Abstract
The recent identification of a high-level-ceftriaxone-resistant (MIC = 2 to 4 μg/ml) isolate of Neisseria gonorrhoeae from Japan (H041) portends the loss of ceftriaxone as an effective treatment for gonococcal infections. This is of grave concern because ceftriaxone is the last remaining option for first-line empirical antimicrobial monotherapy. The penA gene from H041 (penA41) is a mosaic penA allele similar to mosaic alleles conferring intermediate-level cephalosporin resistance (Cephi) worldwide but has 13 additional mutations compared to the mosaic penA gene from the previously studied Cephi strain 35/02 (penA35). When transformed into the wild-type strain FA19, the penA41 allele confers 300- and 570-fold increases in the MICs for ceftriaxone and cefixime, respectively. In order to understand the mechanisms involved in high-level ceftriaxone resistance and to improve surveillance and epidemiology during the potential emergence of ceftriaxone resistance, we sought to identify the minimum number of amino acid alterations above those in penA35 that confer high-level resistance to ceftriaxone. Using restriction fragment exchange and site-directed mutagenesis, we identified three mutations, A311V, T316P, and T483S, that, when incorporated into the mosaic penA35 allele, confer essentially all of the increased resistance of penA41. A311V and T316P are close to the active-site nucleophile Ser310 that forms the acyl-enzyme complex, while Thr483 is predicted to interact with the carboxylate of the β-lactam antibiotic. These three mutations have thus far been described only for penA41, but dissemination of these mutations in other mosaic alleles would spell the end of ceftriaxone as an effective treatment for gonococcal infections.
INTRODUCTION
Neisseria gonorrhoeae is the etiologic agent of the sexually transmitted infection gonorrhea. The World Health Organization estimates that there were 106 million gonococcal infections worldwide in 2008, a 21% increase from the rate in 2005 (1). Gonococcal infections are often asymptomatic, which contributes to the continued transmission of the infection, and if left untreated, gonorrhea can progress to pelvic inflammatory disease, ectopic pregnancy, and infertility. In the absence of a vaccine, effective prevention, diagnosis, and particularly antibiotics are the mainstays for treatment and control of gonococcal infections.
N. gonorrhoeae has shown a remarkable ability to become resistant to nearly every antibiotic used to treat infections (2, 3). Penicillin was introduced in 1943 and was effective for nearly 40 years, but during this time, the MICs of the β-lactam antimicrobials gradually increased (MIC “creep”) until the emergence of chromosomally mediated resistant strains in the mid-1980s, as well as of β-lactamase-producing strains worldwide (4), necessitated its removal as a recommended antibiotic. During this time, resistance to spectinomycin, tetracycline, and erythromycin rendered them unsuitable for treatment of gonococcal infections as well. Fluoroquinolones were introduced in the United States in 1989 as antigonococcal antibiotics, but by 2007, resistance became so widespread that the Centers for Disease Control and Prevention (CDC) removed these antibiotics from the recommended list (5), leaving only the expanded-spectrum cephalosporins ceftriaxone and cefixime. Worryingly, over the most recent decade, the number of strains with increased resistance to ceftriaxone and cefixime has steadily increased (6), and in vitro resistance and treatment failures with cefixime and some with ceftriaxone have been verified in Japan, Europe, and Canada (3, 7). This has prompted the recent revision of CDC (8) and European (9) treatment guidelines so that only ceftriaxone together with azithromycin is now recommended for treatment of uncomplicated gonorrhea. Nevertheless, the threat of widespread resistance to ceftriaxone and possibly untreatable gonorrhea is real, especially in settings where ceftriaxone monotherapy is common and dual antimicrobial therapy is neither feasible nor affordable, and there are few potentially useful antibiotic options in the pipeline (3).
