Abstract
The TonB system couples cytoplasmic membrane proton motive force (pmf) to active transport of diverse nutrients across the outer membrane. Current data suggest that cytoplasmic membrane proteins ExbB and ExbD harness pmf energy. Transmembrane domain (TMD) interactions between TonB and ExbD allow the ExbD C terminus to modulate conformational rearrangements of the periplasmic TonB C terminus in vivo. These conformational changes somehow allow energization of high-affinity TonB-gated transporters by direct interaction with TonB. While ExbB is essential for energy transduction, its role is not well understood. ExbB has N-terminus-out, C-terminus-in topology with three TMDs. TMDs 1 and 2 are punctuated by a cytoplasmic loop, with the C-terminal tail also occupying the cytoplasm. We tested the hypothesis that ExbB TMD residues play roles in proton translocation. Reassessment of TMD boundaries based on hydrophobic character and residue conservation among distantly related ExbB proteins brought earlier widely divergent predictions into congruence. All TMD residues with potentially function-specific side chains (Lys, Cys, Ser, Thr, Tyr, Glu, and Asn) and residues with probable structure-specific side chains (Trp, Gly, and Pro) were substituted with Ala and evaluated in multiple assays. While all three TMDs were essential, they had different roles: TMD1 was a region through which ExbB interacted with the TonB TMD. TMD2 and TMD3, the most conserved among the ExbB/TolQ/MotA/PomA family, played roles in signal transduction between cytoplasm and periplasm and the transition from ExbB homodimers to homotetramers. Consideration of combined data excludes ExbB TMD residues from direct participation in a proton pathway.
INTRODUCTION
In Gram-negative bacteria, the TonB system couples cytoplasmic membrane (CM) proton motive force (pmf) to active transport of ligands across the outer membrane (OM) through TonB-gated transporters (TGTs) (for reviews, see references 1 to 5). The TonB system is composed of three CM proteins, TonB, ExbB, and ExbD, and several different TGTs in the OM that recognize diverse ligands. In Escherichia coli K-12, the TonB system transports iron-siderophore compounds and vitamin B12, while in other Gram-negative bacteria, additional nutrients, including nickel, sucrose, heme, and maltodextrin, are transported (6–9).
TonB and ExbD have similar topologies of one N-terminal transmembrane domain (TMD) and a large periplasmic domain (10, 11). In contrast, ExbB contains three TMDs with a large soluble cytoplasmic loop between TMDs 1 and 2 and a soluble cytoplasmic C-terminal tail domain (12, 13). ExbB appears to be the scaffold upon which TonB and ExbD assemble, since TonB and ExbD are proteolytically unstable in the absence of ExbB, while ExbB stability is independent of TonB and ExbD (13–16; unpublished results).
TonB physically interacts with OM TGTs, connecting CM pmf energy to OM ligand transport (17–20). A proton pathway through ExbB, ExbD, and/or TonB TMD residues has been proposed based on the similar sequence homology to the flagellar pmf-harnessing motor proteins, MotA and MotB (21). Through sequence alignment, two different pathways mediated primarily by ExbB TMD residues were proposed. The first pathway includes residues in TonB: T181 → H2O (near G184) → H20 (TonB) → S16 (TonB) → H2O (near A188) → T148 → H2O (near G144) → D25 (ExbD). The second pathway does not: T181 → S155→ H2O (near G151) → T148 → H2O (near G144) → D25 (ExbD) (21).
We demonstrated that all the residues except H20 in the TonB TMD can be substituted with Ala without significantly decreasing TonB activity (22). Recently, we also showed that the importance of H20 appears to be structural, since the Asn substitution at TonB residue 20, and only Asn, restores full TonB activity (23). These studies removed H20, and thus the TonB TMD, from participation in a proton translocation pathway. However, both ExbB and ExbD are required for conformational response of the TonB C terminus to pmf (24–27).
The predicted proton pathways through ExbB from Zhai and Saier were evaluated by Braun and Herrmann in 2004 (21, 28). They found that individual Ala substitution of predicted ExbB proton pathway residues S155 T148 in TMD2 and T181 in TMD3 were functional, while double T148A/T181A substitution was inactive for unknown reasons. ExbB E176 in TMD3, which is not within the originally proposed proton pathway prediction, was inactive when replaced with Ala but fully active with Asp and, more importantly, Gln substitution, suggesting that E176 was not on a proton translocation pathway (28). In that study, however, other potential TMD residues that could affect ExbB activity, including those within ExbB TMD1, were not evaluated.
The first task for identifying residues on a proton pathway is to define TMD boundaries. In the past, widely different predictions have been made for the locations of ExbB TMD boundaries: residues 16 to 39, 25 to 42, and 23 to 42 for TMD1; residues 128 to 155, 131 to 160, and 141 to 159 for TMD2; residues 162 to 194, 177 to 199, and 172 to 194 for TMD3 (12, 13, 21).
In this study, we evaluated ExbB TMD boundaries with the TOPCONS program that uses a consensus of multiple prediction programs (29). This resulted in significant redefinition of these regions, predicting ExbB TMD1 to consist of residues 22 to 42, TMD2 to consist of residues 132 to 152, and TMD3 to consist of residues 178 to 198. Here we show that block alanine substitution of each half ExbB TMD inactivated ExbB, indicating that all 3 TMDs were essential for function. Subsequent substitution of individual TMD residues identified residues critical for signal transduction or assembly of the energy transduction complex. No residues directly participated in proton translocation. Thus, like the TonB TMD, ExbB TMDs do not appear to be part of a proton translocation pathway.
MATERIALS AND METHODS
Strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Plasmid pKP660 contains ExbB and ExbD, which are expressed from the arabinose promoter of pBAD24 (25). ExbB substitutions were constructed by site-directed mutagenesis PCR using a pKP660 template as previously described (30). Correct sequences of both exbB and exbD genes were confirmed in all mutant plasmids by the Penn State Genomics Core Facility (University Park, PA).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Reference | Arabinose concn (%)b |
|---|---|---|---|
| E. coli strains | |||
| W3110 | F− IN(rrnD-rrnE)1 | 89 | |
| RA1017 | W3110 exbBD::kan ΔtolQRA | 22 | |
| Plasmids | |||
| pBAD24 | Empty vector control | 90 | NAc |
| pKP660 | pBAD24 expressing exbBD from the pBAD promoter | 25 | 0.0003 |
| pKP1459 | exbB(Ala21-31) exbD | Present study | 0.0005 |
| pKP1460 | exbB(Ala32-42) exbD | Present study | 0.0005 |
| pKP1481 | exbB(Ala142-152) exbD | Present study | 0.0005 |
| pKP1482 | exbB(Ala141-131) exbD | Present study | 0.0009 |
| pKP1483 | exbB(Ala176-186) exbD | Present study | 0.0005 |
| pKP1484 | exbB(Ala187-197) exbD | Present study | 0.0008 |
| pKP1461 | exbB(K24A) exbD | Present study | 0.0003 |
| pKP878 | exbB(C25A) exbD | 43 | 0.0003 |
| pKP1474 | exbB(G29A) exbD | Present study | 0.0007 |
| pKP1462 | exbB(S34A) exbD | Present study | 0.0003 |
| pKP1498 | exbB(S34C) exbD | Present study | 0.0003 |
| pKP1499 | exbB(S34T) exbD | Present study | 0.0003 |
| pKP1463 | exbB(T37A) exbD | Present study | 0.0005 |
| pKP1471 | exbB(W38A) exbD | Present study | 0.0004 |
| pKP1475 | exbB(G131A) exbD | Present study | 0.0003 |
| pKP663 | exbB(Y132A) exbD | Present study | 0.0003 |
| pKP1470 | exbB(T135A) exbD | Present study | 0.0003 |
| pKP1612 | exbB(T135C) exbD | Present study | 0.0003 |
| pKP1521 | exbB(T135S) exbD | Present study | 0.0003 |
| pKP1522 | exbB(T135V) exbD | Present study | 0.0005 |
| pKP1476 | exbB(G137A) exbD | Present study | 0.0003 |
| pKP1465 | exbB(S140A) exbD | Present study | 0.0003 |
| pKP1466 | exbB(P141A) exbD | Present study | 0.0003 |
| pKP1477 | exbB(G144A) exbD | Present study | 0.0003 |
| pKP1478 | exbB(G147A) exbD | Present study | 0.0003 |
| pKP1419 | exbB(T148A) exbD | Present study | 0.0003 |
| pKP1422 | exbB(T148C) exbD | Present study | 0.001 |
| pKP1421 | exbB(T148S) exbD | Present study | 0.0005 |
| pKP1437 | exbB(T148V) exbD | Present study | 0.0005 |
| pKP1464 | exbB(W150A) exbD | Present study | 0.0003 |
| pKP1479 | exbB(G151A) exbD | Present study | 0.0003 |
| pKP1327 | exbB(E176A) exbD | Present study | 0.0005 |
| pKP1523 | exbB(E176D) exbD | Present study | 0.0003 |
| pKP1472 | exbB(E176Q) exbD | Present study | 0.0003 |
| pKP1420 | exbB(T181A) exbD | Present study | 0.0003 |
| pKP1422 | exbB(T181S) exbD | Present study | 0.0003 |
| pKP1437 | exbB(T181V) exbD | Present study | 0.0005 |
| pKP1480 | exbB(G184A) exbD | Present study | 0.0003 |
| pKP1473 | exbB(P190A) exbD | Present study | 0.0003 |
| pKP664 | exbB(Y195A) exbD | 43 | 0.0003 |
| pKP1081 | exbB(N196A) exbD | 43 | 0.0006 |
| pKP1520 | exbB(N196D) exbD | Present study | 0.0003 |
| pKP1519 | exbB(N196H) exbD | Present study | 0.0003 |
| pKP1423 | exbB(T148A, T181A) exbD | Present study | 0.0008 |
| pKP1623 | exbB(T148C, T181C) exbD | Present study | 0.0003 |
| pKP1424 | exbB(T148S, T181S) exbD | Present study | 0.0003 |
| pKP1437 | exbB(T148V, T181V) exbD | Present study | 0.0008 |
| pKP1194 | pBAD24 expressing exbD from the pBAD promoter | 91 | 0.05 |
exbB(K24A), exbB with the amino acid change K24A (the K-to-A change at position 24 encoded by exbB). There are multiple amino acid changes in some genes.
Percent arabinose used to achieve chromosomal expression level in M9 minimal medium.
NA, not applicable.
Media and culture conditions.
Luria-Bertani medium (LB), tryptone medium (T), and M9 minimal salts medium were prepared as described previously (31, 32). Agar plates, T-top agar, and liquid cultures were supplemented with 100 μg/ml of ampicillin and plasmid-specific amounts of l-arabinose such that the expression of the mutant proteins from the araBAD promoter on pKP660 was equivalent to wild-type chromosomal levels of expression. M9 salts were supplemented with 1.0% glycerol (wt/vol), 0.4 mg of thiamine/ml, 1 mM MgSO4, 0.5 mM CaCl2, 0.2% Casamino Acids (wt/vol), 40 mg of tryptophan/ml, and 1.85 μM FeCl3. Cultures were grown with aeration at 37°C.
Spot titer assays.
Strains expressing mutant ExbB proteins at chromosomal levels were grown to mid-exponential phase and plated in T-top agar on T-plates containing identical arabinose concentrations and 100 μg/ml ampicillin. The chromosomal level of expression was confirmed by precipitation of cultures with trichloroacetic acid (TCA) at the time of plating and processing them for immunoblot comparison to expression of ExbB from E. coli strain W3110. Fivefold dilutions of colicins and 10-fold dilutions of phage were spotted on the bacterial lawns in triplicate. The plates were incubated ∼18 h at 37°C. Scoring was reported as the reciprocal of the highest dilution that produced clearing (33). For the individual and double Cys substitutions, a low-level disulfide-linked ExbB homodimer was detected only after long exposure of nonreducing anti-ExbB immunoblots; it was judged to be incapable of accounting for the reduced activities observed in vivo (data not shown). Additionally, no evidence of intramolecular cross-links occurring through the native ExbB C25 residue was observed (data not shown).
