using an antibody against alkaline phosphatase confirmed that the gain of alkaline phosphatase activity was not due to additional induction of the PhoAΔ2-22 protein upon disulfide stress (Fig. 2).
Fig 1.
Disulfide bond formation in the cytoplasm of E. coli determined by leaderless alkaline phosphatase activation. PhoAΔ2-22 activity was measured under different stress conditions. Activity is expressed as a percentage of the activity under control conditions. Note the differences in the scales for the 4 mM H2O2- and diamide-treated samples.
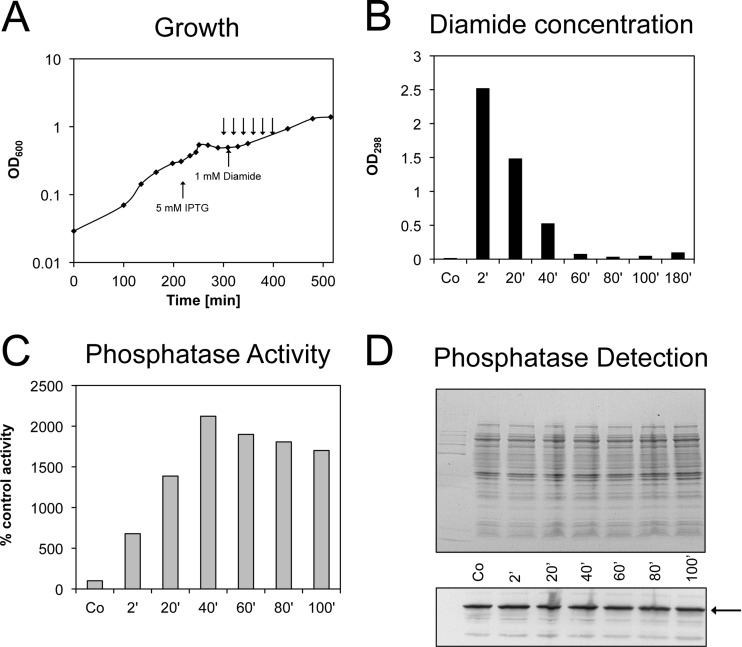
Fig 2.
Disulfide stress upon diamide treatment of E. coli cells grown in MOPS minimal medium. (A) The formation of disulfide bonds in the cytoplasm parallels the diamide concentration in the medium. The arrows in the growth curve represent the time points at which samples were taken to measure the diamide concentration in the medium (B) and PhoAΔ2-22 activity (C). (D) The Coomassie-stained protein gel and Western blot with alkaline phosphatase antibody show that the expression of PhoAΔ2-22 (arrow) is not significantly induced upon addition of diamide to the medium. The incubation times with diamide are indicated. The gain of activity can be attributed only to disulfide bond formation. Co, control.
Disulfide stress induces heat shock proteins in E. coli.
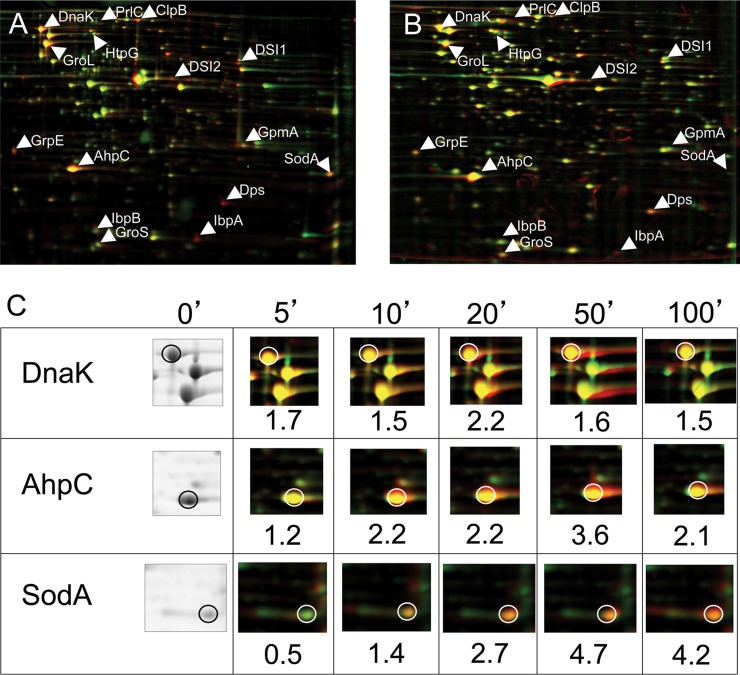
Diamide is a chemical agent introduced by Kosower and Kosower (32) which specifically triggers disulfide formation in thiol-containing compounds. With diamide, we now have a suitable tool to induce disulfide stress without primarily causing other oxidation reactions initiated by classical oxidative stressors. To investigate the cellular response of E. coli toward disulfide stress, we analyzed the protein expression pattern of diamide-stressed cell cultures using 2D gels. We observed the induction of at least 15 different proteins within 10 min of diamide addition. Most of these proteins were identified to be σ32-controlled heat shock proteins like DnaK/J, GroEL/ES, and small heat shock proteins IbpA and IbpB, or oxidative stress proteins, such as SodA, which are expressed in response to OxyR or SoxR activation (Table 3; Fig. 3).
