Abstract
During the past decade or so, there has been a substantial body of work to dissect arboviral evolution and to develop models of adaptation during host switching. Regardless of what species serve as host or vectors, and of the geographic distribution and the mechanisms of replication, arboviruses tend to have slow evolutionary rates in nature. The hypothesis that this is the result of replication in the disparate environments provided by host and vector did not receive solid experimental support in any of the many viral species tested. Instead, it seems that from the virus's point of view, either the two environments are sufficiently similar or one of the environments so dominates viral evolution that there is tolerance for suboptimal adaptation to the other environment. Replication in alternating environments has an unexpected cost in that there is decreased genetic variance that translates into a compromised adaptability for bypassed environments. Arboviruses under strong and continuous positive selection may have unusual patterns of genomic changes, with few or no mutations accumulated in the consensus sequence or with dN/dS values typically consistent with random drift in DNA-based organisms.
Key Words: Arboviruses, Host switching, Adaptation
Genetic Variation in RNA Viruses
RNA-dependent RNA polymerases (RDRPs) and RNA-dependent DNA polymerases (reverse transcriptases) catalyze the replication of RNA viruses. Unlike the DNA-dependent DNA polymerases that mediate replication in the DNA world, RDRPs and reverse transcriptases usually lack 3′ exonuclease activity, and thus mutations incorporated during replication are not corrected [Steinhauer et al., 1992]. The one exception is that of coronaviruses, which are the giants in the world of RNA viruses with genomes up to 30 kb in size and have a relatively low mutation rate due to the presence of 3′–5′ exonuclease activity [Denison et al., 2011]. Furthermore, none of the RNA viruses have the equivalents to any of the systems that correct nonreplicative damage in DNA. The result is a mutation rate in the order of 10−3 to 10−5 substitutions per nucleotide and round of replication, or one mutation per genome [Drake and Holland, 1999].
One area of controversy is whether high mutation rates are the result of selection for increased diversity [Domingo and Holland, 1997] or a byproduct of selection for fast replication [Elena and Sanjuan, 2005], but we should point out that these two hypotheses are not mutually exclusive. Two groups have demonstrated independently that limiting population diversity in poliovirus populations has dramatic negative fitness effects in a mice model, and increasing variation through mutagenesis leads to fitness recovery [Pfeiffer and Kirkegaard, 2005; Vignuzzi et al., 2006; Vignuzzi et al., 2008].
Host cells sometimes provide additional sources of variation that may result in hypermutagenesis through editing [Patterson et al., 1995] or in cells with unbalanced nucleotide pools [Martinez et al., 1994]. The biological significance of these two mechanisms is unclear, but editing has been associated with a rare but fatal disease (subacute sclerosing panencephalitis) characterized by the isolation of defective, hypermutated measles virus [Cattaneo et al., 1986].
For some viruses, recombination is another source or genetic variation. The generation of new genomic sequences always requires coinfection of a single cell by two (or more) different parental virions. Among virologists, the term recombination is usually restricted to genetic exchange during which a new molecule is generated from two or more parental molecules. All RNA virus families may undergo recombination, but, with very few exceptions [Plyusnin et al., 2002], those with negative-stranded genomes can only undergo nonhomologous recombination that results in the generation of defective interfering particles [Holland, 1991]. RNA viruses with positive-sense genomes may undergo homologous recombination. While there are both replicative and nonreplicative mechanisms that result in recombinant genomes, the former is the most common and takes place by template switching or copy choice. Briefly, RDRP with a nascent strand attached leaves the original template and finds an alternative template (with a different sequence if it is to generate a new genomic sequence) to which the nascent strand binds and replication continues. Template switching may occur multiple times during the generation of a single genome and, at least for some viral species, a large fraction of new genomes are recombinant molecules [Kirkegaard and Baltimore, 1986]. Recombination may also result in the acquisition of host cell genomic material. The acquisition of large fragments is unusual, and its significance unknown [Meyers et al., 1989], but some species undergo frequent recombination involving short host sequences [Onafuwa-Nuga and Telesnitsky, 2009].
A second mechanism of genetic exchange among viruses is reassortment, which takes place in segmented viruses. When a cell is coinfected with at least two parental strains with different sequences, each segment from each progenitor is replicated. During the assembly of new virions, segments from different parents become enveloped in a single particle. Influenza pandemics are typically the result of reassortment among different strains, some of human origin some from other animals such as birds or swine [Brockwell-Staats et al., 2009].
Determinants of Survival and Adaptation
From an evolutionary point of view, mutations are classified into three types depending on their effect on fitness: beneficial, neutral, and deleterious. However, mutations can have evolutionary relevance beyond that of their fitness effects.
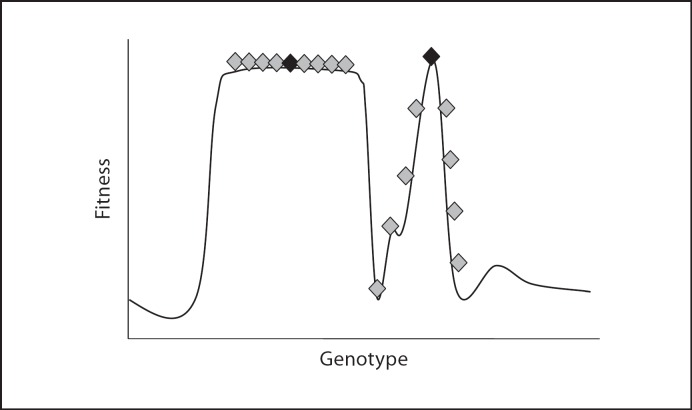
The quasispecies theory [Eigen, 1996; Wilke, 2005] describes the dynamics of populations replicating at a high mutation rate. It is an appropriate framework to describe and explain the evolutionary behavior of RNA viruses [de la Torre and Holland, 1990]. The quasispecies theory predicts selection for increased mutational robustness. Mutational robustness is defined as phenotypic stability in the face of genetic change. Selection for this robustness can result in ‘survival of the flattest’ [Wilke et al., 2001]. Let us consider two fitness peaks, one sharp and one flat (fig. 1) and two progenitors, one at the top of each peak. The progeny of each individual will have an average of one mutation per genome, so the genomic sequences generated will be distributed away from the progenitor's sequence. In the case of the sharp peak, this displacement will result in progeny of lower fitness, but in the case of the flat peak displacement it will result in the occupation of areas where fitness is maintained. As a result, if these two individuals had to compete against one another, the more robust genome would win the competition despite equal initial fitness.
Fig. 1.
Fitness landscapes showing differences in robustness. Two peaks are represented: a flat peak on the left and a sharp peak on the right. Black diamonds represent progenitors that start to replicate and generate the progeny represented by grey diamonds. At high mutation rates, the progeny will be displaced from the site where the progenitor sits. If the peak is flat (high robustness), fitness is maintained; but if the peak is sharp, there is fitness loss (low robustness) and the populations will be outcompeted.