[55Fe]ferrichrome transport.
Initial rates of iron transport supported by ExbB variants were assessed as described previously (33, 34). Strains containing plasmids were subcultured from overnight LB cultures into M9 minimal medium supplemented with 100 μg/ml ampicillin and l-arabinose sufficient to maintain near-chromosomal expression levels. The cells were grown to mid-exponential phase, harvested, and suspended in assay buffer (M9 minimal medium without iron supplementation and with 0.1 mM nitrilotriacetate added to chelate residual free iron). Initial rates of [55Fe]ferrichrome transport were determined in triplicate. Chromosomal levels of mutant ExbB expression were confirmed by TCA precipitation of samples at the time of assay and immunoblotting with anti-ExbB antiserum (35).
In vivo formaldehyde cross-linking.
Strains containing plasmids were subcultured from overnight LB cultures into M9 minimal medium supplemented with 100 μg/ml ampicillin and l-arabinose sufficient to maintain near-chromosomal expression levels. The cells were grown to mid-exponential phase, and 0.5 A550-ml cells were harvested, pelleted at room temperature, and suspended in 938 μl of 100 mM Na2+ phosphate buffer, pH 6.8. The cells were then treated with 1% monomeric paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 15 min at room temperature, harvested, and solubilized at 60°C for 10 min in Laemmli sample buffer (LSB) (36, 37). Rapid formaldehyde cross-linking used for ExbB half-Ala substitutions was performed similarly with the following exceptions. The cells were grown and harvested as described above and suspended in 938 μl of 100 mM Na2+ phosphate buffer, pH 8.0. The cells were treated with 1% paraformaldehyde for 3 min at room temperature. The cross-linking reaction was then quenched with 1% acetic acid. Cross-linked samples were resolved on 13% or 11% SDS-polyacrylamide gels and detected by immunoblotting with anti-ExbB, anti-ExbD, or anti-TonB antibodies (35, 38).
Proteinase K sensitivity.
TonB sensitivity to proteinase K was performed as previously described (26, 39, 40). Spheroplasts were generated as previously described (39), and where indicated, treated with 60 μM protonophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) 5 min prior to proteinase K treatment. Proteinase K at 25 μg ml−1 was added for 2, 5, 10, or 15 min followed by the addition of 1.0 mM PMSF (phenylmethylsulfonyl fluoride) to inactivate the proteinase K. Samples were immediately precipitated with TCA and resolved on 13% SDS-polyacrylamide gels, and TonB was visualized on immunoblots probed with TonB-specific antibodies (38).
RESULTS
Defining ExbB transmembrane domain boundaries.
To address the roles of the three ExbB TMDs in TonB-dependent energy transduction, we began by reassessing their predicted boundaries. The recently described TOPCONS program uses a consensus of multiple different topology prediction algorithms, including evolutionary conservation-based programs and the “biological” hydrophobicity scale from experimentally generated measurements involving Sec61-mediated integration of TMD helices (29). TOPCONS predictions for TMD boundaries were as follows: residues 22 to 42 for TMD1, residues 132 to 152 for TMD2, and residues 178 to 198 for TMD3 (Fig. 1) (http://topcons.cbr.su.se).
Fig 1.
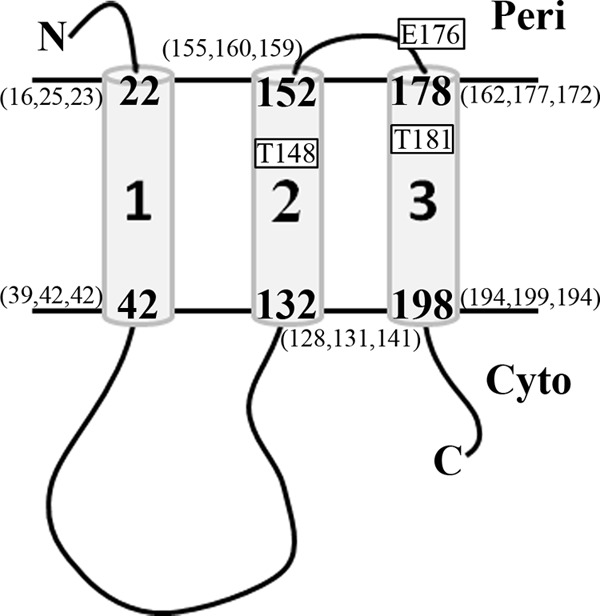
Revision of ExbB TMD boundaries. ExbB topology within the cytoplasmic membrane (parallel black lines) and the TOPCONS-predicted boundaries for TMDs 1, 2, and 3 are shown. The numbers in parentheses in the figure are previous ExbB TMD predictions (12, 13, 21). The majority of ExbB is localized in the cytoplasm (Cyto), as a large cytoplasmic loop and C-terminal tail. The N terminus of ExbB is in the periplasm (Peri). The general locations of the previously identified ExbB E176A TMD mutant and T148A/T181A double mutant are shown (boxes) (28).
Sequence alignments of ExbB sequences with expect scores from e−28 to e−7 showed that the TMD regions are more highly conserved than the predicted soluble domains, as had been noted previously for more closely related species (41, 42) (see Fig. S1 in the supplemental material). Sequence conservation for TMD1, the least conserved TMD, did not indicate clear boundaries but was consistent with the TOPCONS prediction. TMD2 contained conserved Leu (at position 133), Pro (at position 141), Gly (at positions 144-147-151), and Thr (at position 148) residues (Fig. S1). For TMD3, the TOPCONS-predicted periplasmic boundary occurred at residue 178, which omitted Glu 176 and the fully conserved Ala 177 (Fig. S1). Glu 176 was included in these experiments due to its acidic side chain and high level of conservation. TMD3 sequence similarity drops considerably after residue 196, as noted previously (Fig. S1) (43). TMD3 contained conserved Leu (at position 178), Thr (at position 181), Pro (at position 190), and Asn (at position 196) residues (Fig. S1).
All ExbB TMDs are essential for function.
Large segments of the TonB TMD can be simultaneously replaced with Ala residues without affecting its activity (22). To determine rapidly whether any portions of the three ExbB TMDs were dispensable, half of each predicted TMD region (∼11 consecutive codons) was replaced as a block with Ala codons on pKP660, which encodes the exbB-exbD operon under arabinose control. The six resulting plasmids were designated as a group to encode “half-Ala TMD” ExbB proteins. Individually they were designated according to the location of the Ala block substitution, e.g., “TMD1 peri” for substitution of alanines on the periplasmic half of TMD1. The function of each half-Ala TMD was assessed by spot titer assay of sensitivity to colicins B, D, Ia, and M and phage ϕ80, each of which requires the TonB system to enter and subsequently kill E. coli. All of the mutants could be expressed at near-chromosomal levels and were completely tolerant (insensitive) to all agents tested, indicating that each half TMD was essential for ExbB function (Table 2).
Table 2.
ExbB half-alanine substitutions are nonfunctional in spot titer assays
| Mutanta | Ara (%)b | Sensitivityc |
||||
|---|---|---|---|---|---|---|
| Col B | Col D | Col Ia | Col M | ϕ80 | ||
| ΔexbBD ΔtolQRA | NA | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| pExbBD | 0.00005 | S,S,S | S,S,S | S,S,S | S,S,S | S,S,S |
| TMD1, peri | 0.0001 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| TMD1, cyto | 0.0001 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| TMD2, peri | 0.0001 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| TMD2, cyto | 0.0001 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| TMD3, peri | 0.0001 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| TMD3, cyto | 0.0001 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
All mutants were expressed near chromosomal levels in strain RA1017 (ΔexbBD ΔtolRQA). RA1017 contained the empty vector plasmid pBAD24. The corresponding plasmid names for the mutants listed follow: pExbBD, pKP660; TMD1 peri, pKP1459; TMD1 cyto, pKP1460; TMD2 peri, pKP1481; TMD2 cyto, pKP1482; TMD3 peri, pKP1483; TMD3 cyto, pKP1484.
Percentage of l-arabinose (Ara) added to growth media and plates to achieve near chromosomal expression levels. NA, not applicable.
Colicins (colicins [Col] B to M) and phage ϕ80 were spotted in 5-fold and 10-fold dilutions, respectively. Clearing was indicated as follows: S, sensitive to the undiluted agent tested; T, tolerance (insensitivity) to the undiluted agent tested.
To determine whether the half-Ala TMD mutants were dominant, each was expressed at least 100 × greater than the chromosomal level in wild-type strain W3110. Overexpressed half-Ala TMDs from TMDs 2 and 3 significantly reduced the ability of wild-type ExbB to support 55Fe transport to ∼5% or less (see Fig. S2 in the supplemental material). TMD1 peri and cyto were the least impaired, reducing activity to only ∼20% and ∼50% of that of the wild type, respectively. These results suggested that each half-Ala TMD for TMDs 2 and 3 retained significant ability to assemble into a multimeric complex and suggested that TMD1 might play a more direct role in assembly.
Half-Ala TMD mutants prevent formation of the pmf-dependent TonB-ExbD complex.
In vivo interactions in the TonB system can be identified by cross-linking with monomeric formaldehyde (20, 25, 35, 36, 43). Wild-type ExbB cross-links to form homodimers and a homotetrameric complex that also includes ∼85 kDa of an unknown protein(s) known as “ExbB tetramer + X” (36, 43). When expressed in the absence of wild-type ExbB, the least dominant half-Ala TMD1 cyto itself exhibited reduced formation of the ExbB tetramer + X, consistent with the results from the dominance study (see Fig. S3A in the supplemental material). The half-Ala TMD2 cyto also reduced formation of this complex but likely for other reasons than a defect in assembly, since this mutant was dominant. The remaining half-Ala TMD mutants did not reduce tetramer + X formation. Additionally, TMD1 peri and TMD2 cyto cross-linking profiles contained two complexes in the ExbB tetramer + X region, with the lower complex migrating near 130 kDa (Fig. S3A). While the composition of these two complexes was not clear, it is likely that neither complex is a pure ExbB homotetramer, since such a complex would migrate at ∼100 kDa (43). Instead, half-Ala substitution in TMD1 and TMD2 may have altered ExbB conformations to the extent that alternative cross-linkable conformations of ExbB were favored and migrated differently on SDS-polyacrylamide gels.
The uniform inactivity of the half-Ala TMD mutants was almost certainly due to their uniform inability to support formation of the TonB-ExbD formaldehyde cross-link that typifies an active TonB system (25). This pmf-dependent complex was absent from ExbD cross-linking profiles of all the variants (see Fig. S3B in the supplemental material). ExbD coexpressed with TMD2 cyto was present at lower levels, suggesting that its proteolytic stability was reduced. Despite the reduced ExbD levels, an ExbD-ExbB complex was still apparent, suggesting that ExbB-ExbD assembly was not disrupted. Consistent with their decreased ability to form the ExbB tetramer + X complex, ExbB TMD1 cyto and TMD2 cyto mutants significantly eliminated or reduced TonB-ExbB complex detection by anti-TonB antibodies (Fig. S3C).
Identification of important residues in ExbB TMDs.