Table 3.
Proteins upregulated more than 1.5-fold in 2 independent experiments 10 min after addition of 1 mM diamide
| Protein namea | GenBank accession no. | Functiona | Molecular mass (kDa) | No. of peptides matched | Peptide coverage (%) | Regulator |
|---|---|---|---|---|---|---|
| AhpC | EG11384 | Alkyl hydroperoxide reductase, subunit C; reduced by the AhpF subunit | 20.8 | OxyRc | ||
| ClpB | EG10157 | Bichaperone with DnaK for protein disaggregation | 95.6 | 30 | 44 | RpoHd |
| DnaK | EG10241 | Hsp70 molecular chaperone | 69.1 | 27 | 52 | RpoHe |
| Dps | EG11415 | Stress response DNA-binding protein with protective role | 18.7 | 7 | 22 | OxyRc |
| GpmA | EG11699 | Phosphoglycerate mutase, 2,3-bisphosphoglycerate dependent | 28.6 | 9 | 36 | Furf |
| GroEL | EG10599 | Chaperonin Cpn60 | 57.3 | 3 | 12 | RpoHe |
| GroES | EG10600 | Chaperonin Cpn10 | 10.4 | 6 | 61 | RpoHe |
| GrpE | EG10416 | Nucleotide exchange factor for the DnaKJ chaperone | 21.8 | 5 | 25 | RpoHe |
| HtpG | EG10461 | Protein refolding molecular cochaperone Hsp90 | 71.4 | 27 | 45 | RpoHe |
| IbpA | EG11534 | Chaperone, HSP20 family | 15.8 | 5 | 29 | RpoHe |
| IbpB | EG11535 | Chaperone, HSP20 family | 16.0 | 5 | 34 | RpoHe |
| PrlC | EG11441 | Oligopeptidase A, zinc metalloprotease | 77.2 | 16 | 23 | RpoHg |
| SodA | EG10953 | Superoxide dismutase, Mn | 23.1 | 9 | 34 | SoxRh |
| DSI1b | NAi | Spot not identified | NA | |||
| DSI2b | NA | Spot not identified | NA |
Fig 3.
Disulfide stress induces oxidative stress and heat shock genes in E. coli. (A) False-colored overlay of 2D gels of cell extracts of E. coli wild-type (DHB4) cells grown under control conditions (green) and 30 min after addition of 1 mM diamide to the medium (red). Protein spots that are induced upon addition of diamide appear red. Proteins that are upregulated more than 1.5-fold 10 min after addition of diamide are labeled. (B) False-colored overlay of 2D gels of cell extracts of E. coli Δtrx Δgor (WP778) cells grown with DTT in the medium (green) and 30 min after removal of DTT (red). WP778 is a strain that lacks thioredoxin reductase and glutathione oxidoreductase and forms disulfide bonds in the cytoplasm when grown in the absence of DTT. Proteins induced under disulfide stress conditions upon removal of DTT appear red. Proteins upregulated more than 1.5-fold 10 min after addition of diamide are labeled to facilitate comparison. (C) Time course of the induction of selected heat shock and oxidative stress genes upon addition of diamide. Incubation times with diamide and induction factors are indicated.
