Abstract
Ergothioneine (ERG) and mycothiol (MSH) are two low-molecular-weight thiols synthesized by mycobacteria. The role of MSH has been extensively investigated in mycobacteria; however, little is known about the role of ERG in mycobacterial physiology. In this study, quantification of ERG at various points in the growth cycle of Mycobacterium smegmatis revealed that a significant portion of ERG is found in the culture media, suggesting that it is actively secreted. A mutant of M. smegmatis lacking egtD (MSMEG_6247) was unable to synthesize ERG, confirming its role in ERG biosynthesis. Deletion of egtD from wild-type M. smegmatis and an MSH-deficient mutant did not affect their susceptibility to antibiotics tested in this study. The ERG- and MSH-deficient double mutant was significantly more sensitive to peroxide than either of the single mutants lacking either ERG or MSH, suggesting that both thiols play a role in protecting M. smegmatis against oxidative stress and that ERG is able to partly compensate for the loss of MSH.
INTRODUCTION
Glutathione (GSH) is a thiol known for its efficient detoxification of reactive oxygen species, reactive nitrogen species, and free radicals in eukaryotes. Mycobacteria do not synthesize GSH but produce two low-molecular-weight thiols, namely, mycothiol {1-O-[2-(N-acetyl-l-cysteinyl)amido-2-deoxy-α-d-glucopyranosyl]-d-myo-inositol} (MSH) (1–3) and ergothioneine (2-mercaptohistidine trimethylbetaine) (ERG) (4, 5). Four genes are involved in MSH biosynthesis in mycobacteria, namely, mshA, mshB, mshC, and mshD, and mutants harboring deletions in mshB, mshC, and mshD produce different levels of MSH due to the ability of other enzymes to partially compensate for their loss (6, 7). MSH-deficient mutants of Mycobacterium smegmatis show increased sensitivity to oxidative stress, alkylating agents, and a range of antibiotics, including erythromycin, azithromycin, vancomycin, penicillin G, streptomycin, and rifampin, but exhibit increased resistance to isoniazid (INH) and ethionamide (ETH) (8, 9). The MSH-deficient ΔmshA mutant of Mycobacterium tuberculosis requires catalase during in vitro growth, implicating MSH in detoxifying reactive oxygen species (10).
ERG biosynthetic genes (egtA, egtB, egtC, egtD, and egtE) were recently identified in M. smegmatis (11). Although several lines of evidence support the cytoprotective and antioxidative role of ERG in eukaryotes (12), bacteria (13), and, recently, fungi (14), nothing is known of its role in mycobacteria. ERG has also been implicated in modulating the immune response (15) and in the inhibition of metalloenzymes, preventing the copper-induced oxidation of DNA and protein due to its metal-chelating properties (16, 17). Eukaryotes obtain ERG from their diet, and its accumulation in cells is dependent on the activity of a highly specific transporter, OCTN1, since the zwitterionic nature of ERG prevents it from crossing the plasma membrane (18, 19). In an M. smegmatis ΔmshA mutant, which is MSH deficient, the levels of ERG and the organic hydroperoxide resistance (Ohr) protein are elevated, suggesting that ERG may partly compensate for the loss of MSH (20). This may explain the lack of a phenotype for the ΔmshA mutant of M. tuberculosis in mice (10). The ERG biosynthetic genes egtA, egtB, egtC, egtD, and egtE are not predicted to be essential for the in vitro growth of laboratory M. tuberculosis strain H37Rv (21, 22).
In this study, we investigate the role of ERG in M. smegmatis as well as its relationship to MSH in this organism. We confirm the function of egtD in ERG biosynthesis and demonstrate that a significant proportion of ERG synthesized by M. smegmatis is secreted. Furthermore, we show that ERG plays a role in protecting M. smegmatis against peroxides.
MATERIALS AND METHODS
Chemicals.
Antibiotics, cumene hydroperoxide (CuOOH) (80%), Luperox tert-butyl hydroperoxide (TBHP) (70%), Coomassie brilliant blue (CBB), N,N,N′,N′-tetramethylethylenediamine (TEMED), sodium dodecyl sulfate (SDS), ammonium persulfate, Tris base, hydrogen chloride, 30% acrylamide-bisacrylamide, and ERG used in this study were obtained from Sigma-Aldrich. Isoniazid, ethambutol, and kanamycin were dissolved in sterile water, while rifampin was dissolved in dimethyl sulfoxide (DMSO). Working solutions of CuOOH and TBHP were prepared in DMSO and water, respectively. Stock solutions of CuOOH and TBHP were stored at 4°C and shielded from light for a maximum period of 3 months due to instability. The complete protease inhibitor cocktail was obtained from Roche, Mannheim, Germany.