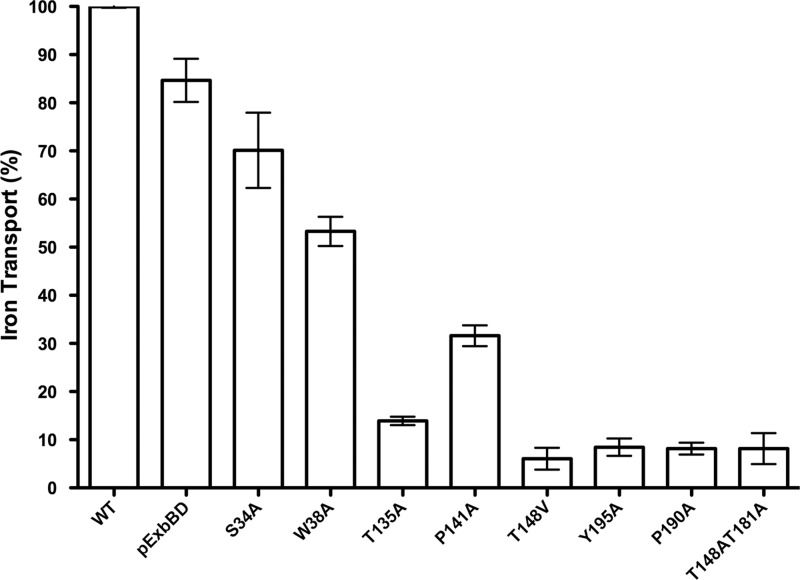
Three roles for ExbB have been proposed: as a scaffold that stabilizes and mediates assembly of TonB and ExbD, as transducer of signals between cytoplasm and periplasm, and as a proton translocator (14–16, 21, 27, 43). To identify residues important for these roles, we made substitutions throughout the three ExbB TMDs based on conservation and theoretical importance for TMD helix packing, signal transduction, or pmf coupling. Selected exbB codons were replaced by Ala codons on pKP660 using site-directed mutagenesis. The ExbB mutants were initially assayed for TonB-dependent 55Fe transport activity. This discriminative assay can resolve the activity level of partial-function mutants relative to that of the wild type (34). Mutants identified as inactive by that criterion could still have low levels of activity. These mutants were additionally tested in spot titers of colicins B, D, Ia, and M and phage ϕ80, which is a more sensitive, but less discriminative, assay for TonB function (34) (Fig. 2 and Table 3). None of the Ala substitutions significantly affected protein stability, as evidenced by the relatively low levels of l-arabinose required to induce their expression at chromosomal levels (Table 1).
Fig 2.
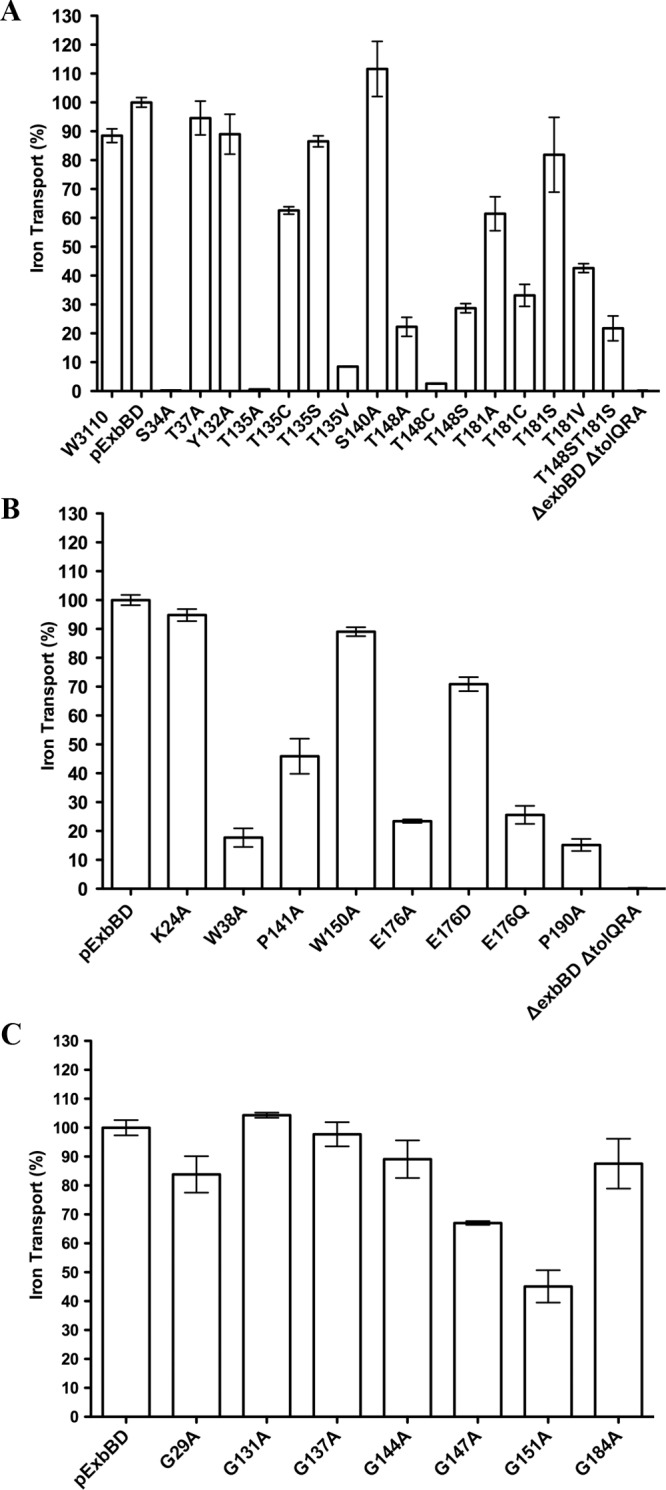
[55Fe]ferrichrome transport supported by ExbB TMD substitutions. ExbB TMD substituted mutants were expressed at chromosomal levels in strain RA1017 (ΔexbBD ΔtolQRA), and the initial rates of [55Fe]ferrichrome transport were measured as described in Materials and Methods. Triplicate samples were taken at all time points, and the initial transport rates were calculated by linear regression. Linear regression slopes were normalized to plasmid-encoded wild-type ExbBD (pExbBD) (100%). Normalized percent activities of at least two independent experiments were averaged, and standard deviations are indicated by the error bars. ExbB TMD substitutions are presented as follows: (A) substitution of hydroxyl residues; (B) substitution of proline, charged, and the remaining polar residues; (C) substitution of glycine residues.
Table 3.
Sensitivity of low activity ExbB substitutions to TonB-dependent colicins and phage ϕ80
| Mutations or substitution(s)a | Ara (%)b | Sensitivityc |
||||
|---|---|---|---|---|---|---|
| Col B | Col D | Col Ia | Col M | ϕ80 | ||
| None (WT) | NA | 8,8,8 | 6,6,6 | 7,7,7 | 6,6,6 | 8,8,8 |
| ΔexbBD ΔtolQRA | NA | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| pExbBD | 0.00005 | 7,7,7 | 4,4,4 | 7,7,7 | 6,6,6 | 7,7,7 |
| S34A | 0 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| S34C | 0.00005 | T,T,T | T,T,T | 2,2,2 | 3,3,3 | 3,3,3 |
| S34T | 0 | T,T,T | T,T,T | U,U,U | T,T,T | 1,1,1 |
| T135A | 0.00005 | 3,3,3 | T,T,T | 4,4,4 | 3,3,3 | 3,3,3 |
| T148A | 0.00005 | 7,7,7 | 2,2,2 | 7,7,7 | 4,4,5 | 3,3,3 |
| T148C | 0.0001 | 3,3,3 | T,T,T | 4,4,4 | T,T,T | T,T,T |
| T148V | 0.00005 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| T181A | 0.00005 | 7,7,7 | 3,3,3 | 7,7,7 | 6,6,6 | 8,8,8 |
| T181C | 0.0001 | 7,7,7 | 3,3,3 | 7,7,7 | 6,6,6 | 8,8,8 |
| T181V | 0.00005 | 7,7,7 | 3,3,3 | 7,7,7 | 3,3,3 | 7,7,7 |
| T148A T181A | 0.00005 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
| T148C T181C | 0.0001 | 3,3,3 | T,T,T | 4,4,4 | T,T,T | 4,4,4 |
| T148V T181V | 0.00005 | T,T,T | T,T,T | T,T,T | T,T,T | T,T,T |
WT indicates strain W3110, and ΔexbBD ΔtolRQA indicates strain RA1017. Both contained the empty vector control from which all plasmids were expressed, pBAD24. All mutants were expressed near chromosomal levels in strain RA1017. The corresponding plasmid names are listed in Table 1.
Percentage of l-arabinose added to growth medium and plates to achieve near chromosomal expression levels. NA, not applicable.
Colicins (Col) and phage ϕ80 were spotted in 5-fold and 10-fold dilutions, respectively. Results are presented as the reciprocal of the highest dilution that provided clearing (8, 5−8, or 10−8 dilution). U, undiluted; T, tolerant (insensitive).
Effects of substitutions at protonatable residues in ExbB TMDs.
Polar residues within TMDs play multiple roles (44, 45). We analyzed Ala substitutions at S34 and T37 (TMD1), Y132, T135, S140, and T148 (TMD2), and T181 (TMD3). Previously, ExbB T148A and T181A were 99% active in iron transport assays; however, steady-state levels of the mutants were not determined (28). Here, Ala substitution at all ExbB hydroxyls within the predicted TMDs were expressed at chromosomally encoded levels.
In these studies, T148A reduced 55Fe transport activity to ∼22%, while ExbB T181A supported intermediate activity (61%) (Fig. 2A). Braun and Herrmann also evaluated double Ala substitutions T148A/T181A and found them completely inactive (28). The inactivity of the T148A/T181A double substitution was confirmed here with spot titers (Table 3). The difference in our results and those of Braun and Herrmann could be due to protein overexpression in their study.
Of the remaining substitutions at hydroxyl residues, only ExbB S34A and T135A eliminated 55Fe transport activity entirely (Fig. 2A), while T135A retained slight activity in the more sensitive spot titer assays (Table 3). The remaining hydroxyl residue substitutions, T37A, Y132A, and S140A, were fully active in 55Fe transport and not considered further (Fig. 2A).
Charged residues play critical roles in proton binding for proton-translocating protein complexes such as H+ Fo-F1 ATPase, M2 viral protein, flagellar motor protein MotB, and bacteriorhodopsin (reviewed in reference 44). Of substitutions at the two charged residues in the ExbB TMDs, K24A in TMD1 was fully functional; ExbB E176A in TMD3 supported ∼25% activity, ruling out an essential role in proton translocation (Fig. 2B). Our observation that additional substitutions of E176 to Asp or Gln supported 70% and 25% activity, respectively, confirmed this conclusion (Fig. 2B). In the Braun and Herrmann study, ExbB E176Q supported 80% iron transport activity (28). Taken together, these results indicated that E176 does not play a key role in proton translocation. Glu residues are known to play important roles in helix-helix interactions (46).
Effects of substitutions at unique function glycine, proline, and tryptophan residues.
Common TMD packing motifs containing Gly residues have been implicated in driving helix assembly (46, 47). ExbB and a closely related homolog, TolQ, both contain a G144 (141)XXG147 (144) XXXG151 (148) packing motif in TMD2 (TolQ positions shown in parentheses). For TolQ TMD2, G141A, G144A, and G148A are completely inactive in assays for pmf-dependent OM stability, TolA-Pal coimmunoprecipitation, and pmf-independent colicin uptake, suggesting that corresponding ExbB residues may also be functionally important (48). However, none of the Ala substitutions of corresponding ExbB TMD2 Gly residues at positions 144, 147, and 151 eliminated activity, supporting ∼90%, 67%, and ∼45% 55Fe transport activity, respectively (Fig. 2C). In addition, Ala substitution of less conserved Gly residues G29 (TMD1) and G131 and G137 (TMD2) and highly conserved G184 (TMD3) were fully functional (Fig. 2C).