To investigate whether the observed induction of heat shock genes was specific for diamide or a feature of general disulfide stress, we analyzed the protein expression of E. coli mutant cells which are exposed to intrinsic disulfide stress. These mutants lack both disulfide-reducing systems due to a deletion in thioredoxin reductase TrxB and glutathione oxidoreductase Gor. The ΔtrxB Δgor double mutant is able to grow only in the presence of DTT in the medium. As shown by a leaderless alkaline phosphatase assay by Prinz et al., these cells accumulate disulfide bonds in the cytoplasm and cease to grow upon removal of DTT from the medium (43). The protein expression pattern of this mutant strain upon removal of DTT looked strikingly similar to the one that we observed under diamide-induced disulfide stress in wild-type cells (Fig. 3). All proteins that were identified to be upregulated more than 1.5-fold 10 min after addition of diamide were upregulated in this strain as well.
Disulfide stress induction of heat shock genes is rpoH dependent.
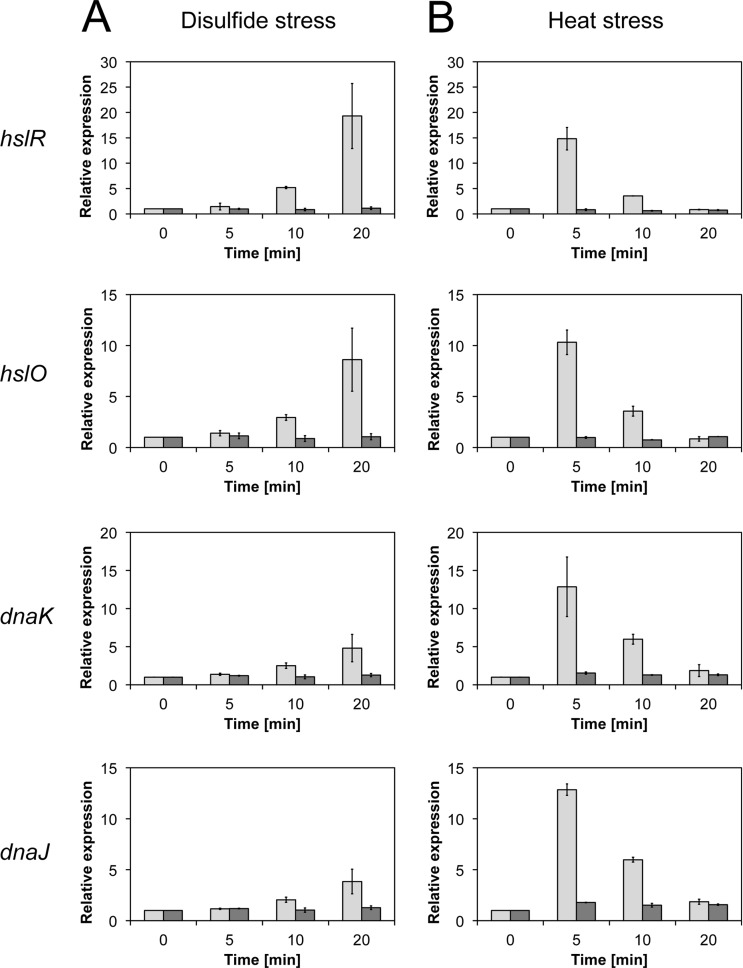
Disulfide stress-mediated induction of heat shock genes has also been observed in the Gram-positive model organism B. subtilis (5) and found to be dependent on the disulfide stress-sensing regulator Spx (9). So far, no specific disulfide stress-sensing regulator is known in E. coli. To investigate whether the heat shock response is directly triggered by oxidative stress in a σ32-dependent manner or requires a specific, yet to be identified regulator, we compared the expression levels of heat shock genes in diamide-treated wild-type and rpoH deletion strains. To directly quantify the induction of heat shock genes upon disulfide stress on the RNA level, we performed quantitative real-time PCR analyses. We selected the genes dnaK and dnaJ, hslR, and hslO, encoding the DnaK/J chaperones, the ribosome-associated heat shock protein Hsp15, and the redox-regulated chaperone Hsp33, respectively (44–46). These genes are organized in two independent transcriptional units, dnaK/J and hslR/O, and are both known to be strongly induced under heat stress conditions (47, 48). Induction of all four genes was observed upon heat stress as well as upon disulfide stress in wild-type E. coli (Fig. 4). As observed before, expression of the heat shock genes peaked within 5 min of heat treatment (Fig. 4B) (49). In contrast, heat shock gene induction upon diamide stress increased gradually over a time period of at least 20 min (Fig. 4A). Comparative analysis in a mutant strain lacking rpoH revealed no significant upregulation of the four genes in response to either heat shock or diamide treatment. These results indicated that σ32 is both required and sufficient for the induction of the disulfide stress-mediated heat shock response in E. coli (Fig. 4).