Mycobacterial strains and culture conditions.
The strains used in this study are indicated in Table S1 in the supplemental material. M. smegmatis was cultured in Difco Middlebrook 7H9 broth (Becton, Dickinson, USA) supplemented with 0.85% NaCl, 0.2% glucose, and 0.05% Tween 80 or on Luria-Bertani agar (Miller, Merck). Escherichia coli strains were cultured in liquid medium containing 1% NaCl, 0.5% Bacto yeast, and 1% Bacto tryptone (Merck) or on Luria-Bertani agar (Miller, Merck). Where appropriate, ampicillin (Ap), kanamycin (Kan), and hygromycin (Hyg) were used in E. coli solid cultures at 50, 50, and 100 μg/ml, respectively, and Hyg or Kan was used in mycobacterial solid cultures at 75 and 50 μg/ml, respectively, while half of these concentrations were used in liquid cultures.
Construction and complementation of mutant strains.
An unmarked egtD (MSMEG_6247) deletion was generated by homologous recombination, as described previously (23). Briefly, upstream (US) and downstream (DS) fragments flanking egtD were amplified from M. smegmatis mc2155 genomic DNA by using the primers listed in Table S2 in the supplemental material. The resulting fragments were cloned individually into the CloneJet vector (Fermentas) and subsequently used to construct the suicide plasmid p2NILegtD. The sacB-lacZ fragment from pGOAL17 or the sacB-hyg-lacZ fragment from pGOAL19 was cloned into the PacI site of p2NILegtD to generate p2NILegtD17 or p2NILegtD19, respectively (23) (see Table S1 in the supplemental material). p2NILegtD17 or p2NILegtD19 was electroporated into M. smegmatis mc2155 or ΔmshA mutant cells, respectively (24), and the egtD deletion was generated in each strain by two-step allelic exchange. A complementation vector was generated by amplifying the region from 2,398 bp upstream of egtD to 1,169 bp downstream of egtD (see Table S2 in the supplemental material) and cloning it into the integrating vector pMV306 (25) (see Table S1 in the supplemental material). The integrity of the allelic-exchange substrates and complementation vector was confirmed by sequencing, and the genotypes of the mutant strains were confirmed by Southern blotting.
Ergothioneine quantification and mycothiol detection.
Intracellular ERG was extracted by using a previously described method, with modifications (26, 27). Liquid M. smegmatis cultures were grown to the specified optical density at 600 nm (OD600), harvested by centrifugation, and resuspended in lysis buffer (40% acetonitrile, 0.25 M perchloric acid). Cells were disrupted by sonication, and the lysate was clarified by centrifugation. The pH of the supernatant was adjusted to between 8 and 10 with potassium carbonate (Sigma-Aldrich) and lyophilized. The dried material was redissolved in a solution containing 25% acetonitrile and 0.05% formic acid and analyzed by ultraperformance liquid chromatography-electrospray ionization-tandem mass spectrometry (UPLC-ESI-MS/MS) (26, 28). UPLC-ESI-MS/MS analysis was performed with a Waters Acquity UPLC system coupled to a Waters Xevo TQ MS system (Waters Corporation, Milford, MA, USA). Compounds were separated on a Waters Acquity BEH phenyl column (100 by 2.1 mm; 1.7 μm) at 50°C using a 1% formic acid (in water) (solvent A)–acetonitrile (solvent B) gradient, starting with 100% solvent A for 1.5 min at a flow rate of 0.3 ml/min. The acetonitrile concentration was increased linearly to 2% over 2 min at a flow rate of 0.4 ml/min and then increased to 100% over 0.3 min at a flow rate of 0.5 ml/min and maintained for 0.2 min. The column was reequilibrated for 2 min (the total run time was 6 min). ERG was analyzed in the ESI-positive mode, and the multiple-reaction-monitoring (MRM) transition m/z 230.1 > 127.1 (cone voltage = 18 V; collision energy = 18 eV) was monitored. The source capillary was at 3.5 kV. The source and desolvation temperatures were 140°C and 400°C, respectively. The desolvation and cone gas flows were 600 and 50 liters/h, respectively. A standard curve was generated for ERG quantification. For quantification of extracellular ERG, the filtered supernatant of the mycobacterial culture was lyophilized and redissolved in a solution containing 25% acetonitrile and 0.05% formic acid and analyzed by UPLC-ESI-MS/MS, as described above. For each time point, the CFU/ml was determined by plating appropriate dilutions of the culture onto solid media. Intracellular and extracellular ERG was expressed as pg/105 CFU for each time point, and the results are representative of at least 3 experiments, expressed as means ± standard deviations of the means. For MSH quantification, samples were treated with 0.02 M dithiothreitol (DTT) prior to analysis. Since no MSH standard is commercially available, we used liquid chromatography–electrospray ionization–high-resolution mass spectrometry (LC-ESI-HRMS) and the known exact m/z value for MSH ([M − H]−) of 485.1441 to detect MSH in the cell lysate of wild-type M. smegmatis, as previously described (29).