Considering that ExbB and TolQ TMD sequences are highly conserved and that TolQR can partially substitute for ExbBD functions in TonB-dependent energy transduction, it is almost certain that ExbB and TolQ TMD functions are mechanistically similar (41, 49, 50). Therefore, it is likely that activity differences between TolQ and ExbB Ala-substituted glycines reflected a disparity in the different assay sensitivities rather than actual mechanistic differences. Additionally, the steady-state levels of the TolQ substitutions in that study are unknown, further complicating activity comparisons to corresponding ExbB substitutions. Given the 100% Gly conservation for distantly related ExbB alignments, glycines at these positions almost certainly comprise an important helix packing motif as Goemaere et al. pointed out (48) (see Fig. S1 in the supplemental material). Ala replacement of the remaining individual Gly residues might be too conservative to highlight their full functional importance.
Proline-kinked α-helices influence TMD helix-helix interactions and can be important for propagating conformational changes during signal transduction (46, 51, 52). In both MotA and TolQ systems, the proline residues are proposed to influence proton translocation functions via regulation of proton flow through helix movements mediated by Pro-kinked helices (48, 53, 54). Substitutions of conserved proline residues P173 and P222 in the corresponding TMD domain of the homologous flagellar motor protein MotA eliminated torque generation (53, 55). Similar to results with MotA P222A, TolQ P187V (ExbB P190A) eliminated Tol-dependent activities (48). In contrast, TolQ P138V (ExbB P141, MotA P173) was partially active.
In these studies, we obtained an intermediate result for Ala substitution at ExbB P141 (TMD2), which supported ∼45% activity in 55Fe transport. While P190A in TMD3 had a more detrimental effect, it still supported ∼15% transport activity (Fig. 2B). The presence of low but detectable activity for ExbB P190A but not the corresponding prolines in MotA and TolQ probably reflected differences in the sensitivities of the assays for the different activities of the three paralogues. These reduced but intermediate activities of substitutions at Pro residues appeared to support a role in TMD helix assembly but did not argue strongly for an absolute role in proton gating.
Tryptophan can play a structural role in TMD anchoring and positioning within the membrane by stabilizing interactions at membrane-water interfaces (45, 56). Tryptophan located within the TMD interior has demonstrated roles in oligomer assembly and in an ion gating mechanism (57, 58). According to our TMD predictions, ExbB W150 in TMD2 was located near the membrane-water interface, and ExbB W38 in TMD1 was located within the TMD boundaries. While ExbB W150A was fully active in 55Fe transport, W38A supported only 18% activity (Fig. 2B). W38 is located on the same helical face as the functionally important S34 residue in TMD1, suggesting that this face was somehow important for ExbB function.
ExbB facilitates, but is not essential for, formation of an initial pmf-independent TonB-ExbD interaction.
To assess effects of ExbB mutants in more mechanistically informative assays, we first need to more fully understand the role of wild-type ExbB. Three stages in the initial energization of TonB at the CM have been identified using the pmf-sensitive assays of TonB proteinase K resistance and formation of TonB-ExbD formaldehyde cross-links (25, 26). In stage I, TonB and ExbD are not detectably associated; TonB is sensitive to proteinase K and does not form a formaldehyde cross-link with ExbD in vivo (26). In stage II, which does not require pmf, TonB and ExbD periplasmic domains assemble such that their interaction prevents proteinase K degradation of TonB residues 1 to 155 (∼23 kDa). When pmf is collapsed, TonB-ExbD interactions are arrested at stage II, demonstrated by essentially complete conversion of all TonBs to the ∼23-kDa proteinase K-resistant fragment, apparently due to protection by ExbD. Only when both pmf and ExbB are present does TonB energization progress from stage II to stage III. In that stage, TonB and ExbD undergo pmf-dependent rearrangement of their conformational relationship such that formaldehyde cross-linkable residues in both proteins are in proximity to cross-link. This shift in the conformational relationship renders the entire length of TonB proteinase K sensitive again.
To determine the role of ExbB in TonB progression from stage I to stage II, the proteinase K sensitivity of TonB in the absence of ExbB was assessed. In E. coli strain RA1017 (ΔexbBD ΔtolQRA) where pExbD was expressed at chromosomal levels, a small but detectable proportion of TonB could form the characteristic proteinase K-resistant fragment after 2 min of treatment with proteinase K, indicating that TonB and ExbD had the capacity to interact in the absence of ExbB (Fig. 3A, middle blot). Because the proportion of TonB in this form did not increase upon the addition of the protonophore CCCP, which stalls TonB at stage II, ExbB was still clearly required for the initial efficient TonB-ExbD stage II assembly (Fig. 3A, compare top and middle blots, +CCCP lanes). These results indicated that, while not absolutely essential for initial stage II TonB-ExbD interactions, ExbB greatly facilitated them. The small proportion of the TonB proteinase K-resistant fragment that forms in the absence of ExbB was still dependent upon ExbD, since TonB, in the absence of ExbB and ExbD, did not form the characteristic ∼23-kDa fragment (Fig. 3A, bottom blot). Instead, a small population of TonB was degraded to an unknown ∼28-kDa fragment as noted previously (26).
Fig 3.
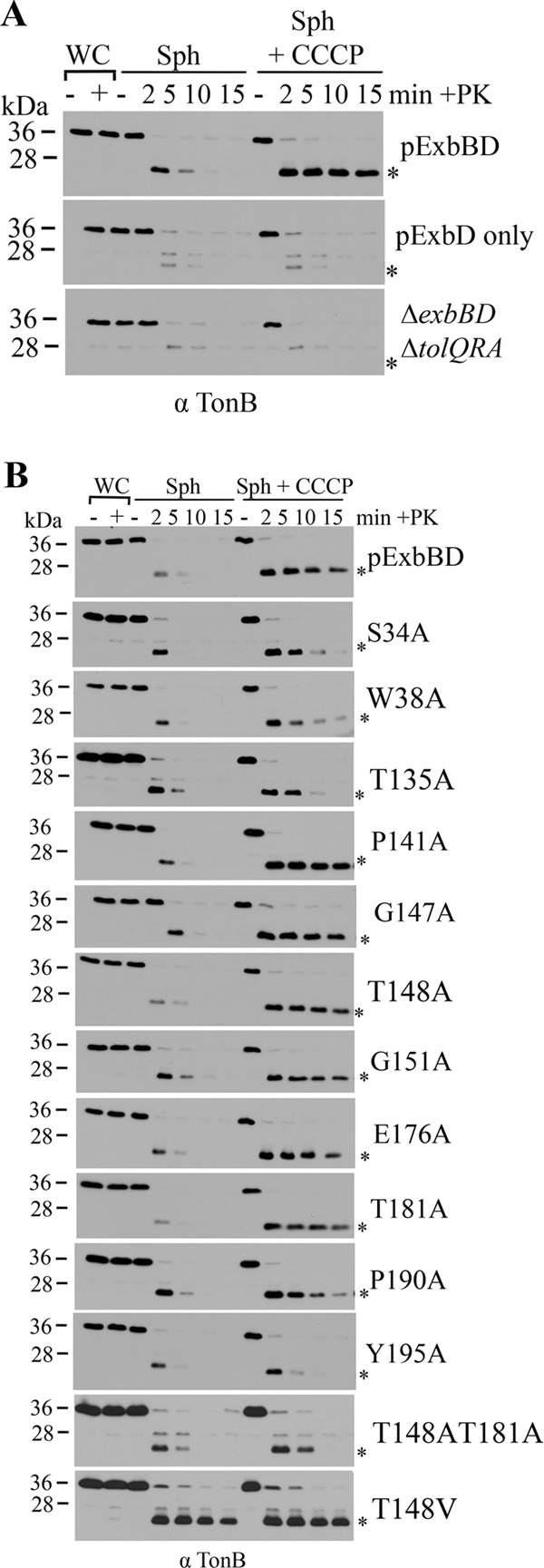
ExbB is required for conformational response of TonB to pmf and efficient TonB-ExbD assembly. Spheroplasts (Sph) or whole cells (WC) were treated with dimethyl sulfoxide (DMSO) (sph) or CCCP and subsequently not treated with proteinase K (−) or treated (+) with proteinase K (+PK) for a time course of 2, 5, 10, and 15 min, followed by the addition of the protease inhibitor (PMSF) and immediate precipitation with trichloroacetic acid (TCA). TCA-precipitated samples were resolved on 13% SDS-polyacrylamide gels and immunoblotted with anti-TonB (α TonB) antibodies. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the blots. Plasmid-expressed wild-type and mutant proteins or the deletion strain are indicated to the right of each blot. The position of the TonB proteinase K-resistant ∼23-kDa fragment is indicated by an asterisk to the right of the blots. (A) ExbB facilitates initial TonB-ExbD assembly. Spheroplasts were generated from the ΔexbBD ΔtolQRA strain (RA1017), expressing pExbBD (pKP660) or pExbD (pKP1194) at chromosomal levels. RA1017 containing the empty vector, pBAD24, is shown in the bottom blot. (B) ExbB TMD mutations affect TonB conformational response to pmf. Spheroplasts were generated in RA1017 expressing plasmid-encoded ExbB TMD mutants at chromosomal levels as well as ExbD. This figure is a composite of representative immunoblots.
Identification of ExbB TMD residues that mimic effects of ExbD D25N on TonB energization.
ExbD and homologous proteins TolR and MotB TMDs all contain a highly conserved protonatable Asp residue which is considered a key component of their respective putative proton pathways (21, 27, 59, 60). The ExbD D25N mutant stalls the TonB-ExbD interaction at the pmf-independent stage II, regardless of whether pmf is present, suggesting protonation of D25 is essential for conformational response to pmf (26). Thus, the behavior of the inactive ExbD D25N substitution in the proteinase K sensitivity assay provides an example where proton translocation is prevented. It is important for the interpretation of our data here to recognize that ExbD D25N also decreases the stability of the TonB-ExbD stage II interaction in the presence of CCCP for unknown reasons. The degradation of TonB stalled at stage II by the ExbD D25N mutation may reflect dissociation of TonB and ExbD D25N or increased accessibility of ExbD D25N and subsequently TonB to proteinase K. In either case, any ExbB TMD substitutions that mimicked the behavior of ExbD D25N in this assay would be candidates to be on a proton translocation pathway.
All ExbB substitution mutants supported the stage II initial assembly of the pmf-independent TonB-ExbD interaction to a much greater extent than seen in the absence of ExbB (compare the levels of the proteinase K-resistant fragment in Fig. 3A, 2-min time point of the middle blot, +CCCP lanes, with Fig. 3B, 2-min time points, +CCCP lanes). There was mutant-specific variability in the stability of the proteinase K-resistant form in the presence of CCCP over time. For those mutants that had the least deleterious effects in the phenotypic assays (P141A, G147A, G151A, E176A, and T181A), the TonB proteinase K-resistant fragment remained stable over the 15-min time course, similar to results in the presence of wild-type ExbB. For ExbB S34A, W38A, T135A, P190A, Y195A, and the T148A/T181A double mutant, the TonB proteinase K-resistant fragment was unstable over the 15-min time course, mimicking the behavior of TonB in the presence of ExbD D25N (Fig. 3B) (26). To confirm that S34A, W38A, T135A, T148A/T181A, and P190A were unable to support a TonB transition from stage II to stage III, their ability to support formation of the pmf-dependent TonB-ExbD formaldehyde cross-linked complex was determined. Consistent with the proteinase K resistance assay results, none of them was able to support formation of the cross-linked complex (see Fig. S4, S5B and C, and S6 in the supplemental material).
The behavior of ExbB W38A, P190A, and Y195A indicated that other factors in addition to proton translocation could affect conversion of TonB from stage II to stage III: tryptophan and proline are not protonatable residues and were likely to play a structural role. Although Y195 is potentially a protonatable residue, Y195F is fully functional, indicating that aromaticity is the only important aspect of the tyrosine residue at that position (43). Thus, this assay could rule out residues as participants in proton translocation by their ability to support a wild-type conformational response of TonB to collapse of pmf. To test participation of S34A, T135A, or T148A/T181A in proton translocation required additional substitutions and assays described below.