Fig 4.
Induction of heat shock genes upon disulfide (A) and heat (B) stress in the wild type (light gray bars) and ΔrpoH mutant (dark gray bars). The expression of the hslR, hslO, dnaJ, and dnaK genes upon disulfide and heat stress depends on σ32, as shown by quantitative real-time PCR. Disulfide stress induces a gradual increase in expression over time, while heat stress causes a transient upregulation within 5 min of stress treatment.
σ32 stabilization upon disulfide stress initiates the heat shock response.
The classical heat shock sigma factor is regulated on three different levels. On the transcriptional level, rpoH, the gene for σ32, is transcribed from at least four different promoters, three of which are dependent on the housekeeping sigma factor σ70 and one of which is dependent on σE (15, 16). A second regulatory mechanism affects the secondary structure of rpoH mRNA. Upon heat shock, the base-pairing pattern in its mRNA changes due to partial melting. This makes the Shine-Dalgarno sequence accessible, which enhances translation initiation (17–20). Yet another regulation of σ32 happens on the level of σ32 protein stability. Under nonstress conditions, σ32 has a very short half-life due to its inactivation with the DnaK chaperone system (24, 50), which mediates σ32's degradation by the protease FtsH (26, 51). Heat shock conditions lead to the unfolding of cellular proteins, which become substrates of DnaK and in turn titrate the DnaK system. This causes the release of σ32 from DnaK and a significant increase in its half-life.
In our disulfide stress experiments, the transcription levels of heat shock genes showed a gradual increase, which was in contrast to the transient induction typical for heat stress. This observation suggested to us a mechanism different from the classical induction of the heat shock response. We hypothesized that σ32 stabilization rather than an RNA thermometer-based posttranscriptional regulation of σ32 might be a more plausible mechanism. To test this hypothesis, we analyzed the steady-state stability of plasmid-encoded σ32 upon disulfide and heat stress. Addition of the protein translation inhibitor chloramphenicol stopped the synthesis of the plasmid-encoded σ32. We then quantified the abundance of σ32 with a specific antibody over time. Under nonstress conditions, σ32 was rapidly degraded with a half-life of 1.5 min (Fig. 5). A shift of E. coli cells from 30°C to 43°C resulted in a considerable increase in the initial amount of σ32. Nevertheless, the half-life of σ32 was slightly shorter than that under control conditions, as observed by others (22). In contrast, disulfide stress caused by diamide and H2O2 (Fig. 5; see Fig. S3 in the supplemental material) significantly stabilized σ32, and no degradation of σ32 was observed during the time course of this experiment. A 20-min paraquat treatment, which induced only low levels of nonnative disulfide bond formation, did increase the half-life of σ32 only slightly to 1.9 min (see Fig. S3 in the supplemental material).
Fig 5.
Analysis of σ32 and LpxC stability upon heat and disulfide stress. Cell cultures containing a σ32 or LpxC expression plasmid were grown to an OD600 of 0.5, and protein expression was induced by the addition of IPTG or arabinose. Cells were either left untreated (control), treated with heat by shifting the cultures from 30°C to 43°C after 20 min of induction, or treated with 1 mM diamide after 5 min of induction. After a total of 25 min of induction, chloramphenicol (Cm) was added (time zero) and the stability of σ32 (A) and LpxC (B) was analyzed by Western blotting using σ32- and LpxC-specific antibodies. t1/2, half-life.
To check if stabilization of σ32 upon disulfide stress involves inhibition of FtsH, we analyzed the stability of the well-characterized FtsH substrate LpxC (Fig. 5B). Under control conditions, LpxC was degraded within 12 min. While heat stress increased the half-life of LpxC approximately 3-fold, diamide stress resulted in complete stabilization. This result indicates that the induction of the heat stress response upon disulfide stress is mediated by a stabilization of σ32 through inhibition of FtsH-dependent degradation.