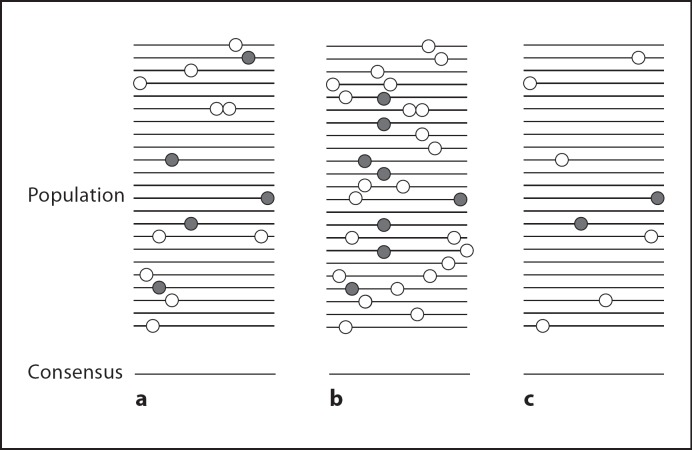
Viral populations are often described by their consensus sequence. The consensus sequence contains at each position the most frequent nucleotide present in the population at that position. However, it is important to realize that at high mutation rates, the consensus sequence may not necessarily be itself a viable viral sequence that is actually present in the population (fig. 2). Yet, the viral sequences (or changes in the sequences) reported in the literature usually refer to the consensus sequence, even if this is slowly changing due to increased use of next-gen sequencing. The quasispecies literature often also uses the term master sequence. The master sequence is the highest-fitness sequence in the population. While the master sequence is a sound concept in theory, in practice it is generally not possible to identify the master sequence in an experimental population. Finally, another term often used in the literature is mutant cloud, which refers to the distribution of mutant genomes in sequence space [Roossinck and Schneider, 2006].
Fig. 2.
Consensus sequence and population diversity. Lines represent genomes and circles indicate mutations. For simplicity, we ignore beneficial variation; grey circles represent neutral mutations and white circles indicate deleterious mutations. The consensus represents the average sequence at each locus and is shown at the bottom. The consensus sequence of a viral progenitor (a) may remain unchanged despite substantial - and biologically significant - increase (b) or decrease (c) in population heterogeneity.
Long-term survival also depends on adaptability, or the ability of a population to increase in fitness. Some authors use the term evolvability [Burch and Chao, 2000], but because evolution may result in fitness stasis when neutral mutations are fixed, or even in fitness losses, we suggest using the term adaptability for the process of adaptation. Adaptability is determined genetically, at least in part, and it may be lost if selection is relaxed, such as it would during repeated bottleneck events. Loss of adaptability results in extinction in the presence of competitors of higher adaptability [Novella, 2004; Novella and Ebendick-Corpus, 2004; Quer et al., 2001; Quer et al., 1996].
The initial period of adaptation to an environment is the result of selection of preexisting variation [Dutta et al., 2008]. At early time points, fitness changes may be substantial without changes in the consensus sequence because a new beneficial allele on its way to fixation is at a frequency that is high enough to have a measurable effect of the population average, but still too low for detection as the dominant nucleotide in that position. In this case, it is only a matter of time until the new allele becomes dominant [Novella and Ebendick-Corpus, 2004]. The history of the strain, including exposure to different hosts, determines what type of beneficial variation is present in a population once the environment changes [Novella et al., 2007].
Arboviruses
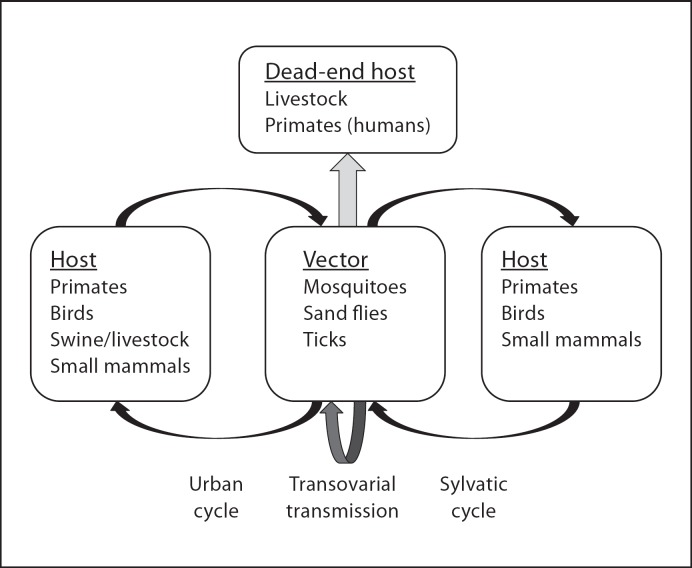
Many viruses cycle between vertebrate hosts and arthropod vectors, including several significant human and veterinary pathogens (table 1). Different species have natural cycles that differ in their complexity (fig. 3). In most cases, but not always, humans and livestock are dead-end hosts and the results of spillover from one of the vectors. Vectors can transmit the virus vertically from females to their eggs, and horizontally to vertebrate hosts during blood meals. There is no horizontal transmission among vectors. Some viral species may go through multiple cycles that are sometimes geographically separated and include more than one vector and more than one vertebrate. One such example is DENV, which can circulate in the wild among primates (sylvatic cycle) and in populated areas among humans (urban cycle). Only some species, such as VSV, feature horizontal transmission among vertebrate hosts. Arthropod-borne viruses, or arboviruses, include members of several families that are characterized by enveloped virions and RNA genomes. We will next provide a brief description of each virus family, but for those interested in additional details, we suggest the relevant chapters of Field's Virology [Burke and Monath, 2001; de Mattos et al., 2001; Griffin, 2001; Lindenbach and Rice, 2001; Nichol, 2001; Rose and Whitt, 2001; Schlesinger and Schlesinger, 2001; Schmaljohn and Hooper, 2001].
Table 1.