Mycobacterial membrane integrity assay.
Mycobacterial cell membrane integrity was determined by flow cytometry using the Live/Dead Baclight bacterial viability and counting kit (Molecular Probes), as previously described (30). Cells harvested at different growth stages were washed with 150 mM NaCl and stained with both SYTO-9 and propidium iodide (PI) to determine the percentage of the population in which the cell membrane was disrupted. Heat-treated mycobacteria (2 h at 80°C) were included as a control for the dead cell population (31, 32).
Oxidative stress and drug susceptibility.
The various M. smegmatis strains were cultured to an OD600 of 1 and then exposed to either 0.6 mM CuOOH or 2 mM TBHP for 2 h. The survival after 2 h of exposure was evaluated by determining the CFU on solid media, and the results are representative of at least 3 experiments, expressed as means ± standard errors of the means. Drug susceptibility was evaluated by the broth microdilution method, as previously described (33). The results are representative of at least 3 repeatable experiments.
SDS-PAGE and mass spectrometry.
Mycobacteria were grown on 7H11 agar plates for 9 days and scraped off the plates with Middlebrook 7H9 liquid medium. The suspension was centrifuged, and the pellet was resuspended in cold (4°C) lysis buffer (10 mM Tris-HCl, 0.1% Tween 80, and the complete protease inhibitor cocktail). Cells were homogenized in tubes containing 0.1-mm beads using a FastPrep-24 (Biospec Products Inc., Bartlesville, United Kingdom) 6 times for 20 s with 1-min intermittent cooling steps on ice (34). Total protein was quantified by using the RCDC protein assay kit (Bio-Rad Laboratories, Hercules, CA), according to manufacturer's instructions. Five micrograms of total protein from each strain was treated with 2-mercaptoethanol and SDS and separated on an SDS-PAGE gel containing a 4% stacking gel (0.124 M Tris-HCl, 0.1% SDS, 0.05% ammonium persulfate, 0.09% TEMED, 4% acrylamide, 0.1% bisacrylamide) and a 15% separating gel (0.375 M Tris-HCl, 0.1% SDS, 15% acrylamide, 0.375% bisacrylamide, 0.05% ammonium persulfate, 0.581 M TEMED). The running buffer (pH ∼8.3) contained 25 mM Tris, 192 mM glycine, and 0.1% SDS. Bands were visualized with the aid of CBB dye (2.5% CBB, 45% methanol, 10% acetic acid), and the identity of MSMEG_0447 (Ohr) was confirmed by mass spectrometry using the Thermo Scientific Easy-nLC II system connected to a LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, Bremen, Germany).
RESULTS AND DISCUSSION
M. smegmatis secretes ergothioneine into the culture medium.
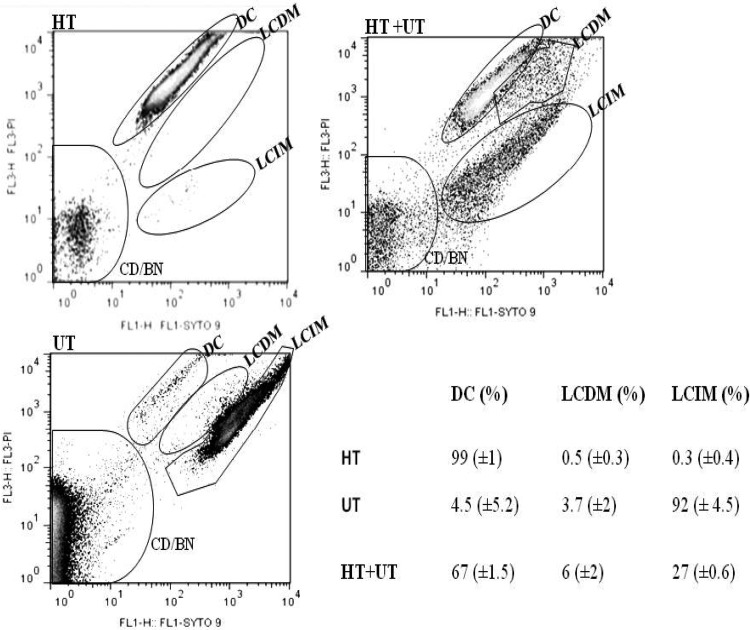
The investigation of extracellular ERG was prompted by a previous observation that ERG was detected in the buffer fluid surrounding resting-cell preparations of M. smegmatis (5). ERG was detected both in the culture media and in the cell lysate of M. smegmatis at the three time points investigated (Table 1). Furthermore, the percentage of extracellular ERG was significantly higher than the percentage of intracellular ERG at all three time points. In order to confirm that the ERG detected in the media was not due to leakage from cells with disrupted cell membranes, the membrane integrity of the cells within the population was determined by flow cytometry (Fig. 1). Dual nucleic acid staining was performed by using the membrane-permeable dye SYTO-9 and the nonpermeable propidium iodide (PI), which is able to enter cells and displace SYTO-9 only when the membrane is damaged (35). Cells were gated into three populations corresponding to their membrane integrity on the basis of PI and SYTO-9 intensities. Cells with the highest PI fluorescence and the lowest SYTO-9 