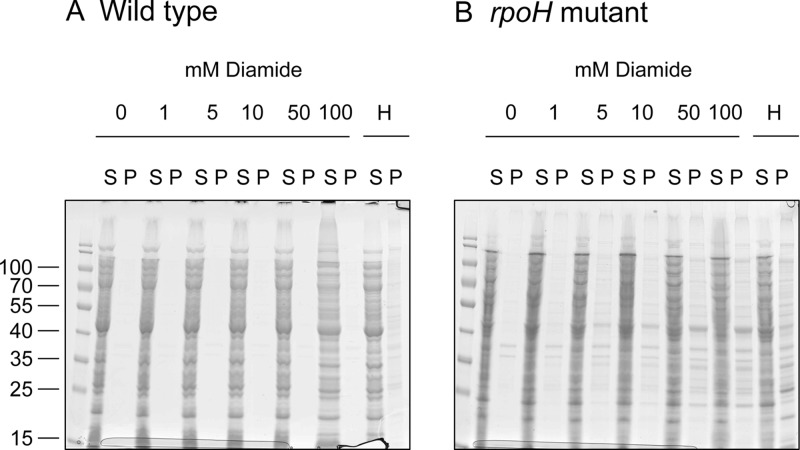
Disulfide stress leads to protein aggregation in the absence of σ32-controlled chaperones.
In addition to a direct FtsH inhibition, the observed stabilization of σ32 under disulfide stress conditions could be caused by titration of the DnaK system. Unfolded proteins, which might accumulate due to the formation of nonnative disulfide bonds, may be recognized and bound by the DnaK system. If that was the case, proteins would aggregate in the absence of DnaK under disulfide stress conditions. We therefore analyzed the solubility of proteins in wild-type and rpoH crude extracts after diamide and heat treatment. In these experiments, the solubility of proteins gradually decreased with increasing diamide concentrations in extracts obtained from the ΔrpoH strain (Fig. 6). This aggregation was not observed in wild-type extracts, indicating that chaperones under the control of σ32, such as DnaK, can prevent disulfide stress-induced protein aggregation. Interestingly, many protein bands that were visible in the pellet fraction of extracts from the rpoH strain after diamide treatment were identical to those that were found after heat treatment. This finding supports our model indicating that a similar set of σ32-controlled chaperones is responsible for the protection of cytoplasmic proteins against heat stress- and disulfide stress-mediated protein aggregation in E. coli.
Fig 6.
Aggregation of proteins from E. coli crude extracts. Extracts from the E. coli wild type (A) and ΔrpoH mutant (B) were treated with diamide (0 to 100 mM) or with heat (H) prior to separation of soluble and insoluble proteins by centrifugation, followed by SDS-PAGE. The solubility of proteins is decreased in the rpoH mutant. Numbers on the left are molecular masses (in kDa). Lanes S, soluble protein; lanes P, insoluble proteins.
DISCUSSION
In this study, we show that disulfide stress, the formation of nonnative disulfide bonds in the cytoplasm, is an integral part of oxidative stress in E. coli. In cells exposed to chemically or genetically induced disulfide stress, we observed a strong upregulation of typical oxidative stress genes under the control of the oxidative stress regulators OxyR and SoxR. These two regulators have cysteines in their redox-sensing centers, which are probably modified under these conditions. OxyR is known to be activated by the formation of an intramolecular disulfide bond (52). Global studies in other organisms have also shown that oxidative stress genes are a major part of the disulfide stress response (5, 53).
In addition, however, we discovered that disulfide stress is a potent inducer of the σ32-dependent heat shock response in E. coli. In our study, we focused on the mechanistic underpinnings of this particular finding. The heat shock response has been shown to be part of the oxidative stress response in other organisms as well. In B. subtilis, a specific part of the heat shock response is affected by diamide-induced disulfide stress (5). The so-called class III heat shock genes under the control of the global regulator CtsR are upregulated in this organism, while other heat shock genes under the control of the CIRCE element and the alternative sigma factor σB are affected to a lesser extent or are not affected at all (5). The induction of class III heat shock genes under these conditions is mediated by the disulfide stress-specific regulator Spx (9).
Unlike in this Gram-positive organism, which uses a specific mechanism that does not rely on an alternative sigma factor, we found that in the Gram-negative bacterium E. coli the heat shock sigma factor σ32 is both required and by itself sufficient for the induction of heat shock genes under disulfide stress. Our experiments show that the σ32-dependent induction of heat shock genes is the consequence of the stabilization of the sigma factor.
This is presumably caused by inhibition of FtsH-dependent degradation under disulfide stress conditions. If direct inactivation of FtsH by disulfide stress was an evolved mechanism, one would expect highly conserved regulatory cysteines in this protease. However, the two cysteines of E. coli FtsH are not highly conserved. Alternatively, substrate proteins with nonnative disulfide bonds could block the active site of FtsH and thus lead to inactivation of the protease. In addition to direct blockage of FtsH, delivery of substrates to the protease by the DnaK system might be disturbed due to binding of unfolded proteins. DnaK might not be able to resolve nonnative disulfide bonds without the help of reducing systems, leading to a longer-lasting and more effective occupation of DnaK under disulfide stress than under heat shock.