Important arboviruses with their natural hosts and vectors
| Virus species | Abbreviation | Natural hosts | Vectors | Distribution |
|---|---|---|---|---|
| Family Rhabdoviridae | ||||
| Vesicular stomatitis virus | VSV | swine | Lutzomyia sand flies | Central and North America |
| Chandipura virus | CHPV | livestock? | Phlebotomus sand flies? | India, Africa |
| Cocal virus | COCV | small mammals, livestock? | Gigantolaelaps mites, Lutzomyia sand flies? | South America |
| Family Togaviridae | ||||
| Eastern equine encephalitis virus | EEEV | birds | Culiseta and Aedes mosquitoes | America |
| Western equine encephalitis virus | WEEV | birds | Culex mosquitoes | America |
| Venezuelan equine encephalitis virus | VEEV | small mammals, horses | Culex, Psophora and Aedes mosquitoes | America |
| Chikungunya virus | CHIKV | primates | Aedes mosquitoes | Subsaharan Africa, South and Southeast Asia |
| Ross River virus | RRV | mammals | Aedes and Culex mosquitoes | Australasia |
| Sindbis virus | SINV | birds | Culex and Culiseta mosquitoes | Eastern Europe, Africa, Southwest Asia, Australia |
| Semliki forest virus | SFV | ? | Aedes mosquitoes | Africa |
| Family Flaviviridae | ||||
| Yellow fever virus | YFV | primates | Aedes mosquitoes | Africa, Central and South America |
| Dengue virus | DENV | primates | Aedes mosquitoes | Tropical areas |
| St. Louis encephalitis virus | SLEV | birds, pigs | Culex mosquitoes | North America |
| Japanese encephalitis virus | JEV | birds, pigs | Culex mosquitoes | East and Southeast Asia |
| Tick-borne encephalitis virus | TBEV | small mammals | Ixodes ticks | Europe |
| West Nile encephalitis virus | WNV | birds | Aedes and Culex mosquitoes | North America, Africa, Europe, Western Asia |
| Family Bunyaviridae | ||||
| California encephalitis virus | CEV | ? | Aedes mosquitoes | North America |
| La Crosse virus | LACV | small mammals, foxes | Aedes mosquitoes | North America |
| Crimean-Congo hemor. fev. virus | CCHV | small mammals | Hyalomma ticks | Africa, Asia, Europe |
| Rift Valley fever | RVFV | livestock | Aedes mosquitoes | Africa |
? = Unproven or unknown hemor.
fev. = hemorrhagic fever.
Fig. 3.
Natural cycles of arboviruses. Different species have one or more of the steps presented in this figure. Human infections are typically the result of transmission during urban cycles or spillover from natural, sylvatic cycles.
Togaviridae
The majority of togaviruses are arboviruses and form the group alphaviruses(the exception is rubella). The viral particle is composed of a single-stranded, positive-sense RNA molecule of about 12 kb with nonstructural proteins in the 5′ half of the genome and structural proteins in the 3′ half of the genome. The RNA is covered by an icosahedral capsid and a membrane that carries viral proteins in its surface, including the receptor-recognition protein. Once the virus attaches and the genome is released into the cell's cytoplasm, it serves as mRNA and produces a polyprotein that gets proteolytically processed and includes nonstructural proteins. One of these proteins is the RDRP that mediates the copying of the genomic positive strand into an antigenomic negative strand, which in turn serves as a template to synthesize a subgenomic RNA for structural polyprotein and a full-length, genomic RNA that will become part of the new crop of virions. The structural polyprotein is also proteolytically processed to generate capsid and envelope (external) proteins. Newly formed capsids with their corresponding genomes acquire their membranes and exit the cell by budding.
Flaviviridae
Virus species in this family are very similar to those in the Togaviridae family. They are composed of a single-stranded, positive-sense RNA genome of about 11 kb inside an icosahedral capsid and surrounded by a membrane envelope that carries viral proteins. However, their genomic structure differs in that the genes for structural proteins are in the 5′ end, while nonstructural genes occupy the 3′ half of the genome. The replication cycle is similar to that described for alphaviruses, except that there is no formation of subgenomic RNA, so the complete genome is translated into a single polyprotein that is processed into unit-size, functional proteins.
Bunyaviridae
Bunyaviruses are the one group of arboviruses that have segmented genomes. There are three single-stranded RNA segments in each viral particle, two of them of negative sense and one ambisense, with one gene of positive and one gene of negative polarity. The genomic size is about 13 kb and the RNA is bound to viral proteins, including the RDRP. After cell attachment and entry, the bunyavirus genome is transcribed and the transcripts are translated into viral proteins, followed by a switch from transcription to replication to generate antigenome intermediates and, from them, some are transcribed and others become new genomic segments that bind viral proteins to form new ribonucleoproteins (RNPs), and these RNPs acquire their membrane and exit by budding.
Rhabdoviridae
The rhabdovirus particle consists of a membrane that carries viral proteins and contains a molecule of single-stranded, negative-sense RNA of about 12 kb and 5–7 individual genes flanked by nontranslated sequences. Several viral proteins, including the RDRP, bind to this genome to form the RNP, which serves as template for transcription and replication. Once the virus binds to a cell and the RNA is delivered into the cytoplasm, the RDRP mediates transcription of monocistronic RNAs that are translated into individual proteins. Next the RDRP switches to a replicative mode and generates full-length, positive-sense molecules that serve as a template to produce full-length, negative-sense progeny. The appropriate new viral proteins bind these genomes, and the progeny RNPs acquire membranes and exit the cell by budding.
Selection for Generalization and Specialization
There is some variation in the literature in the definition and methods to measure the degree of generalization and specialization [Devictor et al., 2010]. In simple terms, generalists are often described as the jack-of-all-trades, master of none. Thus, generalization has a cost in that a generalist cannot compete with a specialist in the environment to which the specialist is adapted. The other side of the coin is that of the specialist, which is highly adapted to one environment but suffers a cost as the potential to perform poorly in other environments [Kassen and Rainey, 2004]. For example, host specificity of phage Φ-6 depends largely on receptor recognition, and the virus seems unable to produce a protein that can recognize multiple bacterial receptors simultaneously, so adaptation to new hosts usually results in loss of recognition of the old host receptor [Ferris et al., 2007]. Some environmental conditions may have an effect on the selection of specialists or generalists, such as interspecies competition [Johnson et al., 2009], coevolution with the host [Hall et al., 2011], or the supply of beneficial variation [Jasmin and Kassen, 2007], which does not seem to be limiting in RNA viruses [Betancourt, 2009; Bollback and Huelsenbeck, 2007; Dutta et al., 2008]. While there are some arguments as to whether true generalists exist in nature [Loxdale et al., 2011], arboviruses are usually considered the ultimate generalists due to their potential to replicate and perform well in a wide variety of hosts [Weaver, 2006]. During the past decade or so, there has been a substantial body of work to analyze the rules governing the evolution of arboviruses in both vectors and hosts.
Viral Evolution during Host Alternation
Because of the alternation between vectors and hosts during their natural cycles, several authors proposed that arboviruses should be under selection for generalization and incur the corresponding costs. The assumptions of this model are that vectors and hosts provide disparate environmental conditions such that there are heavy tradeoffs, and that adaptation to one can only come at the cost of adaptation to the other. In terms of fitness landscapes, the environments that vector and host provide include only a few (and minor) overlapping fitness peaks, so populations that adapt well to one environment become maladapted and disappear during the switch to the other environment, and only populations that could do well enough in both environments would survive (fig. 4a). This hypothesis had the additional appeal of explaining why arboviral genomes are relatively stable in nature [Weaver et al., 1994], at least when compared to other RNA viruses.