fluorescence represent dead cells (DC) in the population, and heat treatment of M. smegmatis resulted in 99% of the cells gating into this subpopulation. The subpopulation with the lowest PI fluorescence and the highest SYTO-9 fluorescence represent living cells with intact membranes (LCIM), and this population was absent following heat treatment of M. smegmatis. A third, intermediate population was observed and is thought to represent cells in the population which were live cells with damaged membranes (LCDM), as described previously (30) (Fig. 1). Analysis of M. smegmatis cells grown to OD600 values of ∼0.5, 1, and >2 revealed that approximately 92%, 87%, and 86% of the culture population was made of LCIM, respectively, while the minority was made of the population of LCDM and DC (Table 2). To further confirm the membrane integrity of the cells within the population, the culture medium was assayed for MSH, which is not normally present extracellularly (36, 37). MSH was analyzed by LC-ESI-HRMS in the cell lysate of wild-type M. smegmatis, as previously described (see Fig. S2A and S2B in the supplemental material) (29). The identity of the ion was verified by its absence in the ΔmshA mutant, which is MSH deficient (see Fig. S2D in the supplemental material). No MSH was detected in the culture media from wild-type M. smegmatis (see Fig. S2C in the supplemental material), supporting the flow cytometry data. Therefore, while a small percentage of ERG present in the media may originate from cells with disrupted membranes, the observation that the largest proportion of ERG (∼81% at OD600 values of 0.5 and 1) is found in the media while the majority of cells display an intact membrane (∼90% at OD600 values of 0.5 and 1) implies that ERG is secreted by M. smegmatis. Preliminary data for ERG quantification in M. tuberculosis (H37Rv) suggest a similar trend in slow-growing mycobacteria (see Table S3 in the supplemental material) but require further validation.
Table 1.
Ergothioneine amounts in mc2155 and the ΔmshA strain
| Strain | Mean amt of ERG (pg/105 CFU) ± SDa |
|||||
|---|---|---|---|---|---|---|
| OD600 ∼ 0.5 |
OD600 ∼ 1 |
OD600 > 2 |
||||
| IN | EX | IN | EX | IN | EX | |
| mc2155 | 4.07 ± 0.66 | 17.03 ± 3.5 | 1.74 ± 0.30 | 7.6 ± 1.45 | 0.83 ± 0.80 | 12.66 ± 2.65 |
| ΔmshA | 7.50 ± 3.5 | 44.30 ± 4.0 | 6.3 ± 2.61 | 39 ± 10.5 | 1.62 ± 0.48 | 37.61 ± 2.55 |
IN, intracellular; EX, extracellular.
Fig 1.
Characterization of mycobacterial integrity by flow cytometry. The program Flowjo (version 7/9) was used to analyze the population distribution of mycobacteria, which is represented by a pseudocolor plot. Using automatic and manual gating options, we could count the number of events per population and therefore estimate the representative percentage of each population. When the untreated (UT) sample was stained with SYTO-9 and propidium iodide (PI), the analyses indicated that 92% of the population was LCIM, while 3.7% was LCDM and 4.5% was DC. In the heat-treated (HT) sample, 99% of the population was indicated to be dead, while 0.5% was LCDM and 0.2% was LCIM. When equal amounts of the heat-treated and untreated cells were mixed, 67% of the population was indicated to be dead, while 27% was LCIM and 3.7% was LCDM. This shows that this technique was sensitive enough to detect cells with damaged membranes even in a population mixture, implying consistency of the results showing that in an actively growing wild-type M. smegmatis culture at an OD600 of ∼0.5, the membrane integrity of ∼92% of the cells is still intact. DC, dead cells; CD/BN, cellular debris and background noise; HT+UT, mixture of an equal ratio of the untreated and heat-treated samples.