The fundamental difference in the regulation of genes under disulfide stress in these Gram-positive and Gram-negative model organisms is reflected in the regulation of target genes. In B. subtilis hslO, the gene for the redox-regulated chaperone Hsp33 is under direct control of the disulfide stress regulator Spx (9). In E. coli, hslO is under σ32 control. Hsp33 is a chaperone that is activated only upon a posttranslational modification involving the formation of disulfide bonds in the regulatory C-terminal domain (45). Hsp33 needs this disulfide bond formation to gain activity. Hsp33 has been shown to take over the role of the DnaK system under oxidative heat stress conditions (54).
Some alphaproteobacteria possess two or more RpoH homologs which may have overlapping and specialized functions (55, 56). RpoHII from Rhodobacter sphaeroides has recently been shown to be involved in the defense against singlet oxygen, while it does not seem to play a major role in the heat shock response (57). It is tempting to speculate that in this organism gene duplication allowed the divergent evolution of one RpoH with a broader role into two separate heat shock and oxidative stress regulators.
In E. coli, the upregulation of heat shock genes by oxidative stress is regulated by σ32 alone. This σ32-dependent regulatory mechanism is distinct from heat shock-mediated gene regulation. Under oxidative stress, the formation of nonnative disulfide bonds in the cytoplasm triggers protein unfolding. The unfolded proteins then bind to the DnaK system, which prevents delivery of σ32 to FtsH. Inactivation of FtsH could additionally lead to the lasting stabilization of σ32.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael J. Gray for his help in setting up the RT-PCR experiments.
This work was supported by a Boehringer Ingelheim travel grant to A.M., by National Institutes of Health grant GM065318 to U.J., a grant from the German Research Foundation (DFG; SFB 642, ATP- and GTP-Dependent Membrane Processes) to F.N., and a grant from the NRW-Rückkehrerprogramm of the German state of North Rhine-Westphalia to L.I.L.
Footnotes
Published ahead of print 12 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00127-13.
REFERENCES
- 1. Storz G, Tartaglia LA, Ames BN. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189–194 [DOI] [PubMed] [Google Scholar]
- 2. Mongkolsuk S, Helmann JD. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9–15 [DOI] [PubMed] [Google Scholar]
- 3. Delaunay A, Isnard AD, Toledano MB. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liebeke M, Pöther D-C, van Duy N, Albrecht D, Becher D, Hochgräfe F, Lalk M, Hecker M, Antelmann H. 2008. Depletion of thiol-containing proteins in response to quinones in Bacillus subtilis. Mol. Microbiol. 69:1513–1529 [DOI] [PubMed] [Google Scholar]
- 5. Leichert LIO, Scharf C, Hecker M. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakano S, Erwin KN, Ralle M, Zuber P. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498–510 [DOI] [PubMed] [Google Scholar]
- 7. Kallifidas D, Thomas D, Doughty P, Paget MSB. 2010. The σR regulon of Streptomyces coelicolor A32 reveals a key role in protein quality control during disulphide stress. Microbiology (Reading, Engl.) 156:1661–1672 [DOI] [PubMed] [Google Scholar]
- 8. Chi BK, Albrecht D, Gronau K, Becher D, Hecker M, Antelmann H. 2010. The redox-sensing regulator YodB senses quinones and diamide via a thiol-disulfide switch in Bacillus subtilis. Proteomics 10:3155–3164 [DOI] [PubMed] [Google Scholar]
- 9. Rochat T, Nicolas P, Delumeau O, Rabatinová A, Korelusová J, Leduc A, Bessières P, Dervyn E, Krásny L, Noirot P. 2012. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 40:9571–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fargier E, Mac Aogáin M, Mooij MJ, Woods DF, Morrissey JP, Dobson ADW, Adams C, O'Gara F. 2012. MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J. Bacteriol. 194:3502–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arsène F, Tomoyasu T, Bukau B. 2000. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 55:3–9 [DOI] [PubMed] [Google Scholar]
- 13. Craig EA, Gross CA. 1991. Is Hsp70 the cellular thermometer? Trends Biochem. Sci. 16:135–140 [DOI] [PubMed] [Google Scholar]
- 14. Straus DB, Walter WA, Gross CA. 1987. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature 329:348–351 [DOI] [PubMed] [Google Scholar]