Fig. 4.
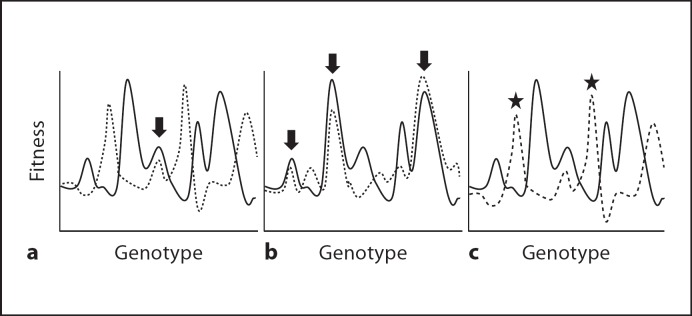
Fitness landscape during host switch. Solid lines and dotted lines represent the landscapes provided by two different environments. The original proposal (a) was that vector and host (or different hosts) provide landscapes with only a few and minor overlapping peaks where the virus could survive in both environments (indicated by the arrow). Experimental results indicate that this is not the case and, instead, in some cases there are multiple or major peaks (arrows in b) or one of the environments - typically the vector - dominates the evolution of the virus, which climbs fitness peaks in such environment (indicated by asterisks in c) even though these correspond to fitness valleys in the other environment.
The basic experimental test of this hypothesis consists of following regimes of repeated passages on each host and comparing the results with those observed in regimes where the virus was allowed to replicate back and forth between host and vector. Progeny of virus evolved under these conditions is tested for changes in fitness, and their genomes are sequenced to determine the number and type of mutation accumulated in the process. Some groups measure fitness in competition assays similar to those developed by Holland et al. [1991], using a genetically-marked, surrogate WT and following changes in the ratio between the two competitors. The genetic marker may result in a phenotypic change that allows the formation of an identifiable plaque, in which case numbers represent live virus. In other cases, the marker allows the specific amplification of each competitor's gene fragment [Troyer et al., 2008] or the fragment is amplified for both competitors, but one of the fragments has a restriction site [Vignuzzi et al., 2006; Weaver et al., 1999]. In these last two cases, numbers represent genomes, which may or may not correspond to live virus. Finally, some groups use viral titer or viral growth curves as a surrogate of fitness [Ciota et al., 2007a; Ciota et al., 2008; Llewellyn et al., 2002; Moutailler et al., 2011; Vasilakis et al., 2009].
Cost of Specialization and Generalization
Continuous passages of viruses in a single host or vector type always result in viral adaptation to that environment when fitness is measured in competition [Coffey and Vignuzzi, 2011; Greene et al., 2005; Mandl et al., 2001; Novella et al., 1995a, b; Novella et al., 1999; Novella et al., 2004; Smith-Tsurkan et al., 2010; Turner et al., 2010; Weaver et al., 1999]. Using virus production as a surrogate, most of the time there is evidence of adaptation [Chen et al., 2003; Ciota et al., 2007b; Ciota et al., 2008; Ciota et al., 2009; Coffey et al., 2008; Cooper and Scott, 2001; Llewellyn et al., 2002; Moutailler et al., 2011; Romanova et al., 2007; Vasilakis et al., 2009], but occasionally there are no measurable changes [Ciota et al., 2007c]. It is possible that continuous passages result in no fitness changes if the virus is already highly adapted to the environment. It is also possible that measuring virus production is not sensitive enough to detect existing differences [Novella and Ebendick-Corpus, 2004], and it would be interesting to see whether competitions render the same results.
The first unexpected result observed during adaptation to single hosts or vectors was that tradeoffs were not always predictable, not even outside host taxonomic groups. Tradeoffs for VSV replicating in sand fly (LL-5) or mammalian cells (Baby Hamster Kidney, BHK-21) are asymmetric and only evident when the virus establishes persistent infection of the sand fly cells [Novella et al., 1995a; Zarate and Novella, 2004]. Acute infection of BHK-21 or LL-5 cells results in adaptation to the bypassed host [Novella et al., 1999]. Studies using other mammalian cell lines confirmed that tradeoffs are unpredictable and less common than originally thought [Smith-Tsurkan et al., 2010; Turner et al., 2010], and suggest that they are not the main cost of specialization. However, virus adaptation to the bypassed cell lines is usually not as substantial as adaptation to the environment in question, so there is a cost due to the shape of fitness landscapes: a genotype is at high fitness peaks in both environments, but the peak in the bypassed environment is shorter than the peak in the environment where the virus is adapting [Smith-Tsurkan et al., 2010]. For EEEV, tradeoffs are usually observed between mammalian and insect cell lines, but not among different mammalian host cells lines or among different mosquito cell lines [Weaver et al., 1999]. Tradeoffs for other arboviral species where these studies exist are unpredictable. For instance, vector-adapted RRV, SFV, VSV and RRV [Novella et al., 1995a; Peleg, 1971; Singh et al., 1971; Taylor and Marshall, 1975] replicate poorly in mammalian environments, but WNV and SLEV do not lose fitness in bypassed avian cells [Ciota et al., 2008; Ciota et al., 2009]. A simple explanation for the lack of expected tradeoffs could be that cell culture does not provide the environmental complexity found in live vectors and hosts, which would include additional components such as antiviral molecules. However, some of these experiments use in vivoinfection and the results are consistent [Ciota et al., 2008; Jerzak et al., 2008; Vasilakis et al., 2009]. Plasmodium parasites, which cause malaria and are also transmitted by mosquitoes, also have the ability to perform well in multiple avian hosts [Hellgren et al., 2009], suggesting that this ability is not restricted to arboviruses.
These results are important for predicting and modeling the emergence of new diseases from their natural reservoirs. Emergence requires some degree of contact between the old host and new host, but if the frequency of the old host is high and tradeoffs are significant, there will be little or no selection to promote niche expansion. Thus, emergence is most likely at intermediate frequencies of the original host [Benmayor et al., 2009]. However, in the absence of tradeoffs we would expect a direct correlation between the frequency of the old host and probability of emergence.