ExbB S34A and W38A are assembly mutants that define a TMD1 cyto face important for TonB-ExbB interactions.
Residues whose only role was related to proton translocation would not be expected to impair ExbB assembly functions and thus would be dominant. Assessment of dominance of ExbB S34A, W38A, T135A, T148A, Y195A, P190A, and the double mutant T148A/T181A was carried out as for the half-Ala TMD mutants above in wild-type strain W3110.
ExbB S34A did not reduce 55Fe transport lower than ∼70% of chromosomally encoded ExbB activity compared to overexpressed wild-type ExbB, which reduced 55Fe transport to ∼85% of chromosomally encoded activity (Fig. 4). Thus, ExbB S34A was not dominant, suggesting that it could not assemble into a complex with wild-type ExbB. Likewise, ExbB W38A exhibited only moderate dominance. ExbB S34 and W38 reside on the same helical face of TMD1 cyto. Consistent with that, the half-Ala TMD1 cyto was the least dominant of the half-Ala TMDs, reducing activity to ∼45% (see Fig. S2 in the supplemental material). The ability of assembly mutants S34A and W38A to form the ExbB tetramer + X complex was surprisingly not impaired, suggesting that there were subsequent essential configurations of wild-type ExbB tetramer in which they could not participate during energy transduction (Fig. S7A).
Fig 4.
ExbB S34A and W38A TMD mutants are not dominant. ExbB mutants were coexpressed with ExbD using 0.01% l-arabinose in wild-type (WT) strain W3110. All mutant proteins were overexpressed >100-fold compared to chromosomal levels, based on serial dilutions and immunoblot analysis (data not shown). Initial rates of [55Fe]ferrichrome transport were measured from triplicate samples and normalized to the results for strain W3110 (100%). Averaged percentages of at least two independent experiments are shown.
Where ExbB S34A and W38A appeared to be impaired was in their ability to cross-link to TonB in vivo. S34A eliminated the TonB-ExbB complex, while W38A reduced it (Fig. 5). These data are consistent with the iron transport results where S34A was completely inactive and W38A had about 20% iron transport activity. Even though ExbB S34A did not stabilize TonB in a half-life study (data not shown), it clearly still maintained some sort of interaction because it supported greater formation of the TonB-ExbD stage II complex than the absence of ExbB did (Fig. 3B). This implied that S34A altered but did not abolish TonB-ExbB interaction through the TMDs. ExbB W38A consistently gave rise to high levels of the tetramer + X complex as well as a high level of background complex formation, possibly suggesting that it was delayed in moving out of the tetramer + X conformation (see Fig. S7A in the supplemental material).
Fig 5.
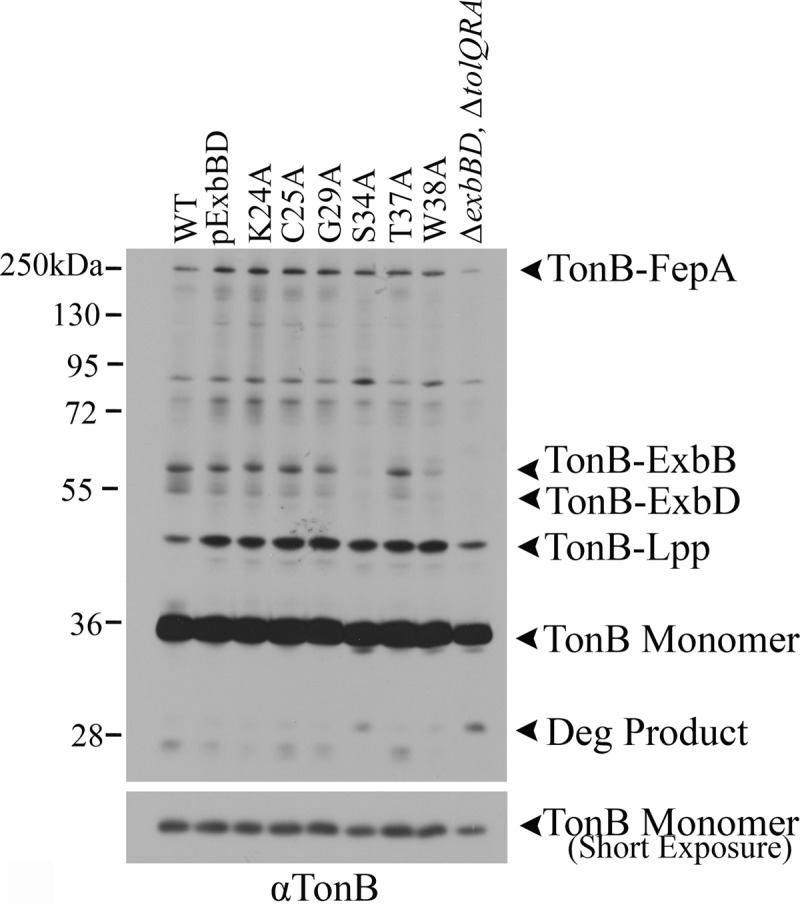
ExbB S34A and W38A reduce the TonB-ExbB formaldehyde cross-linked complex. ExbB TMD1 mutants were expressed at chromosomal levels in strain RA1017 (ΔexbBD ΔtolQRA), and parent strain W3110 (WT) served as the wild-type chromosomal control. The strain or mutations are indicated above the lanes. Cultures grown to mid-exponential phase were cross-linked with monomeric formaldehyde and solubilized in gel sample buffer at 60°C. Samples were resolved on 11% SDS-polyacrylamide gels and immunoblotted with anti-TonB antibody. The positions and compositions of complexes are indicated to the right of the blot. The positions of molecular mass standards (in kilodaltons) are indicated to the left of the blot. A shorter exposure of the same immunoblot is shown in the bottom blot for comparison of monomer levels.
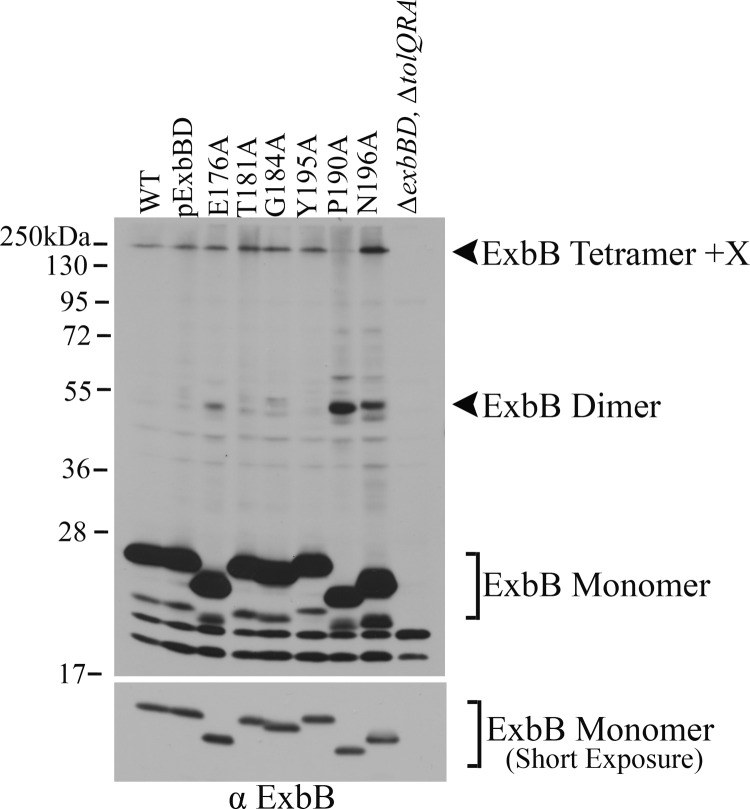
ExbB P190 and P141 mediate efficient transition from ExbB homodimers to ExbB tetramer + X.
ExbB appears to be a dimer of dimers, which subsequently interacts with an unknown protein to form the ExbB tetramer + X (43). ExbB P141A and P190A exhibited dominant-negative phenotypes with P190A being more dominant of the two, reducing wild-type activity to ∼10%. Both ExbB P141A and P190A exhibited an increase in ExbB homodimer formation, suggesting that they could not easily transition from dimer to tetramer. For P190A, the ExbB tetramer + X complex was essentially absent (Fig. 6; see Fig. S7B in the supplemental material). It is important to note that failure to form tetramer + X complex is not proof that it no longer exists, only that the relationships between cross-linkable residues in that complex has changed. In this case, however, since a correspondingly high level of dimer complex was found instead, the data suggested that ExbB P190A existed primarily as a dimer, with dominance due to preventing normal tetramerization of wild-type ExbB. These results removed ExbB P141 and P190 from playing a direct role in proton translocation and were consistent with the iron transport results where activity was reduced but not absent in the Ala substitutions.
Fig 6.
ExbB P190A reduces ExbB tetramer + X complex formation. ExbB TMD3 mutants were expressed at chromosomal levels in strain RA1017 (ΔexbBD ΔtolQRA), and parent strain W3110 (WT) served as the wild-type chromosomal control. The strain or mutations are indicated above the lanes. Cultures grown to mid-exponential phase were cross-linked with monomeric formaldehyde and solubilized in gel sample buffer at 60°C. Samples were resolved on 13% SDS-polyacrylamide gels and immunoblotted with anti-ExbB antibody. The positions and compositions of complexes are indicated to the right of the blot. The positions of molecular mass standards are indicated to the left of the blot. A shorter exposure of the same immunoblot is shown in the bottom blot for comparison of monomer levels.
Substitution analysis of ExbB T135 and the T148/T181 combination.
Potential direct roles of important polar ExbB TMD residues in proton translocation were further investigated by determining which other near-cognate residues could functionally substitute. Both Ser and Thr are found on proton translocation pathways, whereas Cys, which has a similar overall structure, is not (44). Cysteine can form hydrogen bonds (61). Valine has a methyl group instead of the hydroxyl group of threonine and thus has a somewhat similar shape while being nonpolar. Substitution of ExbB T135 with cysteine and serine supported 62% and 86% activity in 55Fe transport, respectively. Even substitution with valine retained 8% activity (Fig. 2A). The fact that all the T135C/S/V substitutions increased activity relative to T135A (0%) indicated that hydrophobicity, size, and shape, or ability to form a hydrogen bond was important at this position, rather than the ability to translocate protons.
Additional single substitutions at ExbB T148 and T181 further supported the contention, based on the Ala substitutions, that neither residue was directly on a proton translocation pathway. Because the T148S substitution, which could conceivably contribute to a proton pathway, did not restore significant activity over and above the T148A substitution (28% versus 23% in the 55Fe transport assay [Fig. 2A]), there appeared to be a preference for Thr at that position that was not due to a requirement for proton translocation. ExbB T148C and T148V substitutions resulted in ∼3% and 0% activity, respectively. In contrast to T148, all T181 substitutions were active. ExbB T181 Ser, Cys, and Val substitutions supported 81%, 33%, and 42% 55Fe transport, respectively, demonstrating that T181 was not directly a part of a proton translocation pathway (Fig. 2A).
The T148A/T181A double mutant was completely inactive. This was not due to a requirement for T181 as shown above. Double substitutions of T148/T181 to Cys, Ser, or Val residues were assayed for 55Fe transport and/or in spot titers (Fig. 2A and Table 3). The double Ser substitution retained ∼21% 55Fe transport activity. Since ExbB T181S supported 81% activity, this was not an additive effect but instead reflected the activity of the least active substitution of the pair, T148S. This result suggested that the two residues were not acting synergistically. The ExbB T148C/T181C substitution supported nearly full sensitivity to colicin Ia in spot titers, whereas the requirement of the TonB system for pmf is absolute (Table 3). Taken together, these results argue against the idea that T148 or T181 plays any direct role in proton translocation. Consistent with the effect of the single T148V substitution, the ExbB T148V/T181V mutant was completely inactive (Table 3).