Chromosomally mediated resistance to penicillin is conferred by multiple resistance determinants that are mutated versions of endogenous genes. These determinants are penA, which encodes penicillin-binding protein 2 (PBP2), an essential transpeptidase (TPase) involved in cell division; mtrR, which increases the expression of the MtrC-MtrD-MtrE efflux pump (10, 11); penB, encoding mutations in the constriction loop of PorB1b, the major outer membrane porin (12–14); ponA, encoding a mutated variant of the other essential PBP in the gonococcus, PBP1 (15); and a nontransformable resistance determinant involved in high-level resistance whose genetic identity is unknown (15). Each of these determinants increases resistance incrementally (2- to 6-fold), but together, the determinants increase resistance to penicillin by 400-fold (15, 16).
The resistance determinants harbored by strains with intermediate resistance to cephalosporin (Cephi) are very similar, with the major difference residing in the penA gene. Whereas most penicillin-resistant strains have a penA gene with 4 to 8 mutations relative to a wild-type penA gene (17), Cephi strains harbor mosaic penA genes that contain upwards of 60 to 70 mutations, which have arisen through DNA recombination with multiple Neisseria species penA genes (18, 19). The initial Cephi strains emerged in Japan and then spread to other parts of the world (3, 18–22).
Very recently, a strain (referred to as H041) isolated in Japan from a female sex worker was found to have very high MICs of both ceftriaxone (MIC = 2 to 4 μg/ml) and cefixime (MIC = 8 μg/ml) and was resistant to most other antibiotics (23, 24). This strain contains a novel mosaic penA allele (referred to as penA41) with 61 amino acid differences compared to a wild-type allele, with several of these mutations being unique among the ∼40 mosaic penA alleles reported thus far. The penA41 allele confers resistance to both ceftriaxone and cefixime well above the current breakpoints (0.25 μg/ml for both), even when transferred to a wild-type strain, with MICs 5- to 10-fold higher in strains with additional resistance determinants (i.e., mtrR and penB) (24).
Knowing the identity of the amino acid alterations that directly result in the high-level cephalosporin resistance conferred by the mosaic penA41 allele is critical for both understanding the mechanisms involved in PBP2 remodeling and monitoring the resistance potential of new strains that appear certain to emerge in the near future. In this study, we set out to identify the mutations in penA41 that are responsible for the marked increase in resistance to the expanded-spectrum cephalosporins relative to a standard mosaic penA allele. We identified three novel mutations, A311V, V316P, and T483S, that, when incorporated into the mosaic penA35 allele, are responsible for essentially all of the additional resistance conferred by penA41. Two of these mutations, A311V and T316P, are located near the active-site nucleophile Ser310, in a region previously shown to harbor mutations that increase resistance (25), whereas the remaining mutation, T483S, is in a different location in the structure of PBP2, where it may interact with the β-lactam carboxylate.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
FA19 is a wild-type penicillin- and cephalosporin-susceptible strain that we have used extensively as a genetic recipient for antimicrobial resistance determinants (26). FA6140 is a penicillin-resistant, cephalosporin-susceptible strain isolated in Durham, NC, in 1986 that contains all of the known penicillin resistance determinants found in clinical isolates (27). Strains were grown on GC broth (GCB) agar plates containing supplements I and II (28) in a 4% CO2–96% air atmosphere at 37°C.
Chimeric penA construction and site-directed mutagenesis.
To identify the regions containing amino acid mutations conferring resistance, we utilized a plasmid (pUC18us-penA35-REs) containing the penA35 gene (mosaic penA allele XXIX) (24, 25) and 300 bp of downstream sequence into which silent restriction sites were incorporated within the coding sequence, splitting the gene into 6 modules (mod0 to mod5) (25). The corresponding modules from penA41 (mosaic penA allele C [24, 29]) were created by amplifying the modules from penA41 with primers harboring the appropriate silent restriction sites at their 5′ ends and replacing the corresponding modules in pUC18us-penA35-REs. After verification of the incorporated sequence, the plasmid was used to transform FA19, as described below.