It is not surprising, given the frequent lack of tradeoffs, that alternation between vector cells and host cells does not result in evolutionary stasis, and fitness generally increases in both cell types [Novella et al., 1999; Novella et al., 2004; Smith-Tsurkan et al., 2010]. Even in the instances where tradeoffs were significant, replication back and forth in two environments results in fitness gains in both environments [Cooper and Scott, 2001; Greene et al., 2005; Weaver et al., 1999], suggesting the frequent existence of common fitness peaks in two or more hosts or vectors (fig. 4b). Interestingly, the first evidence of stasis came from experiments in which VSV was alternated between persistent LL-5 cell infection and acute BHK-21 cell infection. Persistent infections, which take place when insect vectors are infected, are characterized by slow, but continuous viral replication, with minimal or no cytopathic effect (cell damage). Under such conditions, heavy tradeoffs are observed when the virus is tested in mammalian cells, and evolution during alternation is very similar to that in the vector alone, with dramatic fitness increases followed by stasis in LL-5 vector cells, and fitness loss in BHK-21 mammalian cells [Zarate and Novella, 2004]. Similar experiments with VEEE [Coffey et al., 2008] and DENV [Vasilakis et al., 2009] also indicate that the vector is the most important determinant of the evolutionary trajectories of the virus and the contribution of the vertebrate is minimal, thus allowing these populations to be placed in low fitness peaks (fig. 4c). One possible explanation for this predominance, at least in vivo, is the potential effect of bottlenecks, which are likely to occur during the infection of mosquitoes (resulting in less than 100% infection rates) [Coffey et al., 2008] and may delay the speed of adaptation [Dutta et al., 2008].
Molecular Evolution
We do not know how fitness changes in natural arbovirus populations, and studies of evolution in nature are based on sequence analysis and some epidemiology. Measurement of the speed of mutation accumulation shows that arboviruses tend to evolve at a slower pace than other RNA viruses [Hanley and Weaver, 2008], although still much more rapidly than DNA-based organisms. The results presented in the previous section clearly indicate that tradeoffs operating during switching between vectors and hosts do not limit the speed of fitness increases. It was still possible that other mechanisms operating during alternation could promote genetic stasis, but results from sequence analysis show otherwise.
When measuring mutation accumulation, in vitro and in vivo experiments consistently produce similar results. In most cases, when alternation results in as much fitness increase as single host/vector replication, alternation results in the accumulation of at least as many mutations as during viral evolution in the fastest evolving cell type [Coffey et al., 2008; Greene et al., 2005; Jerzak et al., 2008; McCurdy et al., 2011; Novella et al., 1999; Novella et al., 2004; Remold et al., 2008, Vasilakis et al., 2009]. In most of the remaining cases, alternation results in genomic patterns that are similar to those promoted by replication in the vector alone [Presloid et al., 2008; Zarate and Novella, 2004]. Consistent with this predominance, tick-borne arboviruses tend to spend much of their life in their respective vectors, and they also show particularly slow rates of genomic evolution [Gould et al., 2003]. There are a few other reports that show different patterns of mutation accumulation and are difficult to interpret due the low number of mutations [Moutailler et al., 2011; Weaver et al., 1999].
Sequence analyses have also helped us to understand the molecular basis of specific and nonspecific host adaptation and fitness costs. Extended receptor recognition is a common mechanism leading to niche expansion. Unlike in some phages, eukaryotic viruses’ external glycoproteins can often acquire the ability to recognize new receptors without losing recognition of old receptors [Baranowski et al., 2001; Mandl et al., 2001; Smit et al., 2002]. Nonspecific adaptation can be important for human health because sometimes these new viral properties result in the ability to use dead-end hosts or new vectors for amplification and transmission, respectively, and thus promote emergence of epidemic strains [Anishchenko et al., 2006; Tsetsarkin et al., 2011]. It is important to note that emergence is highly dependent on the specific sequences of circulating genotypes, because epistasis may impose limitations on the number and type of mutations that a strain may acquire [Tsetsarkin et al., 2011]. The compactness of RNA virus genomes dictates extensive epistasis that can be observed experimentally [Novella and Ebendick-Corpus, 2004; Remold et al., 2008; Sanjuan et al., 2004].
Two mechanisms can operate to produce a tradeoff. The first is mutation accumulation, which consists of the acquisition of mutations that are neutral in the environment where the organism is adapting (for instance, during hitchhiking), but deleterious in a bypassed environment [Elena and Lenski, 2003]. A second is antagonistic pleiotropy, which occurs when a mutation that is beneficial in the adaptive environment is, at the same time, deleterious in a bypassed environment [Levins, 1968]. Some results clearly show a contribution of antagonistic pleiotropy to tradeoffs [Presloid et al., 2008], but there is little or no evidence supporting that mutation accumulation has a role to explain tradeoffs [Smith-Tsurkan et al., 2010].
The types of mutations accumulated during adaptation were unexpected in many cases. Researchers use the relative frequency of synonymous and nonsynonymous mutations to deduce the type of selective forces that operate during evolution (after correction for synonymous sites and nonsynonymous sites, respectively). An excess of nonsynonymous sites (dN/dS >1) is considered to reflect positive selection, frequencies similar to that expected by random mutation (dN/dS ∼1) indicate random drift and excess in synonymous mutation (dN/dS <1) represents negative selection [Nei, 1987]. The assumption for this interpretation is that because proteins retain their sequence if the mutation is synonymous, selection cannot operate. However, in the case of RNA viruses the genomic sequence itself may be the target of selection, for instance, by carrying encapsidation signals [reviewed in Novella, 2003]. In the case of flaviviruses, codon usage seems to be a target for selection, and different dinucleotides are more or less frequently used depending on the host [Lobo et al., 2009]. Because of this additional layer to selection, regimes in which positive selection is the main or only driver of evolution (as demonstrated by fitness trajectories) frequently result in dN/dS values close to 1 [Novella et al., 2004]. Unfortunately, many studies do not even include results of synonymous mutations as they are assumed to be of no value, and other reports either have insufficient data or the data are not tested for statistical significance. However, convergence and parallel evolution of synonymous sites in different viral species support the idea that synonymous mutations are under selection [reviewed in Novella, 2003]. Overall, these results suggest that to conclude the type of selection (or lack thereof) operating on arboviral populations, dN/dS should be used with a great deal of caution and in light of the particular viral function(s) under investigation.
Quasispecies Clouds and Adaptability
Fitness changes do not always correlate with changes in the genomic consensus sequence. As indicated earlier, the simplest explanation is that the beneficial mutations are still on their way to fixation. However, changes in the composition of the quasispecies can have profound effects on the ability of a virus population to survive or undergo extinction [Grande-Perez et al., 2005]. Several studies report WNV and SLEV cloud size changes [reviewed in Ciota et al., 2011]. Both viruses show limited fixation of mutations during fitness changes, but characteristics and differential changes in cloud size [Ciota et al., 2007a, c; Fitzpatrick et al., 2010]. SLEV tends to be highly homogeneous in cell culture [Ciota et al., 2007a], and natural isolates seem to lose genetic heterogeneity with time (fig. 2c) [Ciota et al., 2011]. In contrast, WNV increases its mutant cloud size during adaptation (fig. 2b). There is a correlation between mutant cloud size and success in nature, so the SLEV has a relatively high genetic stability, low prevalence and is more restricted in its geographic distribution, while WNV, which was first brought to the USA in 1999, has spread across the country in just a few years and has a higher prevalence. Whether the relationship is just a correlation or cause-effect remains to be seen, but some plant viruses show similar correlation between host range quasispecies cloud size [Roossinck and Schneider, 2006; Schneider and Roossinck, 2001].