- 15. Erickson JW, Vaughn V, Walter WA, Neidhardt FC, Gross CA. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1:419–432 [DOI] [PubMed] [Google Scholar]
- 16. Fujita N, Ishihama A. 1987. Heat-shock induction of RNA polymerase σ32 synthesis in Escherichia coli: transcriptional control and a multiple promoter system. Mol. Gen. Genet. 210:10–15 [DOI] [PubMed] [Google Scholar]
- 17. Nagai H, Yuzawa H, Yura T. 1991. Interplay of two cis-acting mRNA regions in translational control of σ32 synthesis during the heat shock response of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:10515–10519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morita M, Kanemori M, Yanagi H, Yura T. 1999. Heat-induced synthesis of σ32 in Escherichia coli: structural and functional dissection of rpoH mRNA secondary structure. J. Bacteriol. 181:401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. 1999. Translational induction of heat shock transcription factor σ32: evidence for a built-in RNA thermosensor. Genes Dev. 13:655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamath-Loeb AS, Gross CA. 1991. Translational regulation of σ32 synthesis: requirement for an internal control element. J. Bacteriol. 173:3904–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grossman AD, Straus DB, Walter WA, Gross CA. 1987. σ32 synthesis can regulate the synthesis of heat shock proteins in Escherichia coli. Genes Dev. 1:179–184 [DOI] [PubMed] [Google Scholar]
- 22. Tilly K, Spence J, Georgopoulos C. 1989. Modulation of stability of the Escherichia coli heat shock regulatory factor sigma. J. Bacteriol. 171:1585–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanemori M, Nishihara K, Yanagi H, Yura T. 1997. Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of σ32 and abnormal proteins in Escherichia coli. J. Bacteriol. 179:7219–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gamer J, Multhaup G, Tomoyasu T, McCarty JS, Rüdiger S, Schönfeld HJ, Schirra C, Bujard H, Bukau B. 1996. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor σ32. EMBO J. 15:607–617 [PMC free article] [PubMed] [Google Scholar]
- 25. Guisbert E, Herman C, Lu CZ, Gross CA. 2004. A chaperone network controls the heat shock response in E. coli. Genes Dev. 18:2812–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herman C, Thévenet D, D'Ari R, Bouloc P. 1995. Degradation of σ32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. U. S. A. 92:3516–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Derman AI, Prinz WA, Belin D, Beckwith J. 1993. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 262:1744–1747 [DOI] [PubMed] [Google Scholar]
- 29. Brickman E, Beckwith J. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and ϕ80 transducing phages. J. Mol. Biol. 96:307–316 [DOI] [PubMed] [Google Scholar]
- 30. Hiniker A, Bardwell JCA. 2004. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J. Biol. Chem. 279:12967–12973 [DOI] [PubMed] [Google Scholar]
- 31. Wong C, Sridhara S, Bardwell JC, Jakob U. 2000. Heating greatly speeds Coomassie blue staining and destaining. Biotechniques 28:426–428, 430,, 432 [DOI] [PubMed] [Google Scholar]
- 32. Kosower NS, Kosower EM. 1995. Diamide: an oxidant probe for thiols. Methods Enzymol. 251:123–133 [DOI] [PubMed] [Google Scholar]
- 33. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 34. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 35. Bore E, Hébraud M, Chafsey I, Chambon C, Skjaeret C, Moen B, Møretrø T, Langsrud Ø, Rudi K, Langsrud S. 2007. Adapted tolerance to benzalkonium chloride in Escherichia coli K-12 studied by transcriptome and proteome analyses. Microbiology (Reading, Engl.) 153:935–946 [DOI] [PubMed] [Google Scholar]
- 36. Obrist M, Narberhaus F. 2005. Identification of a turnover element in region 2.1 of Escherichia coli σ32 by a bacterial one-hybrid approach. J. Bacteriol. 187:3807–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Führer F, Langklotz S, Narberhaus F. 2006. The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol. Microbiol. 59:1025–1036 [DOI] [PubMed] [Google Scholar]
- 38. Ferianc P, Farewell A, Nyström T. 1998. The cadmium-stress stimulon of Escherichia coli K-12. Microbiology (Reading, Engl.) 144:1045–1050 [DOI] [PubMed] [Google Scholar]