Individual mutations were incorporated into penA35 by using overlap-extension PCR (30) with pUC18us-penA35-REs as a template. The outside primers spanned the silent restriction sites of the module being mutagenized, and after the second PCR amplification, the mutagenized module was digested with the appropriate restriction enzymes and replaced by the corresponding fragment from pUC18us-penA35-REs. For the A311V and T316P mutations, which were very close to the BamHI restriction site at codon 309, the mutations were introduced into the BamHI-containing primer, and the amplified fragment was used to replace the corresponding fragment in the plasmid. The resulting plasmids were verified by DNA sequencing and used in transformation experiments, as described below.
Transformation experiments.
DNA transformations in N. gonorrhoeae were performed essentially as described previously (16). Briefly, 900 μl of piliated FA19 at an optical density at 600 nm (OD600) of 0.18 in GCB with supplements I and II (28), 10 mM MgCl2, and 20 mM bicarbonate was added to 2 μg of plasmid in 100 μl of 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and the cells and DNA were incubated in a 37°C incubator in a 4% CO2–96% air atmosphere for 5 h. At the end of the incubation, 300 μl of the mixture was removed, and the cells were pelleted, resuspended in 50 μl GCB, and plated onto GCB plates containing 0.05 to 1.0 μg/ml cefixime. After overnight incubation, multiple clones from each transformation were passaged, and frozen stocks were made.
To test for correct recombination, several colonies from each passaged clone were resuspended in H2O and boiled for 10 min, and cell debris was pelleted by centrifugation. The supernatants (2 μl) were subsequently used to amplify the penA gene with Taq polymerase, and PCR products were purified and sequenced with sense primers that covered the coding sequence from codon 48 to the end of the gene.
MIC measurements.
Two to four verified clones from each individual transformation were passaged on GCB plates and resuspended at an OD600 of 0.18. GCB plates containing <2-fold changes in concentrations of ceftriaxone and cefixime were poured on the day of the experiment, and agar dilution MICs were determined as previously described (16, 25). Aliquots (5 μl; ∼50,000 CFU) of each clone were spotted onto the antibiotic-containing plates, and the plates were incubated overnight. The next morning, growth was scored, in which growth was defined as >5 colonies growing in the spot. The MIC determinations were repeated a minimum of three times, and the values for each mutant were averaged.
Kinetic analysis of PBP2 acylation by β-lactam antibiotics.
PBP2 mutant proteins were purified and used to determine the k2/Ks values for their acylation rates with penicillin, ceftriaxone, and cefixime, as described previously (25, 31). The reaction of β-lactam antibiotics with PBP2 is denoted by the equation , where E · S is the noncovalent enzyme-antibiotic complex, E-S′ is the acyl-enzyme complex, and P is the hydrolyzed antibiotic. k2/Ks constants, which are a direct measure of the ability of an antibiotic to inhibit a PBP (32), were calculated from first-order rates of acylation of purified, soluble PBP2 variants by [14C]penicillin G (Moravek, Brea, CA), as previously described (25, 31, 33). Graphs of PBP2-[14C]penicillin G complex formation versus time were obtained by incubating 32 μg of protein with 25 to 125 μM [14C]penicillin G, aliquots of ∼5 μg were removed at 15-s intervals, precipitated with 5% trichloroacetic acid, and filtered over Whatman GC-A filters, and the filters were submitted to scintillation counting. The k2/Ks values of nonradioactive cephalosporin antibiotics were derived by determining the concentration of the cephalosporins that inhibited 50% of the binding of a known amount of [14C]penicillin G (32). The k2/Ks values were then determined by using the equation , where [PenG] is the concentration of [14C]penicillin G used in the reaction and [Ceph]0.5 is the concentration of cephalosporin antibiotic that inhibits the binding of [14C]penicillin G by 50%.
RESULTS
The mosaic penA allele from H041.