Provided with a sufficient supply of beneficial variation, one might expect that host/vector alternation may promote higher genotypic (and phenotypic) variance, which, as discussed, may promote adaptation to new environments [Jasmin and Kassen, 2007; Kassen and Rainey, 2004]. Surprisingly, this does not seem to be the case. Alternation of CHIKV between mammalian and mosquito cells results in lower genotypic variance and compromised adaptability in new environments [Coffey and Vignuzzi, 2011]. Similar studies on VSV suggest that viruses with a history of host switching fare worse (as measured by virus production) in new thermal environments than viruses with a history of replication in single host [Alto and Turner, 2010]. Results from our laboratory also failed to show increased phenotypic variance in VSV during alternation between different mammalian host cells or between mammalian and sand fly host cells compared to VSV replicating in single cell types [unpubl. data].
Acknowledgements
This work was supported by NIH grant R01 AI065960.
References
- Alto BW, Turner PE. Consequences of host adaptation for performance of vesicular stomatitis virus in novel thermal environments. Evol Ecol. 2010;24:299–315. [Google Scholar]
- Anishchenko M, Bowen RA, Paessler S, Austgen L, Greene IP, Weaver SC. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc Natl Acad Sci USA. 2006;103:4994–4999. doi: 10.1073/pnas.0509961103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowski E, Ruiz-Jarabo CM, Domingo E. Evolution of cell recognition by viruses. Science. 2001;292:1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- Benmayor R, Hodgson DJ, Perron GG, Buckling A. Host mixing and disease emergence. Curr Biol. 2009;19:764–767. doi: 10.1016/j.cub.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt AJ. Genomewide patterns of substitution in adaptively evolving populations of the RNA bacteriophage Ms2. Genetics. 2009;181:1535–1544. doi: 10.1534/genetics.107.085837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback JP, Huelsenbeck JP. Clonal interference is alleviated by high mutation rates in large populations. Mol Biol Evol. 2007;24:1397–1406. doi: 10.1093/molbev/msm056. [DOI] [PubMed] [Google Scholar]
- Brockwell-Staats C, Webster RG, Webby RJ. Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza a (H1N1) Influenza Other Respi Viruses. 2009;3:207–213. doi: 10.1111/j.1750-2659.2009.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch CL, Chao L. Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature. 2000;406:625–628. doi: 10.1038/35020564. [DOI] [PubMed] [Google Scholar]
- Burke DS, Monath TP. Flaviviruses. In: Knipe DM, Howley PM, editors. Fields Virology. ed 4. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1043–1126. [Google Scholar]
- Cattaneo R, Schmid A, Rebmann G, Baczko K, Ter Meulen V, Bellini WJ, Rozenblatt S, Billeter MA. Accumulated measles virus mutations in a case of subacute sclerosing panencephalitis: interrupted matrix protein reading frame and transcription alteration. Virology. 1986;154:97–107. doi: 10.1016/0042-6822(86)90433-2. [DOI] [PubMed] [Google Scholar]
- Chen WJ, Wu HR, Chiou SS. E/NS1 modifications of dengue 2 virus after serial passages in mammalian and/or mosquito cells. Intervirology. 2003;46:289–295. doi: 10.1159/000073208. [DOI] [PubMed] [Google Scholar]
- Ciota AT, Jia Y, Payne AF, Jerzak G, Davis LJ, Young DS, Ehrbar D, Kramer LD. Experimental passage of St. Louis encephalitis virus in vivo in mosquitoes and chickens reveals evolutionarily significant virus characteristics. PLoS One. 2009;4:e7876. doi: 10.1371/journal.pone.0007876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Koch EM, Willsey GG, Davis LJ, Jerzak GV, Ehrbar DJ, Wilke CO, Kramer LD. Temporal and spatial alterations in mutant swarm size of St. Louis encephalitis virus in mosquito hosts. Infect Genet Evol. 2011;11:460–468. doi: 10.1016/j.meegid.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Lovelace AO, Jia Y, Davis LJ, Young DS, Kramer LD. Characterization of mosquito-adapted West Nile virus. J Gen Virol. 2008;89:1633–1642. doi: 10.1099/vir.0.2008/000893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Lovelace AO, Jones SA, Payne A, Kramer LD. Adaptation of two flaviviruses results in differences in genetic heterogeneity and virus adaptability. J Gen Virol. 2007a;88:2398–2406. doi: 10.1099/vir.0.83061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Lovelace AO, Ngo KA, Le AN, Maffei JG, Franke MA, Payne AF, Jones SA, Kauffman EB, Kramer LD. Cell-specific adaptation of two flaviviruses following serial passage in mosquito cell culture. Virology. 2007b;357:165–174. doi: 10.1016/j.virol.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Ngo KA, Lovelace AO, Payne AF, Zhou Y, Shi PY, Kramer LD. Role of the mutant spectrum in adaptation and replication of West Nile virus. J Gen Virol. 2007c;88:865–874. doi: 10.1099/vir.0.82606-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci USA. 2008;105:6970–6975. doi: 10.1073/pnas.0712130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey LL, Vignuzzi M. Host alternation of Chikungunya virus increases fitness while restricting population diversity and adaptability to novel selective pressures. J Virol. 2011;85:1025–1035. doi: 10.1128/JVI.01918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Scott TW. Differential evolution of eastern equine encephalitis virus populations in response to host cell type. Genetics. 2001;157:1403–1412. doi: 10.1093/genetics/157.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre JC, Holland JJ. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J Virol. 1990;64:6278–6281. doi: 10.1128/jvi.64.12.6278-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mattos CA, de Mattos CC, Rupprecht CE. Rhabdoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. ed 4. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1245–1278. [Google Scholar]
- Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devictor V, Clavel J, Julliard R, Lavergne S, Mouillot D, Thuiller W, Venail P, Villéger S, Moquet N. Defining and measuring ecological specialization. J Appl Ecol. 2010;47:15–25. [Google Scholar]
- Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta RN, Rouzine IM, Smith SD, Wilke CO, Novella IS. Rapid adaptive amplification of preexisting variation in an RNA virus. J Virol. 2008;82:4354–4362. doi: 10.1128/JVI.02446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M. On the nature of virus quasispecies. Trends Microbiol. 1996;4:216–218. doi: 10.1016/0966-842X(96)20011-3. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Elena SF, Sanjuan R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J Virol. 2005;79:11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MT, Joyce P, Burch CL. High frequency of mutations that expand the host range of an RNA virus. Genetics. 2007;176:1013–1022. doi: 10.1534/genetics.106.064634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick KA, Deardorff ER, Pesko K, Brackney DE, Zhang B, Bedrick E, Shi PY, Ebel GD. Population variation of West Nile virus confers a host-specific fitness benefit in mosquitoes. Virology. 2010;404:89–95. doi: 10.1016/j.virol.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EA, de Lamballerie X, Zanotto PM, Holmes EC. Origins, evolution, and vector/host coadaptations within the genus flavivirus. Adv Virus Res. 2003;59:277–314. doi: 10.1016/s0065-3527(03)59008-x. [DOI] [PubMed] [Google Scholar]