Role of ExbB T148.
The inactive or reduced activity of individual T148 substitutions described above suggested that T148 was more important than T181 for ExbB activity. To define that role, T148C/V substitutions were analyzed using the more mechanistically informative assays applied to the single substitutions. T148V concomitantly reduced ExbB tetramer + X and homodimer complex detection in formaldehyde cross-linking. T148V/T181V reduced ExbB tetramer + X detection, with what appeared to be a compensatory increase in ExbB homodimers (see Fig. S5A in the supplemental material). Double T148/T181 Ala and Cys substitutions did not affect ExbB multimer formation, suggesting that the phenotype of the Val substitutions reflected structural interference. The structural interference did not prevent ExbB T148V from being highly dominant (Fig. 4). ExbB T148V also had a unique phenotype in the proteinase K sensitivity assays that assess TonB-ExbD interactions (Fig. 3B). While the rest of the inactive ExbB TMD mutants had a TonB proteinase K profile similar to that of ExbD D25N, T148V produced a TonB proteinase K profile in which the TonB proteinase K-resistant fragment was present at nearly 100% in the cell population, even in the presence of pmf (spheroplasts without added CCCP). ExbB T148V also did not reduce the stability of the TonB proteinase K-resistant fragment under any circumstance. Thus, ExbB T148V appeared to keep TonB permanently stalled in stage II and stabilized its contact with ExbD.
ExbB S34A is an assembly mutant.
Although S34A was entirely inactive, S34 in the first TMD was unlikely to be on a proton pathway. ExbB S34A was not dominant in the 55Fe transport assay and prevented formation of the TonB-ExbB formaldehyde cross-link that reflects interactions through the TMDs (Fig. 5) (40). These data suggested that S34 played a structural role in allowing correct assembly. Additional S34 substitutions supported this conclusion. S34C/T restored very low but detectable activity in spot titers, suggesting that both size and polarity are important at S34 (Table 3). Confirming the role for ExbB S34A in assembly with TonB, both ExbB S34C and S34V restored a low but detectable level of the TonB-ExbB formaldehyde cross-linked complex (Fig. 7).
Fig 7.
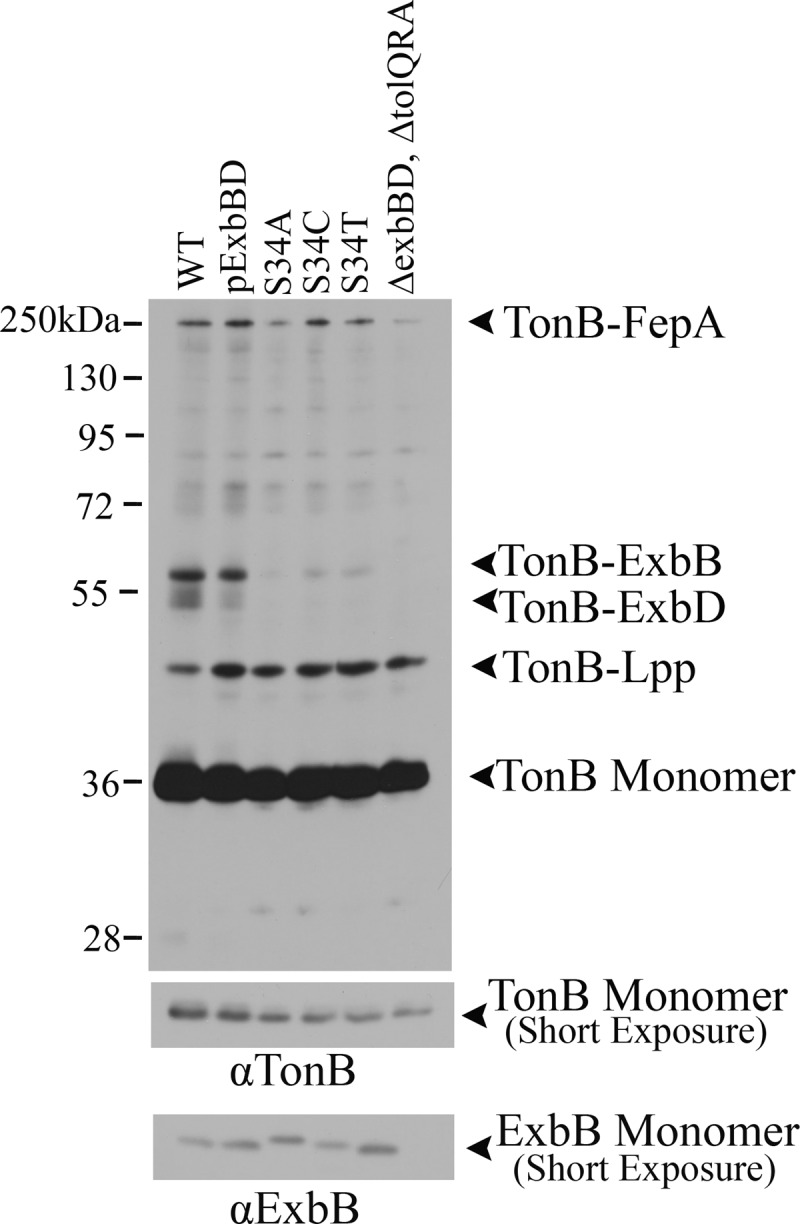
Conservative S34 substitutions restore low but detectable levels of the TonB-ExbB formaldehyde cross-linked complex. ExbB S34 substitutions were expressed at chromosomal levels in strain RA1017 (ΔexbBD ΔtolQRA), and parent strain W3110 (WT) served as the wild-type chromosomal control. The strain or mutations are indicated above the lanes. Cultures grown to mid-exponential phase were cross-linked with monomeric formaldehyde and solubilized in gel sample buffer at 60°C. Samples were resolved on 11% SDS-polyacrylamide gels and immunoblotted with anti-TonB antibody. The positions and compositions of complexes are indicated to the right of the blot. The positions of molecular mass standards are indicated to the left of the blot. A shorter exposure of the same immunoblot is shown in the bottom blot for comparison of TonB monomer levels. For ExbB monomer levels, the same samples were resolved on a 13% SDS-polyacrylamide gel and immunoblotted with anti-ExbB antibody.
DISCUSSION
New TMD predictions for the ExbB/TolQ/MotA/PomA family.
The members of the ExbB/TolQ/MotA/PomA family are integral CM proteins. ExbB and TolQ have three TMDs with the N terminus in the periplasm. MotA and PomA have four TMDs, with the “extra” TMD occurring at the N terminus, and the N-terminal residue located in the cytoplasm. There is a high degree of homology among the C-terminal two TMDs of all four proteins (Fig. 8). That and the fact that they are all in multiprotein complexes that require pmf as the energy source unite them as a family. However, the E. coli ExbB, TolQ, and MotA proteins and the Vibrio alginolyticus MotA orthologue PomA have dissimilar published TMD predictions (ExbB [12, 13, 21], TolQ [62, 63], MotA [64–66], and PomA [67]). Because all four proteins are likely to have similar mechanisms of action, we subjected ExbB, TolQ, MotA, and PomA to analysis with TOPCONS. The results significantly altered the previous predictions for the last two TMDs of each protein and brought them into greater congruency with the ExbB TMD predictions (see Fig. S8 in the supplemental material). On the basis of that information, we aligned the last two TMDs of each protein based on six shared highly conserved residues: those corresponding to ExbB G137, G144, G151, and P141 in the second to last TMD and those corresponding to ExbB T181 and G184 in the last TMD (Fig. 8). For MotA, residues corresponding to ExbB P141, G144, T181, and G184 are absolutely conserved among over 50 comparisons in many different species (68).
Fig 8.
Sequence alignment of ExbB, TolQ, MotA, and PomA. ExbB, TolQ, MotA, and PomA sequences were aligned using Clustal omega multiple-sequence alignment program (http://www.ebi.ac.uk/Tools/msa/clustalo/). Charged residues are shown on a gray background. Predicted ExbB TMDs from Fig. S1 in the supplemental material are indicated by a black bar above the sequence. The dashed bar indicates the predicted location of the MotA/PomA TMD1 (64). ExbB (NCBI accession no. YP_491200.1), TolQ (YP_489017.1), and MotA (YP_490152.1) sequences are from Escherichia coli K-12 strain W3110. The PomA sequence is from Vibrio alginolyticus (GenBank accession no. BAA20284.1).The importance of glycines in alignment of transmembrane domains among these proteins has been recognized previously (59, 92). Gaps introduced to maximize alignment are indicated by dashes in the sequences. Asterisks, colons, and periods indicate identical, conserved, and semiconserved residues, respectively.
Glycine residues in TMDs drive helix assembly and are thus important markers by which conserved TMDs can be aligned. ExbB, TolQ, and PomA all have a G144XXG147 XXXG151 motif in the second to last TMD (ExbB residues 132 to 152). Such glycine zipper motifs are highly conserved and play a role in helix packing (47). MotA lacks the central Gly residue (G147 in ExbB) and in its place carries the most conservative replacement, Ala, which is least likely to interfere in packing. All 4 second to last TMDs have an additional Gly residue (Gly137 in ExbB). We suggest that this redefinition of the boundaries of the last two TMDs is an accurate one, reflecting the high degree of residue conservation and current best identifications of transmembrane domain helices. It is important to note that while our predictions are for 21-residue TMDs, longer TMDs cannot be not ruled out. However, wherever the TMD boundaries lie, they will almost certainly be based upon the high degree of residue conservation.
By significantly redefining the TMD boundaries, the TOPCONS predictions for MotA and TolQ may alter interpretations of previous studies. Previously defined TMD boundaries place MotA P173 (corresponding to P141 in ExbB) at the cytoplasmic edge of the second to last TMD, where it was proposed to interact with the essential pmf-responsive MotB residue Asp32, also at the cytoplasmic edge of the TMD (68). The redefined TMD boundaries for MotA place the conserved P173 in the center of the TMD, where it may be sufficiently distant from MotB D32 that the proposed interaction does not occur. This relocation suggests that prolines corresponding to MotA P173 serve structural roles and are not directly involved in proton translocation as previous studies have suggested (53, 54). A nonessential structural role for the P173 in MotA is consistent with our findings that the corresponding ExbB P141A retained 45% activity. In addition, ExbB P141A supported formation of both stage II and stage III TonB-ExbD interactions.
In studies of PomA T186I and MotA T209W mutants, which correspond to ExbB T181, were both inactive (65, 69). Our results suggested that the observed inactivity was likely due to the nature of the substitutions; the corresponding ExbB T181 was highly tolerant to substitutions with Ala, Ser, Val, and Cys residues. To the extent that the four proteins have similar mechanisms, the conserved Thr in the second to last PomA and MotA TMDs does not appear to be important for anything other than optimal helix packing (46).
ExbB TMDs do not directly participate in a proton translocation pathway.
We mutated all the residues with side chains not predicted to serve a strictly structural purpose in the three ExbB TMDs, with the goal of testing their role in proton translocation as part of TonB-dependent energy transduction (Fig. 9). Our results exclude the possibility that ExbB TMD residues have any residue-specific role in proton translocation. First, substitutions of residues that are on a proton pathway should strongly decrease activity. Few of the ExbB Ala substitutions tested had severe effects on iron transport activity. Where strong decreases in iron transport activity were observed, several different assays coupled with analysis of further substitutions provided explanations that ruled out a direct role in proton translocation, as they also did for the moderately inactive substitutions. Instead, as described below, ExbB TMDs play roles in assembly of the ExbB tetramer + X complex, interaction with TonB, presumably through its TMD, and correct assembly of the TonB-ExbD complex and signal transduction between cytoplasm and periplasm. The role in signal transduction is consistent with the positioning of ExbB and TonB-ExbD residues on opposite sides of the cytoplasmic membrane, providing a means of communication between cytoplasm and periplasm (43).