Table 2.
Estimation of the proportions of different mycobacterial populations in a culture at different growth stages by flow cytometry
| OD600 | Mean % of mycobacterial population ± SD |
||
|---|---|---|---|
| DC (%) | LCDM (%) | LCIM (%) | |
| ∼0.5 | 4.5 ± 5.2 | 3.7 ± 2 | 92 ± 4.5 |
| ∼1 | 2.6 ± 0.84 | 10.1 ± 2.17 | 87.22 ± 2.06 |
| >2 | 2.5 ± 1.6 | 11.2 ± 2.5 | 86.3 ± 4.02 |
ERG exists as a thione in solution at neutral pH, and as a result, it is relatively resistant to oxidation (38). It is therefore better suited to function extracellularly than MSH, since it does not require an enzyme to maintain it in the reduced form (39). While MSH is not detected extracellularly, a previous study utilizing MSH-deficient mutants demonstrated that M. smegmatis is able to actively transport MSH into cells (36), and MSMEG_1642 was proposed as a possible MSH transporter. Since ERG is unable to diffuse across membranes (18, 40), an ERG transporter would also be required in M. smegmatis. In humans, the OCTN1 transporter is required for ERG transport into cells and involves cotransport of Na+ (18). This leads us to speculate that the ERG transporter in mycobacteria may be a member of the monovalent cation:proton antiporter family or the H+/Na+-translocating F-type, V-type, and A-type ATPase superfamily, which both utilize Na+ as a substrate (41). Since ERG has anti-inflammatory properties (15), the secretion of this molecule by M. tuberculosis may play a role in modulating the immune response by the host during infection and therefore warrants further investigation.
A previous study found that ERG levels were elevated in MSH-deficient mutants of M. smegmatis, suggesting that ERG potentially compensates for the loss of MSH in this organism (20). Our analysis revealed a similar trend, with a 2- to 5-fold increase in intracellular and extracellular ERG levels observed for the M. smegmatis ΔmshA mutant (Table 1), which appears to be lower than what was previously reported, but this may be due to the different methods used for thiol quantification. The previous study utilized a method based on high-performance liquid chromatography (HPLC) separation and fluorescent detection of thiols conjugated to monobromobimane (mBBr) (42, 43), whereas we utilized UPLC-ESI-MS/MS to quantify ERG.
egtD is dispensable for in vitro growth but is required for ergothioneine biosynthesis in M. smegmatis.
The recent annotation of the ERG biosynthetic genes in mycobacteria (11) allowed us to investigate the role of ERG in mycobacterial physiology by generating an ERG-deficient mutant of M. smegmatis using allelic-exchange mutagenesis. An unmarked deletion of egtD, the first gene in ERG biosynthesis, was generated in mc2155 and the ΔmshA mutant and confirmed by Southern blotting (see Fig. S1 in the supplemental material). Loss of ERG biosynthesis in the ΔegtD single mutant and the ΔmshA ΔegtD double mutant and restoration of ERG biosynthesis in the complemented strains were confirmed by UPLC-ESI-MS/MS (Table 3). No growth defect was associated with deletion of egtD in the wild-type strain (data not shown), and the extended lag phase (9) and sensitivity to the inoculum size (20) observed previously for the ΔmshA mutant were not exacerbated by the loss of ERG in this mutant. Since the ERG level in the ΔmshA mutant is elevated relative to the level in the wild-type strain, we compared the levels of MSH in the ERG-deficient single mutant and the wild type via LC-ESI-HRMS. Relative comparison of the MSH peak areas revealed no significant difference in the MSH levels between the two strains (see Table S4 and Fig. S3 in the supplemental material). Therefore, while the ERG level was elevated in the MSH-deficient mutant, the MSH level was unchanged in the ERG-deficient mutant. This implies that the wild-type MSH level may be adequate to compensate for the loss of intracellular ERG or that compensation occurs by another mechanism.
Table 3.
Amount of ergothioneine detected in different fractions in various M. smegmatis strains at an OD600 of 1
| Fraction | ERG concn (pg/105 CFU) ± SD |
|||||
|---|---|---|---|---|---|---|
| mc2155 | ΔmshA | ΔegtD | ΔmshA ΔegtD | ΔegtDca | ΔmshA ΔegtDca | |
| Intracellular | 1.74 ± 0.3 | 6.3 ± 2.6 | 0 | 0 | 0.88 ± 0.3 | 2.2 ± 0.5 |
| Extracellular | 7.6 ± 1.5 | 39 ± 10.5 | 0 | 0 | 4.3 ± 2 | 16 ± 5.1 |
ΔegtDc, complemented egtD mutant (ΔegtD attB::pMV306D; see Table S1 in the supplemental material); ΔmshA ΔegtDc, complemented ΔmshA ΔegtD mutant (ΔmshA ΔegtD::pMV306D; see Table S1 in the supplemental material).