- Grande-Perez A, Gomez-Mariano G, Lowenstein PR, Domingo E. Mutagenesis-induced, large fitness variations with an invariant arenavirus consensus genomic nucleotide sequence. J Virol. 2005;79:10451–10459. doi: 10.1128/JVI.79.16.10451-10459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene IP, Wang E, Deardorff ER, Milleron R, Domingo E, Weaver SC. Effect of alternating passage on adaptation of sindbis virus to vertebrate and invertebrate cells. J Virol. 2005;79:14253–14260. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DE. Alphaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. ed 4. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 917–962. [Google Scholar]
- Hall AR, Scanlan PD, Buckling A. Bacteria-phage coevolution and the emergence of generalist pathogens. Am Nat. 2011;177:44–53. doi: 10.1086/657441. [DOI] [PubMed] [Google Scholar]
- Hanley KA, Weaver SC. Arbovirus evolution. In: Domingo E, Parrish CR, Holland JJ, editors. Origin and Evolution of Viruses. London: Academic Press; 2008. pp. 351–382. [Google Scholar]
- Hellgren O, Perez-Tris J, Bensch S. A jack-of-all-trades and still a master of some: prevalence and host range in avian malaria and related blood parasites. Ecology. 2009;90:2840–2849. doi: 10.1890/08-1059.1. [DOI] [PubMed] [Google Scholar]
- Holland JJ. Defective Viral Genomes. In: Knipe D, editor. Fundamental Virology. New York: Raven Press; 1991. pp. 151–165. [Google Scholar]
- Holland JJ, de la Torre JC, Clarke DK, Duarte E. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol. 1991;65:2960–2967. doi: 10.1128/jvi.65.6.2960-2967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin JN, Kassen R. Evolution of a single niche specialist in variable environments. Proc Biol Sci. 2007;274:2761–2767. doi: 10.1098/rspb.2007.0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak GV, Brown I, Shi PY, Kramer LD, Ebel GD. Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology. 2008;374:256–260. doi: 10.1016/j.virol.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KP, Malenke JR, Clayton DH. Competition promotes the evolution of host generalists in obligate parasites. Proc Biol Sci. 2009;276:3921–3926. doi: 10.1098/rspb.2009.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassen R, Rainey PB. The ecology and genetics of microbial diversity. Annu Rev Microbiol. 2004;58:207–231. doi: 10.1146/annurev.micro.58.030603.123654. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levins R. Evolution in Changing Environments. Princeton: Princeton University Press; 1968. [Google Scholar]
- Lindenbach BD, Rice CM. Flaviviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. ed 4. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 991–1042. [Google Scholar]
- Llewellyn ZN, Salman MD, Pauszek S, Rodriguez LL. Growth and molecular evolution of vesicular stomatitis serotype New Jersey in cells derived from its natural insect-host: evidence for natural adaptation. Virus Res. 2002;89:65–73. doi: 10.1016/s0168-1702(02)00113-2. [DOI] [PubMed] [Google Scholar]
- Lobo FP, Mota BE, Pena SD, Azevedo V, Macedo AM, Tauch A, Machado CR, Franco GR. Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS One. 2009;4:e6282. doi: 10.1371/journal.pone.0006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loxdale HD, Lushai G, Harvey JA. The evolutionary improbability of ‘generalism’ in nature, with special reference to insects. Biol J Linn Soc Lond. 2011;103:1–18. [Google Scholar]
- Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J Virol. 2001;75:5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MA, Vartanian JP, Wain-Hobson S. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc Natl Acad Sci USA. 1994;91:11787–11791. doi: 10.1073/pnas.91.25.11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy K, Joyce J, Hamilton S, Nevins C, Sosna W, Puricelli K, Rayner JO. Differential accumulation of genetic and phenotypic changes in Venezuelan equine encephalitis virus and Japanese encephalitis virus following passage in vitro and in vivo. Virology. 2011;415:20–29. doi: 10.1016/j.virol.2011.03.030. [DOI] [PubMed] [Google Scholar]
- Meyers G, Rumenapf T, Thiel HJ. Ubiquitin in a togavirus. Nature. 1989;341:491. doi: 10.1038/341491a0. [DOI] [PubMed] [Google Scholar]
- Moutailler S, Roche B, Thiberge JM, Caro V, Rougeon F, Failloux AB. Host alternation is necessary to maintain the genome stability of Rift Valley fever virus. PLoS Negl Trop Dis. 2011;5:e1156. doi: 10.1371/journal.pntd.0001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Evolutionary Change of Nucleotide Sequences. In: Nei M, editor. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. pp. 64–110. [Google Scholar]
- Nichol ST. Bunyaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. ed 4. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1603–1634. [Google Scholar]
- Novella IS. Contributions of vesicular stomatitis virus to the understanding of RNA virus evolution. Curr Opin Microbiol. 2003;6:399–405. doi: 10.1016/s1369-5274(03)00084-5. [DOI] [PubMed] [Google Scholar]
- Novella IS. Negative effect of genetic bottlenecks on the adaptability of vesicular stomatitis virus. J Mol Biol. 2004;336:61–67. doi: 10.1016/j.jmb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Novella IS, Clarke DK, Quer J, Duarte EA, Lee CH, Weaver SC, Elena SF, Moya A, Domingo E, Holland JJ. Extreme fitness differences in mammalian and insect hosts after continuous replication of vesicular stomatitis virus in sandfly cells. J Virol. 1995a;69:6805–6809. doi: 10.1128/jvi.69.11.6805-6809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella IS, Duarte EA, Elena SF, Moya A, Domingo E, Holland JJ. Exponential increases of RNA virus fitness during large population transmissions. Proc Natl Acad Sci USA. 1995b;92:5841–5844. doi: 10.1073/pnas.92.13.5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella IS, Ebendick-Corpus BE. Molecular basis of fitness loss and fitness recovery in vesicular stomatitis virus. J Mol Biol. 2004;342:1423–1430. doi: 10.1016/j.jmb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Novella IS, Ebendick-Corpus BE, Zarate S, Miller EL. Emergence of mammalian cell-adapted vesicular stomatitis virus from persistent infections of insect vector cells. J Virol. 2007;81:6664–6668. doi: 10.1128/JVI.02365-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella IS, Hershey CL, Escarmis C, Domingo E, Holland JJ. Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J Mol Biol. 1999;287:459–465. doi: 10.1006/jmbi.1999.2635. [DOI] [PubMed] [Google Scholar]