Fig 9.
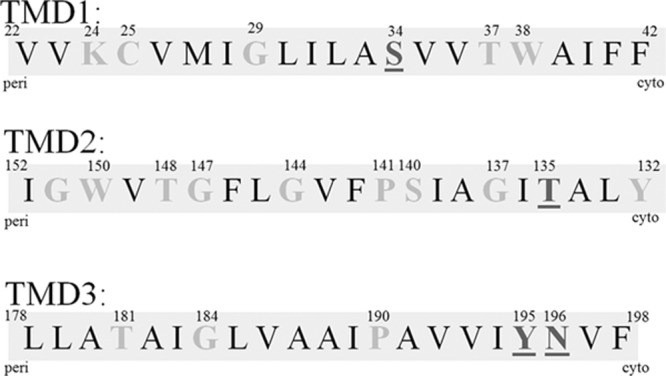
Summary of residue substitutions in ExbB transmembrane domains. Predicted ExbB TMDs are depicted as shaded sequences. Gray residues within each sequence indicate the positions of Ala substitutions. All substituted residues are shown in gray: light gray residues were functional, and dark gray, underlined residues were nonfunctional when substituted to alanine. The corresponding amino acid numbers are listed above the residues. ExbB Y195A and N196A substitutions were previously reported (43).
Role of ExbB TMD1.
ExbB TMD1 is the least conserved among the corresponding TMDs of the ExbB/TolQ/MotA/PomA family. The properties of the half-Ala cyto substitution and the two inactive substitutions, S34 and W38, within that region indicated that ExbB interacts with TonB through this TMD. The primary sequences of the corresponding TMDs among the family therefore likely reflect different additional TMDs through which each family member interacts in energy transduction, outer membrane integrity, and flagellar rotation (Fig. 8).
The inactive ΔVal17 deletion in the single TonB TMD is suppressed by ExbB mutations in TMD1: V35E, V36D, and A39E (39, 40). On the basis of previous definitions of ExbB TMD1 boundaries, these residues were thought to be near the cytoplasmic boundary of TMD1. However, the redefinition of the boundaries in this study places them well within the cytoplasmic half of TMD1. While the ExbB suppressors are at a depth similar relative to that of W38 and S34 in TMD1, ExbB V36 and A39 are located on the opposite face and V35 is closer to the W38 face (see Fig. S9A in the supplemental material). The suppressors somehow reestablish ExbB TMD1 interactions with the TonB TMD. It is notable that the suppressors all exchange the negatively charged Asp or Glu for the native Ala or Val residues (39, 40). Similar suppressors exist between TonB/ExbB paralogues TolA/TolQ, where a TolQ TMD1 face corresponding to ExbB S34 supports similar conserved functional interactions (70, 71).
Role of ExbB TMD2.
All individual ExbB TMD2 Ala substitutions formaldehyde cross-linked with TonB and with ExbD and could form the ExbB homodimer and tetramer + X complex, thus eliminating key roles in assembly. Most exhibited small to moderate decreases in ExbB activity. This TMD appeared to play a role in signal transduction, because ExbB T135A, the only inactive Ala substitution in TMD2, did not support the stage III pmf-dependent interaction between TonB and ExbD. In addition, the T148V substitution caused TonB-ExbD to stably stall at the pmf-independent stage II. Because the half-Ala TMD2 cyto reduced ExbD stability, there appeared to be a role in ExbD interaction as well.
Role of ExbB TMD3.
With one exception, all individual ExbB TMD3 substitutions formaldehyde cross-linked with TonB and ExbD and could form the ExbB homodimer and tetramer + X. The exception was ExbB P190A, which had ∼15% activity and appeared to be stalled in a homodimeric state. Thus, the kink induced by P190 appeared to be important for the transition from homodimer to tetramer + X. Consistent with that idea, ExbB P141A in TMD2 led to a more modest decrease in activity, but like ExbB P190A, accumulated higher levels of formaldehyde cross-linked homodimers. In the reevaluated TMD boundaries, P141 and P190 were located at similar depths in the membrane (Fig. 9). Prolines are known to play important roles in signaling mediated by transmembrane domains (51). Residue N196 was previously shown to be important for signal transduction (43).
Idiosyncratic profile of reduced activity ExbB TMD mutants in colicin uptake.
We have hypothesized that the conformation of the disordered TonB C terminus is modulated by ExbD protein such that it recognizes ligand-bound transporters by induced fit. Different Cys substitutions in the TonB C terminus have assay-dependent and residue-specific (idiosyncratic) phenotypes in a variety of assays that measure sensitivity to colicins and phage and ability to transport Fe-ferrichrome (72, 73). The C terminus of TolA may be similar (74).
It is logical that the C terminus of TonB would interact differentially with different transporters and colicin ligands, all of which have different amino acid sequences. It is not logical that mutations in ExbB would have idiosyncratic phenotypes, because ExbB supposedly interacts only with TonB and ExbD in the energy transduction complex and not with the transporter proteins to which energy is transduced. Nonetheless, idiosyncratic phenotypes were observed for some ExbB substitutions in colicin sensitivity assays. For example, ExbB T148C/T181C was nearly insensitive to colicin D and completely insensitive to colicin M while retaining significant sensitivity to colicin B and nearly wild-type sensitivity to colicin Ia (Table 3). Colicins B and D both use the outer membrane transporter FepA as a receptor; however, colicin D exerts its effects as a tRNase in the cytoplasm, whereas colicin B kills cells by creating a pore in the cytoplasmic membrane. Colicin Ia, which uses the transporter Cir as a receptor, also kills by creation of a cytoplasmic membrane protein pore. It seems unlikely that ExbB T148C/T181C differentially alters TonB conformations and transporter recognition. Amino acid sequence homologies and functional cross talk between ExbB and TolQ raise the possibility that ExbB could be directly and differentially involved in direct interaction with colicins in the cytoplasmic membrane. This idea is based on the finding that after colicin A was osmotically introduced into the periplasmic space, its killing activity in the cytoplasmic membrane depended on the presence of TolQ (75). Potential ExbB-colicin interaction has not been explored for ExbB substitutions.
How does the TonB system harness pmf?
While the use of cytoplasmic membrane pmf energy for TonB-dependent transport across the OM has been well established, the mechanism by which pmf is harnessed remains unknown (1, 39, 76–78).
The results of our experiments here ruled out direct participation of ExbB TMD residues in proton translocation and instead supported a role in signal transduction, a homodimer-to-homotetramer transition, and direct interaction with the TonB TMD. In helical wheel projections of ExbB TMD2 and TMD3, conserved small residues line one face of each TMD from residues in G147 to A134 in TMD2 and T181 to A187 in TMD3, which could hypothetically form part of an aqueous channel through which protons would move (see Fig. S9B and C in the supplemental material). We think that it is more likely that it represents a helix packing motif. Although an aqueous channel model has been proposed for proton translocation in the Mot system, it has not been experimentally explored (68). Repositioning of the TMD boundaries in this study also raised questions about that model.
The TonB TMD retains TonB in the cytoplasmic membrane but has no direct role in harnessing and responding to pmf (23, 79–81). ExbD thus emerges as the only protein with a TMD residue—an essential conserved Asp25—that somehow responds to pmf. ExbD mediates initial stages of TonB energization, as ExbD transits from a homodimer to two different conformations of the TonB-ExbD heterodimer (25, 26, 80).
It is not clear how a single Asp would, by itself, harness pmf and use it to transmit conformational changes to the TonB C terminus. However, perhaps ExbD does not function alone in this capacity. It has been known for years that unidentified proteins are essential for TonB activity (82, 83). At least one of these proteins is characterized by a short half-life, and because they have not turned up in genetic selections, they are likely to be either redundant or essential (84, 85). Both ExbD and ExbB formaldehyde cross-link to unidentified proteins in vivo (25, 36, 43). The most abundant ExbB in vivo formaldehyde cross-linked complex contains an ExbB tetramer linked to an unknown protein or proteins of ∼85 kDa. In this study, the two substitutions that disrupted formation of the tetramer + X complex, P190A and T148V, also rendered ExbB nonfunctional. The ExbB tetramer + X complex does not require TonB or ExbD to form, suggesting that it is a stable and abundant complex (36). It is not known through which domain of ExbB this complex forms. Likewise, ExbD cross-links in vivo to a protein of ∼18 kDa (25). This cross-link grows more abundant when ExbD is not active.
What might be the function of the unknown proteins? It is possible that an unidentified protein could contribute TMDs to form a more complete proton translocator (86). Alternatively, for the ExbD paralogue, TolR, it has been shown that rotation of the helices is important for function (87). If the ExbD helices also need to rotate, perhaps this is accomplished by the interaction of ExbB cytoplasmic domains with unidentified cytoplasmic proteins. It has been recently shown that the thrombopoietin receptor has multiple dimeric conformations achieved through rotation of helices (88). For any of these models, it is likely that the identification of unknown proteins will be important for future progress in understanding the mechanism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bryan Ferlez for construction and initial characterization of ExbB S34A.
This work was supported by a National Institute of General Medical Sciences grant GM42146 to K.P.
Footnotes
Published ahead of print 19 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00017-13.