Ergothioneine deficiency does not alter M. smegmatis susceptibility to tested antibiotics.
The altered susceptibility of MSH-deficient mycobacterial mutants to antibiotics prompted us to explore the susceptibility of ERG-deficient strains to antibiotics. The susceptibilities of the ΔegtD mutant and the ΔmshA ΔegtD mutant to isoniazid, ethambutol, and kanamycin did not differ significantly from those of their respective parental strains (Table 4), demonstrating that ERG does not play a role in protecting M. smegmatis against these compounds.
Table 4.
MICs for various M. smegmatis strains
| Strain | MIC (μg/ml)a |
||||
|---|---|---|---|---|---|
| Kanamycin | Isoniazid | Ethionamide | Rifampin | Ethambutol | |
| mc2155 | 0.5–1 | 5–20 | 12.5–25 | 0.3–0.6 | 0.4–0.75 |
| ΔmshA | ND | 312–625 | 375–400 | 0.3–0.6 | 0.4–0.75 |
| ΔegtD | 0.5–1 | 5–20 | 25–50 | 0.3–0.6 | 0.4–0.75 |
| Complemented ΔegtD | ND | ND | 12.5–25 | ND | ND |
| ΔmshA ΔegtD | ND | 312–625 | 400–425 | 0.3–0.6 | 0.4–0.75 |
ND, not determined.
Ergothioneine protects M. smegmatis against peroxide.
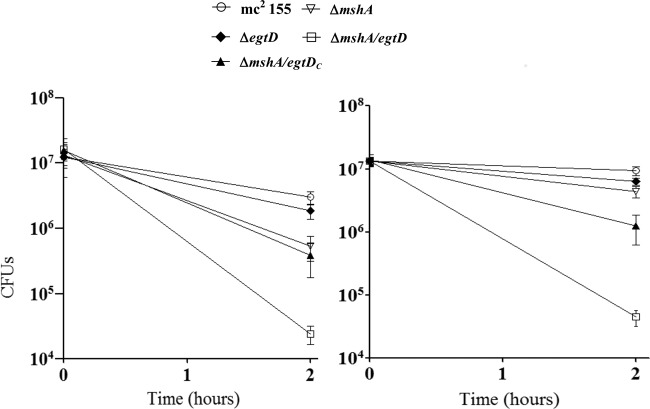
A marginal increase in susceptibility to TBHP and CuOOH was observed for both the ΔmshA and the ΔegtD single mutants, while a marked increase in susceptibility was observed for the ΔmshA ΔegtD double mutant (Fig. 2). This demonstrates that both ERG and MSH are involved in protecting M. smegmatis against peroxide and that the loss of either low-molecular-weight thiol is potentially compensated for by the presence of the other. This compensation appears, at least in the MSH-deficient mutant, to involve increasing the amount of the remaining thiol molecule (20). A previous study demonstrated that MSMEG_0447, annotated Ohr (organic hydroperoxide resistance protein), is highly induced in the ΔmshA mutant and that overexpression of Ohr in M. smegmatis increases resistance to CuOOH (20). We therefore investigated the Ohr expression level in all strains using SDS-PAGE (see Fig. S4 in the supplemental material). The Ohr expression level was elevated in the ΔmshA and ΔmshA ΔegtD mutants relative to levels in the wild-type strain and the ΔegtD mutant. This indicates that the Ohr expression level is not influenced by the loss of ERG. In addition, the increased sensitivity of the ERG- and MSH-deficient double mutant to peroxides, despite the elevated Ohr level, suggests that ERG also plays a role in protecting M. smegmatis against peroxide. Despite the increased sensitivity of the MSH-deficient M. tuberculosis ΔmshA mutant to oxidative stress, it survives in the mouse model of infection (10). Considering the fact that M. smegmatis Ohr has no homolog in M. tuberculosis (20), we speculate that ERG may compensate for the loss of MSH in this organism, and the lack of a phenotype of the MSH-deficient M. tuberculosis mutant in the mouse model is potentially due to compensation by ERG; however, this remains to be demonstrated.
Fig 2.
Survival responses of M. smegmatis strains to oxidative stress generated by TBHP (left) and CuOOH (right). Mycobacteria were grown to the exponential phase and exposed to oxidative stress generated by either 2 mM TBHP or 0.6 mM CuOOH. The data were analyzed by using GraphPad Prism version 5.01 and are represented as means with standard errors of the means. Two-way analysis of variance followed by a Bonferroni test was performed. The double mutant (ΔmshA ΔegtD) was more sensitive to oxidative stress than any other strain used in this study; it was significantly (P < 0.01) sensitive to oxidative stress generated by CuOOH (right) relative to the wild type and the ΔegtD mutant.