- 39. Greenberg JT, Demple B. 1989. A global response induced in Escherichia coli by redox-cycling agents overlaps with that induced by peroxide stress. J. Bacteriol. 171:3933–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sone M, Kishigami S, Yoshihisa T, Ito K. 1997. Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272:6174–6178 [DOI] [PubMed] [Google Scholar]
- 41. Leichert LI, Jakob U. 2004. Protein thiol modifications visualized in vivo. PLoS Biol. 2:e333. 10.1371/journal.pbio.0020333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bus JS, Gibson JE. 1984. Paraquat: model for oxidant-initiated toxicity. Environ. Health Perspect. 55:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prinz WA, Aslund F, Holmgren A, Beckwith J. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272:15661–15667 [DOI] [PubMed] [Google Scholar]
- 44. Korber P, Stahl JM, Nierhaus KH, Bardwell JC. 2000. Hsp15: a ribosome-associated heat shock protein. EMBO J. 19:741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jakob U, Muse W, Eser M, Bardwell JC. 1999. Chaperone activity with a redox switch. Cell 96:341–352 [DOI] [PubMed] [Google Scholar]
- 46. Bukau B, Horwich AL. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366 [DOI] [PubMed] [Google Scholar]
- 47. Zhou YN, Kusukawa N, Erickson JW, Gross CA, Yura T. 1988. Isolation and characterization of Escherichia coli mutants that lack the heat shock sigma factor sigma 32. J. Bacteriol. 170:3640–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. 2006. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20:1776–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bukau B. 1993. Regulation of the Escherichia coli heat-shock response. Mol. Microbiol. 9:671–680 [DOI] [PubMed] [Google Scholar]
- 50. Tatsuta T, Joob DM, Calendar R, Akiyama Y, Ogura T. 2000. Evidence for an active role of the DnaK chaperone system in the degradation of σ32. FEBS Lett. 478:271–275 [DOI] [PubMed] [Google Scholar]
- 51. Tatsuta T, Tomoyasu T, Bukau B, Kitagawa M, Mori H, Karata K, Ogura T. 1998. Heat shock regulation in the ftsH null mutant of Escherichia coli: dissection of stability and activity control mechanisms of σ32 in vivo. Mol. Microbiol. 30:583–593 [DOI] [PubMed] [Google Scholar]
- 52. Zheng M, Aslund F, Storz G. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718–1721 [DOI] [PubMed] [Google Scholar]
- 53. Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol. Microbiol. 42:1007–1020 [DOI] [PubMed] [Google Scholar]
- 54. Winter J, Ilbert M, Graf PCF, Ozcelik D, Jakob U. 2008. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell 135:691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Narberhaus F, Krummenacher P, Fischer HM, Hennecke H. 1997. Three disparately regulated genes for σ32-like transcription factors in Bradyrhizobium japonicum. Mol. Microbiol. 24:93–104 [DOI] [PubMed] [Google Scholar]
- 56. Oke V, Rushing BG, Fisher EJ, Moghadam-Tabrizi M, Long SR. 2001. Identification of the heat-shock sigma factor RpoH and a second RpoH-like protein in Sinorhizobium meliloti. Microbiology 147:2399–2408 [DOI] [PubMed] [Google Scholar]
- 57. Nuss AM, Glaeser J, Klug G. 2009. RpoH(II) activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J. Bacteriol. 191:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397–413 [DOI] [PubMed] [Google Scholar]
- 59. Narberhaus F, Balsiger S. 2003. Structure-function studies of Escherichia coli RpoH (σ32) by in vitro linker insertion mutagenesis. J. Bacteriol. 185:2731–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rudd KE. 2000. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 28:60–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kitagawa M, Wada C, Yoshioka S, Yura T. 1991. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (σ32). J. Bacteriol. 173:4247–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chuang SE, Blattner FR. 1993. Characterization of twenty-six new heat shock genes of Escherichia coli. J. Bacteriol. 175:5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vassinova N, Kozyrev D. 2000. A method for direct cloning of fur-regulated genes: identification of seven new Fur-regulated loci in Escherichia coli. Microbiology 146:3171–3182 [DOI] [PubMed] [Google Scholar]
- 64. Conlin CA, Miller CG. 2000. opdA, a Salmonella enterica serovar Typhimurium gene encoding a protease, is part of an operon regulated by heat shock. J. Bacteriol. 182:518–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zheng M, Storz G. 2000. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 59:1–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.