REFERENCES
- 1. Krewulak KD, Vogel HJ. 2011. TonB or not TonB: is that the question? Biochem. Cell Biol. 89:87–97 [DOI] [PubMed] [Google Scholar]
- 2. Kuehl CJ, Crosa JH. 2010. The TonB energy transduction systems in Vibrio species. Future Microbiol. 5:1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64:43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zimbler DL, Penwell WF, Gaddy JA, Menke SM, Tomaras AP, Connerly PL, Actis LA. 2009. Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 22:23–32 [DOI] [PubMed] [Google Scholar]
- 5. Cornelissen CN, Hollander A. 2011. TonB-dependent transporters expressed by Neisseria gonorrhoeae. Front. Microbiol. 2:117. 10.3389/fmicb.2011.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schauer K, Gouget B, Carriere M, Labigne A, de Reuse H. 2007. Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63:1054–1068 [DOI] [PubMed] [Google Scholar]
- 7. Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. 2007. Heme acquisition by hemophores. Biometals 20:603–613 [DOI] [PubMed] [Google Scholar]
- 8. Lohmiller S, Hantke K, Patzer SI, Braun V. 2008. TonB-dependent maltose transport by Caulobacter crescentus. Microbiology (Reading, Engl.) 154:1748–1754 [DOI] [PubMed] [Google Scholar]
- 9. Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denance N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. 10.1371/journal.pone.0000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roof SK, Allard JD, Bertrand KP, Postle K. 1991. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J. Bacteriol. 173:5554–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kampfenkel K, Braun V. 1992. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174:5485–5487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kampfenkel K, Braun V. 1993. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 268:6050–6057 [PubMed] [Google Scholar]
- 13. Karlsson M, Hannavy K, Higgins CF. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389–396 [DOI] [PubMed] [Google Scholar]
- 14. Fischer E, Günter K, Braun V. 1989. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J. Bacteriol. 171:5127–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmer BMM, Thomas MG, Larsen RA, Postle K. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177:4742–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pramanik A, Zhang F, Schwarz H, Schreiber F, Braun V. 2010. ExbB protein in the cytoplasmic membrane of Escherichia coli forms a stable oligomer. Biochemistry 49:8721–8728 [DOI] [PubMed] [Google Scholar]
- 17. Gresock MG, Savenkova MI, Larsen RA, Ollis AA, Postle K. 2011. Death of the TonB shuttle hypothesis. Front. Microbiol. 2:206. 10.3389/fmicb.2011.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cadieux N, Kadner RJ. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. U. S. A. 96:10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogierman M, Braun V. 2003. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J. Bacteriol. 185:1870–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skare JT, Ahmer BMM, Seachord CL, Darveau RP, Postle K. 1993. Energy transduction between membranes - TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302–16308 [PubMed] [Google Scholar]
- 21. Zhai YF, Heijne W, Saier MH., Jr 2003. Molecular modeling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim. Biophys. Acta 1614:201–210 [DOI] [PubMed] [Google Scholar]
- 22. Larsen RA, Deckert GE, Kastead KA, Devanathan S, Keller KL, Postle K. 2007. His20 provides the sole functionally significant side chain in the essential TonB transmembrane domain. J. Bacteriol. 189:2825–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Swayne C, Postle K. 2011. Taking the Escherichia coli TonB transmembrane domain “offline”? Nonprotonatable Asn substitutes fully for TonB His20. J. Bacteriol. 193:3693–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Held KG, Postle K. 2002. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J. Bacteriol. 184:5170–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ollis AA, Manning M, Held KG, Postle K. 2009. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol. Microbiol. 73:466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ollis AA, Postle K. 2012. ExbD mutants define initial stages in TonB energization. J. Mol. Biol. 415:237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Braun V, Gaisser S, Herrmann C, Kampfenkel K, Killman H, Traub I. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Braun V, Herrmann C. 2004. Point mutations in transmembrane helices 2 and 3 of ExbB and TolQ affect their activities in Escherichia coli K-12. J. Bacteriol. 186:4402–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bernsel A, Viklund H, Hennerdal A, Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vakharia-Rao H, Kastead KA, Savenkova MI, Bulathsinghala CM, Postle K. 2007. Deletion and substitution analysis of the Escherichia coli TonB Q160 region. J. Bacteriol. 189:4662–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Shedlovsky A, Brenner S. 1963. A chemical basis for the host-induced modification of T-even bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 50:300–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Postle K. 2007. TonB system, in vivo assays and characterization. Methods Enzymol. 422:245–269 [DOI] [PubMed] [Google Scholar]
- 34. Larsen RA, Chen GJ, Postle K. 2003. Performance of standard phenotypic assays for TonB activity, as evaluated by varying the level of functional, wild-type TonB. J. Bacteriol. 185:4699–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Higgs PI, Larsen RA, Postle K. 2002. Quantitation of known components of the Escherichia coli TonB-dependent energy transduction system: TonB, ExbB, ExbD, and FepA. Mol. Microbiol. 44:271–281 [DOI] [PubMed] [Google Scholar]
- 36. Higgs PI, Myers PS, Postle K. 1998. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180:6031–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 38. Larsen RA, Myers PS, Skare JT, Seachord CL, Darveau RP, Postle K. 1996. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178:1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larsen RA, Thomas MG, Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809–1824 [DOI] [PubMed] [Google Scholar]
- 40. Larsen RA, Thomas MT, Wood GE, Postle K. 1994. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol. Microbiol. 13:627–640 [DOI] [PubMed] [Google Scholar]
- 41. Eick-Helmerich K, Braun V. 1989. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J. Bacteriol. 171:5117–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tourasse NJ, Li WH. 2000. Selective constraints, amino acid composition, and the rate of protein evolution. Mol. Biol. Evol. 17:656–664 [DOI] [PubMed] [Google Scholar]
- 43. Jana B, Manning M, Postle K. 2011. Mutations in the ExbB cytoplasmic carboxy terminus prevent energy-dependent interaction between the TonB and ExbD periplasmic domains. J. Bacteriol. 193:5649–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Decoursey TE. 2003. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83:475–579 [DOI] [PubMed] [Google Scholar]
- 45. Ulmschneider MB, Sansom MS, Di Nola A. 2005. Properties of integral membrane protein structures: derivation of an implicit membrane potential. Proteins 59:252–265 [DOI] [PubMed] [Google Scholar]
- 46. Senes A, Engel DE, DeGrado WF. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14:465–479 [DOI] [PubMed] [Google Scholar]
- 47. Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU. 2005. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 102:14278–14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goemaere EL, Cascales E, Lloubes R. 2007. Mutational analyses define helix organization and key residues of a bacterial membrane energy-transducing complex. J. Mol. Biol. 366:1424–1436 [DOI] [PubMed] [Google Scholar]
- 49. Brinkman KK, Larsen RA. 2008. Interactions of the energy transducer TonB with noncognate energy-harvesting complexes. J. Bacteriol. 190:421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Braun V, Herrmann C. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261–268 [DOI] [PubMed] [Google Scholar]
- 51. Sansom MS, Weinstein H. 2000. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol. Sci. 21:445–451 [DOI] [PubMed] [Google Scholar]
- 52. von Heijne G. 1991. Proline kinks in transmembrane alpha-helices. J. Mol. Biol. 218:499–503 [DOI] [PubMed] [Google Scholar]
- 53. Braun TF, Poulson S, Gully JB, Empey JC, Van Way S, Putnam A, Blair DF. 1999. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 181:3542–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nakamura S, Morimoto YV, Kami-ike N, Minamino T, Namba K. 2009. Role of a conserved prolyl residue (Pro173) of MotA in the mechanochemical reaction cycle of the proton-driven flagellar motor of Salmonella. J. Mol. Biol. 393:300–307 [DOI] [PubMed] [Google Scholar]
- 55. Zhou J, Blair DF. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428–439 [DOI] [PubMed] [Google Scholar]
- 56. Braun P, von Heijne G. 1999. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry 38:9778–9782 [DOI] [PubMed] [Google Scholar]
- 57. Rasmussen A, Rasmussen T, Edwards MD, Schauer D, Schumann U, Miller S, Booth IR. 2007. The role of tryptophan residues in the function and stability of the mechanosensitive channel MscS from Escherichia coli. Biochemistry 46:10899–10908 [DOI] [PubMed] [Google Scholar]
- 58. Pielak RM, Chou JJ. 2011. Influenza M2 proton channels. Biochim. Biophys. Acta 1808:522–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cascales E, Lloubes R, Sturgis JN. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795–807 [DOI] [PubMed] [Google Scholar]
- 60. Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou P, Tian F, Lv F, Shang Z. 2009. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins 76:151–163 [DOI] [PubMed] [Google Scholar]
- 62. Vianney A, Lewin TM, Beyer WF, Lazzaroni JC, Portalier R, Webster RE. 1994. Membrane topology and mutational analysis of the TolQ protein of Escherichia coli required for the uptake of macromolecules and cell envelope integrity. J. Bacteriol. 176:822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kampfenkel K, Braun V. 1993. Membrane topologies of the TolQ and TolR proteins of Escherichia coli: inactivation of TolQ by a missense mutation in the proposed first transmembrane segment. J. Bacteriol. 175:4485–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dean GE, Macnab RM, Stader J, Matsumura P, Burks C. 1984. Gene sequence and predicted amino acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 159:991–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou JD, Fazzio RT, Blair DF. 1995. Membrane topology of the MotA protein of Escherichia coli. J. Mol. Biol. 251:237–242 [DOI] [PubMed] [Google Scholar]
- 66. Blair DF, Berg HC. 1991. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J. Mol. Biol. 221:1433–1442 [DOI] [PubMed] [Google Scholar]
- 67. Li N, Kojima S, Homma M. 2011. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 16:985–999 [DOI] [PubMed] [Google Scholar]
- 68. Kim EA, Price-Carter M, Carlquist WC, Blair DF. 2008. Membrane segment organization in the stator complex of the flagellar motor: implications for proton flow and proton-induced conformational change. Biochemistry 47:11332–11339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kojima S, Kuroda M, Kawagishi I, Homma M. 1999. Random mutagenesis of the pomA gene encoding a putative channel component of the Na(+)-driven polar flagellar motor of Vibrio alginolyticus. Microbiology (Reading, Engl.) 145:1759–1767 [DOI] [PubMed] [Google Scholar]
- 70. Germon P, Clavel T, Vianney A, Portalier R, Lazzaroni JC. 1998. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J. Bacteriol. 180:6433–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang XY, Goemaere EL, Seddiki N, Celia H, Gavioli M, Cascales E, Lloubes R. 2011. Mapping the interactions between Escherichia coli TolQ transmembrane segments. J. Biol. Chem. 286:11756–11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ghosh J, Postle K. 2004. Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol. Microbiol. 51:203–213 [DOI] [PubMed] [Google Scholar]
- 73. Postle K, Kastead KA, Gresock MG, Ghosh J, Swayne CD. 2010. The TonB dimeric crystal structures do not exist in vivo. mBio 1(5):e00307–10. 10.1128/mBio.00307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Karlsson F, Malmborg-Hager AC, Borrebaeck CA. 2006. Escherichia coli TolA tolerates multiple amino-acid substitutions as revealed by screening randomized variants for membrane integrity and phage receptor function. FEMS Microbiol. Lett. 259:81–88 [DOI] [PubMed] [Google Scholar]
- 75. Bourdineaud J-P, Howard SP, Lazdunski C. 1989. Localization and assembly into the Escherichia coli envelope of a protein required for entry of colicin A. J. Bacteriol. 171:2458–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bradbeer C. 1993. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175:3146–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Klebba PE. 2003. Three paradoxes of ferric enterobactin uptake. Front. Biosci. 8:s1422–s1436 [DOI] [PubMed] [Google Scholar]
- 78. Braun V. 2006. Energy transfer between biological membranes. ACS Chem. Biol. 1:352–354 [DOI] [PubMed] [Google Scholar]
- 79. Jaskula JC, Letain TE, Roof SK, Skare JT, Postle K. 1994. Role of the TonB amino terminus in energy transduction between membranes. J. Bacteriol. 176:2326–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ollis AA, Postle K. 2011. The same periplasmic ExbD residues mediate in vivo interactions between ExbD homodimers and ExbD-TonB heterodimers. J. Bacteriol. 193:6852–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ollis AA, Postle K. 2012. Identification of functionally important TonB-ExbD periplasmic domain interactions in vivo. J. Bacteriol. 194:3078–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Postle K. 1990. TonB and the gram-negative dilemma. Mol. Microbiol. 4:2019–2025 [DOI] [PubMed] [Google Scholar]
- 83. Skare JT, Postle K. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 5:2883–2890 [DOI] [PubMed] [Google Scholar]
- 84. Mann BJ, Holroyd CD, Bradbeer C, Kadner RJ. 1986. Reduced activity of TonB-dependent functions in strains of Escherichia coli. FEMS Lett. 33:255–260 [Google Scholar]
- 85. Skare JT, Roof SK, Postle K. 1989. A mutation in the amino terminus of a hybrid TrpC-TonB protein relieves overproduction lethality and results in cytoplasmic accumulation. J. Bacteriol. 171:4442–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. 2012. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc. Natl. Acad. Sci. U. S. A. 109:16696–16701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang XY, Goemaere EL, Thome R, Gavioli M, Cascales E, Lloubes R. 2009. Mapping the interactions between Escherichia coli Tol subunits: rotation of the TolR transmembrane helix. J. Biol. Chem. 284:4275–4282 [DOI] [PubMed] [Google Scholar]
- 88. Matthews EE, Thevenin D, Rogers JM, Gotow L, Lira PD, Reiter LA, Brissette WH, Engelman DM. 2011. Thrombopoietin receptor activation: transmembrane helix dimerization, rotation, and allosteric modulation. FASEB J. 25:2234–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hill CW, Harnish BW. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:7069–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ollis AA, Kumar A, Postle K. 2012. The ExbD periplasmic domain contains distinct functional regions for two stages in TonB energization. J. Bacteriol. 194:3069–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.