Conclusion.
A significant portion of the ERG synthesized by M. smegmatis is secreted, suggesting that it has a unique extracellular function, unlike MSH, which is found only inside the cell. Both ERG and MSH function to protect M. smegmatis from the toxic effect of peroxides, and the loss of either thiol is partly compensated for by the presence of the other.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yossef Av-Gay for kindly donating the M. smegmatis ΔmshA mutant, John Michie for his technical support with flow cytometry, Suereta Fortuin and Dominique Anderson for their technical support with SDS-PAGE, Ruzayda van Aarde and Victoria Pichler for their technical support with Southern blotting, and Salome Smit (Proteomics Laboratory, LCMS Unit, Central Analytical Facility, Stellenbosch University) for performing the proteomic analyses.
Footnotes
Published ahead of print 29 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02572-12.
REFERENCES
- 1. Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, Cohen G, Davies J, Fahey RC, Davis C. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spies HS, Steenkamp DJ. 1994. Thiols of intracellular pathogens. Identification of ovothiol A in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Eur. J. Biochem. 224:203–213 [DOI] [PubMed] [Google Scholar]
- 3. Newton GL, Bewley CA, Dwyer TJ, Horn R, Aharonowitz Y, Cohen G, Davies J, Faulkner DJ, Fahey RC. 1995. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur. J. Biochem. 230:821–825 [DOI] [PubMed] [Google Scholar]
- 4. Genghof DS, Van Damme O. 1964. Biosynthesis of ergothioneine and hercynine by mycobacteria. J. Bacteriol. 87:852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Genghof DS, Van Damme O. 1968. Biosynthesis of ergothioneine from endogenous hercynine in Mycobacterium smegmatis. J. Bacteriol. 95:340–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rawat M, Johnson C, Cadiz V, Av-Gay Y. 2007. Comparative analysis of mutants in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 363:71–76 [DOI] [PubMed] [Google Scholar]
- 7. Newton GL, Buchmeier N, Fahey RC. 2008. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 72:471–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newton GL, Unson MD, Anderberg SJ, Aguilera JA, Oh NN, delCardayre SB, Av-Gay Y, Fahey RC. 1999. Characterization of Mycobacterium smegmatis mutants defective in 1-d-myo-inosityl-2-amino-2-deoxy-alpha-d-glucopyranoside and mycothiol biosynthesis. Biochem. Biophys. Res. Commun. 255:239–244 [DOI] [PubMed] [Google Scholar]
- 9. Rawat M, Newton GL, Ko M, Martinez GJ, Fahey RC, Av-Gay Y. 2002. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46:3348–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vilchèze C, Av-Gay Y, Attarian R, Liu Z, Hazbón MH, Colangeli R, Chen B, Liu W, Alland D, Sacchettini JC, Jacobs WRJ. 2008. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis. Mol. Microbiol. 69:1316–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seebeck FP. 2010. In vitro reconstitution of Mycobacterial ergothioneine biosynthesis. J. Am. Chem. Soc. 132:6632–6633 [DOI] [PubMed] [Google Scholar]
- 12. Cheah IK, Halliwell B. 2012. Ergothioneine; antioxidant potential, physiological function and role in disease. Biochim. Biophys. Acta 1822:784–793 [DOI] [PubMed] [Google Scholar]
- 13. Hartman Z, Hartman PE. 1987. Interception of some direct-acting mutagens by ergothioneine. Environ. Mol. Mutagen. 10:3–15 [DOI] [PubMed] [Google Scholar]
- 14. Bello MH, Barrera-Perez V, Morin D, Epstein L. 2012. The Neurospora crassa mutant NcΔEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 49:160–172 [DOI] [PubMed] [Google Scholar]
- 15. Rahman I, Gilmour PS, Jimenez LA, Biswas SK, Antonicelli F, Aruoma OI. 2003. Ergothioneine inhibits oxidative stress- and TNF-alpha-induced NF-kappa B activation and interleukin-8 release in alveolar epithelial cells. Biochem. Biophys. Res. Commun. 302:860–864 [DOI] [PubMed] [Google Scholar]
- 16. Zhu B, Mao L, Fan R, Zhu J, Zhang Y, Wang J, Kalyanaraman B, Frei B. 2011. Ergothioneine prevents copper-induced oxidative damage to DNA and protein by forming a redox-inactive ergothioneine-copper complex. Chem. Res. Toxicol. 24:30–34 [DOI] [PubMed] [Google Scholar]
- 17. Hanlon DP. 1971. Interaction of ergothioneine with metal ions and metalloenzymes. J. Med. Chem. 14:1084–1087 [DOI] [PubMed] [Google Scholar]