- Novella IS, Zarate S, Metzgar D, Ebendick-Corpus BE. Positive selection of synonymous mutations in vesicular stomatitis virus. J Mol Biol. 2004;342:1415–1421. doi: 10.1016/j.jmb.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JB, Thomis DC, Hans SL, Samuel CE. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology. 1995;210:508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- Peleg J. Growth of viruses in arthropod cell cultures: applications. I. Attenuation of Semliki Forest (SF) virus in continuously cultured Aedes Aegypti mosquito cells (Peleg) as a step in production of vaccines. Curr Top Microbiol Immunol. 1971;55:155–161. doi: 10.1007/978-3-642-65224-0_26. [DOI] [PubMed] [Google Scholar]
- Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A, Kukkonen SK, Plyusnina A, Vapalahti O, Vaheri A. Transfection-mediated generation of functionally competent Tula hantavirus with recombinant S RNA segment. EMBO J. 2002;21:1497–1503. doi: 10.1093/emboj/21.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presloid JB, Ebendick-Corpus BE, Zarate S, Novella IS. Antagonistic pleiotropy involving promoter sequences in a virus. J Mol Biol. 2008;382:342–352. doi: 10.1016/j.jmb.2008.06.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quer J, Hershey CL, Domingo E, Holland JJ, Novella IS. Contingent neutrality in competing viral populations. J Virol. 2001;75:7315–7320. doi: 10.1128/JVI.75.16.7315-7320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quer J, Huerta R, Novella IS, Tsimring L, Domingo E, Holland JJ. Reproducible nonlinear population dynamics and critical points during replicative competitions of RNA Virus quasispecies. J Mol Biol. 1996;264:465–471. doi: 10.1006/jmbi.1996.0654. [DOI] [PubMed] [Google Scholar]
- Remold SK, Rambaut A, Turner PE. Evolutionary genomics of host adaptation in vesicular stomatitis virus. Mol Biol Evol. 2008;25:1138–1147. doi: 10.1093/molbev/msn059. [DOI] [PubMed] [Google Scholar]
- Romanova L, Gmyl AP, Dzhivanian TI, Bakhmutov DV, Lukashev AN, Gmyl LV, Rumyantsev AA, Burenkova LA, Lashkevich VA, Karganova GG. Microevolution of tick-borne encephalitis virus in course of host alternation. Virology. 2007;362:75–84. doi: 10.1016/j.virol.2006.12.013. [DOI] [PubMed] [Google Scholar]
- Roossinck MJ, Schneider WL. Mutant clouds and occupation of sequence space in plant RNA viruses. Curr Top Microbiol Immunol. 2006;299:337–348. doi: 10.1007/3-540-26397-7_12. [DOI] [PubMed] [Google Scholar]
- Rose JK, Whitt MA. Rhabdoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. ed 4. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1221–1244. [Google Scholar]
- Sanjuan R, Moya A, Elena SF. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci USA. 2004;101:15376–15379. doi: 10.1073/pnas.0404125101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S, Schlesinger MJ. Togaviridae: the viruses and their replication. In: Knipe DM, Holwey PM, editors. Fields Virology. ed 4. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 895–916. [Google Scholar]
- Schmaljohn CS, Hooper JW. Bunyaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. ed 4. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1581–1602. [Google Scholar]
- Schneider WL, Roossinck MJ. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J Virol. 2001;75:6566–6571. doi: 10.1128/JVI.75.14.6566-6571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KR, Bhat UK, Paul SD. Adaptation of Singh's Aedes Albopictus (Atc-15) cell line to media with calf and goat sera and their susceptibility to infection with some arboviruses. Indian J Med Res. 1971;59:31–34. [PubMed] [Google Scholar]
- Smit JM, Waarts BL, Kimata K, Klimstra WB, Bittman R, Wilschut J. Adaptation of alphaviruses to heparan sulfate: interaction of Sindbis and Semliki forest viruses with liposomes containing lipid-conjugated heparin. J Virol. 2002;76:10128–10137. doi: 10.1128/JVI.76.20.10128-10137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Tsurkan SD, Wilke CO, Novella IS. Incongruent fitness landscapes, not tradeoffs, dominate the adaptation of vesicular stomatitis virus to novel host types. J Gen Virol. 2010;91:1484–1493. doi: 10.1099/vir.0.017855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122:281–288. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- Taylor WP, Marshall ID. Adaptation studies with Ross River virus: laboratory mice and cell cultures. J Gen Virol. 1975;28:59–72. doi: 10.1099/0022-1317-28-1-59. [DOI] [PubMed] [Google Scholar]
- Troyer RM, Garver KA, Ranson JC, Wargo AR, Kurath G. In vivo virus growth competition assays demonstrate equal fitness of fish rhabdovirus strains that co-circulate in aquaculture. Virus Res. 2008;137:179–188. doi: 10.1016/j.virusres.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci USA. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Morales NM, Alto BW, Remold SK. Role of evolved host breadth in the initial emergence of an RNA virus. Evolution. 2010;64:3273–3286. doi: 10.1111/j.1558-5646.2010.01051.x. [DOI] [PubMed] [Google Scholar]
- Vasilakis N, Deardorff ER, Kenney JL, Rossi SL, Hanley KA, Weaver SC. Mosquitoes put the brake on arbovirus evolution: experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog. 2009;5:e1000467. doi: 10.1371/journal.ppat.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- Weaver SC. Evolutionary influences in arboviral disease. In: Domingo E, editor. Quasispecies: Concept and Implications for Virology. Curr Topics Microbiol Immunol. New York: Springer; 2006. pp. 285–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Brault AC, Kang W, Holland JJ. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol. 1999;73:4316–4326. doi: 10.1128/jvi.73.5.4316-4326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Hagenbaugh A, Bellew LA, Netesov SV, Volchkov VE, Chang GJ, Clarke DK, Gousset L, Scott TW, Trent DW, et al. A comparison of the nucleotide sequences of Eastern and Western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology. 1994;202:1083. doi: 10.1006/viro.1994.1445. [DOI] [PubMed] [Google Scholar]
- Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol Biol. 2005;5:44. doi: 10.1186/1471-2148-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412:331–333. doi: 10.1038/35085569. [DOI] [PubMed] [Google Scholar]
- Zarate S, Novella IS. Vesicular stomatitis virus evolution during alternation between persistent infection in insect cells and acute infection in mammalian cells is dominated by the persistence phase. J Virol. 2004;78:12236–12242. doi: 10.1128/JVI.78.22.12236-12242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]