- 18. Gründemann D, Harlfinger S, Golz S, Geerts A, Lazar A, Berkels R, Jung N, Rubbert A, Schömig E. 2005. Discovery of the ergothioneine transporter. Proc. Natl. Acad. Sci. U. S. A. 102:5256–5261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gründemann D. 2012. The ergothioneine transporter controls and indicates ergothioneine activity—a review. Prev. Med. 54(Suppl):S71–S74. 10.1016/j.ypmed.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 20. Ta P, Buchmeier N, Newton GL, Rawat M, Fahey RC. 2011. Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J. Bacteriol. 193:1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84 [DOI] [PubMed] [Google Scholar]
- 22. Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(Part 8):1969–1975 [DOI] [PubMed] [Google Scholar]
- 24. Newton GL, Koledin T, Gorovitz B, Rawat M, Fahey RC, Av-Gay Y. 2003. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA). J. Bacteriol. 185:3476–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 26. Ey J, Schömig E, Taubert D. 2007. Dietary sources and antioxidant effects of ergothioneine. J. Agric. Food Chem. 55:6466–6474 [DOI] [PubMed] [Google Scholar]
- 27. Steenkamp DJ, Vogt RN. 2004. Preparation and utilization of a reagent for the isolation and purification of low-molecular-mass thiols. Anal. Biochem. 325:21–27 [DOI] [PubMed] [Google Scholar]
- 28. Markova NG, Karaman-Jurukovska N, Dong KK, Damaghi N, Smiles KA, Yarosh DB. 2009. Skin cells and tissue are capable of using L-ergothioneine as an integral component of their antioxidant defense system. Free Radic. Biol. Med. 46:1168–1176 [DOI] [PubMed] [Google Scholar]
- 29. Holsclaw CM, Muse WB, III, Carroll KS, Leary JA. 2011. Mass spectrometric analysis of mycothiol levels in wild-type and mycothiol disulfide reductase mutant Mycobacterium smegmatis. Int. J. Mass Spectrom. 305:151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lehtinen J, Nuutila J, Lilius E. 2004. Green fluorescent protein-propidium iodide (GFP-PI) based assay for flow cytometric measurement of bacterial viability. Cytometry A 60:165–172 [DOI] [PubMed] [Google Scholar]
- 31. Tell LA, Foley J, Needham ML, Walker RL. 2003. Comparison of four rapid DNA extraction techniques for conventional polymerase chain reaction testing of three Mycobacterium spp. that affect birds. Avian Dis. 47:1486–1490 [DOI] [PubMed] [Google Scholar]
- 32. Kremer L, Guérardel Y, Gurcha SS, Locht C, Besra GS. 2002. Temperature-induced changes in the cell-wall components of Mycobacterium thermoresistibile. Microbiology. 148:3145–3154 [DOI] [PubMed] [Google Scholar]
- 33. Wallace RJJ, Nash DR, Steele LC, Steingrube V. 1986. Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J. Clin. Microbiol. 24:976–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. U. S. A. 108:680–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stocks SM. 2004. Mechanism and use of the commercially available viability stain, BacLight. Cytometry A 61:189–195 [DOI] [PubMed] [Google Scholar]
- 36. Bzymek KP, Newton GL, Ta P, Fahey RC. 2007. Mycothiol import by Mycobacterium smegmatis and function as a resource for metabolic precursors and energy production. J. Bacteriol. 189:6796–6805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newton GL, Av-Gay Y, Fahey RC. 2000. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry. 39:10739–10746 [DOI] [PubMed] [Google Scholar]
- 38. Heath H, Toennies G. 1958. The preparation and properties of ergothioneine disulphide. Biochem. J. 68:204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hartman PE. 1990. Ergothioneine as antioxidant. Methods Enzymol. 186:310–318 [DOI] [PubMed] [Google Scholar]
- 40. Paul BD, Snyder SH. 2010. The unusual amino acid L-ergothioneine is a physiologic cytoprotectant. Cell Death Differ. 17:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wolfe LM, Mahaffey SB, Kruh NA, Dobos KM. 2010. Proteomic definition of the cell wall of Mycobacterium tuberculosis. J. Proteome Res. 9:5816–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fahey RC, Newton GL. 1987. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 143:85–96 [DOI] [PubMed] [Google Scholar]
- 43. Newton GL, Dorian R, Fahey RC. 1981. Analysis of biological thiols: derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography. Anal. Biochem. 114:383–387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.