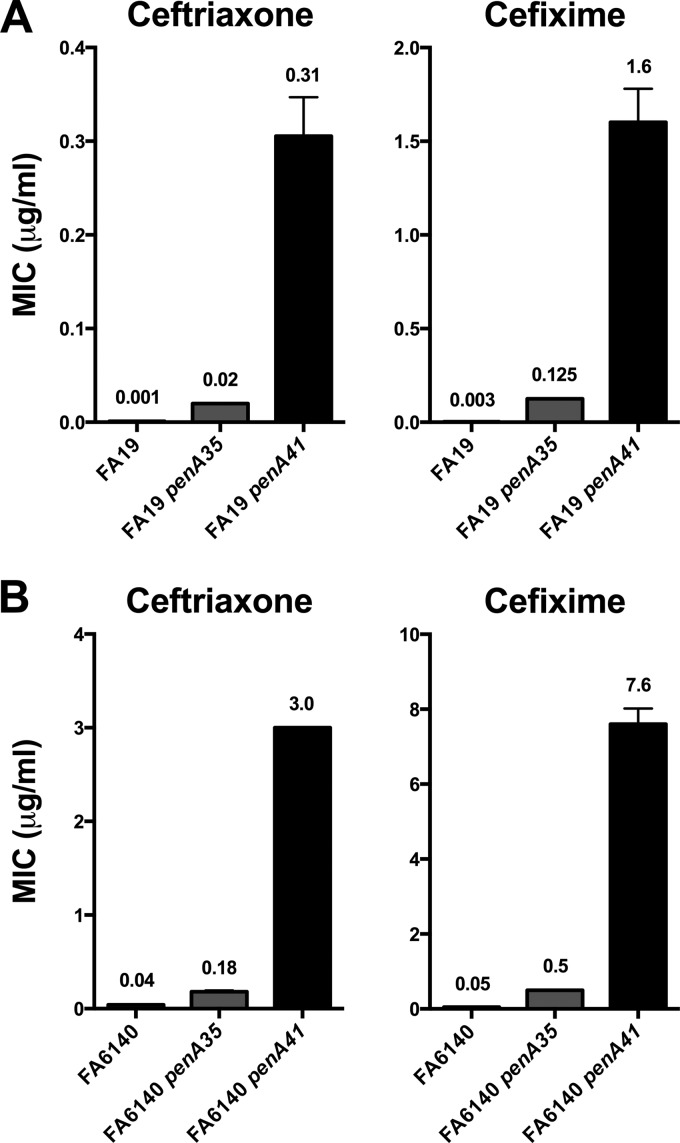
The penA gene from the high-level-cephalosporin-resistant isolate H041 (MICs of 2 to 4 μg/ml and 8 μg/ml for ceftriaxone and cefixime, respectively [24]) is a mosaic allele that encodes a PBP2 variant with 61 amino acid alterations compared to wild-type PBP2 and 13 alterations compared to PBP2 from the intermediate-level-cephalosporin-resistant strain 35/02 (PBP235/02) (Fig. 1A). Twelve of the 13 mutations relative to 35/02 are located in the penicillin-binding domain (amino acids 240 to 581). The penA alleles from 35/02 and H041 were transformed into either a wild-type strain (FA19) or a penicillin-resistant, cephalosporin-susceptible strain containing all of the known penicillin resistance determinants (FA6140), and the MICs of ceftriaxone and cefixime were determined (Fig. 2). Compared to FA19, the MICs of ceftriaxone for the FA19 transformants containing penA35 and penA41 increased 20-fold and 300-fold, respectively, while the MICs of cefixime for the same transformants increased 45-fold and 570-fold, respectively. For FA6140, the MICs of ceftriaxone increased 4.5-fold and 75-fold in the transformants containing penA35 and penA41, respectively, while the MICs of cefixime increased 10-fold and 170-fold, respectively (Fig. 2). These data highlight the dramatic increases in resistance to expanded-spectrum cephalosporins conferred by the penA41 gene.
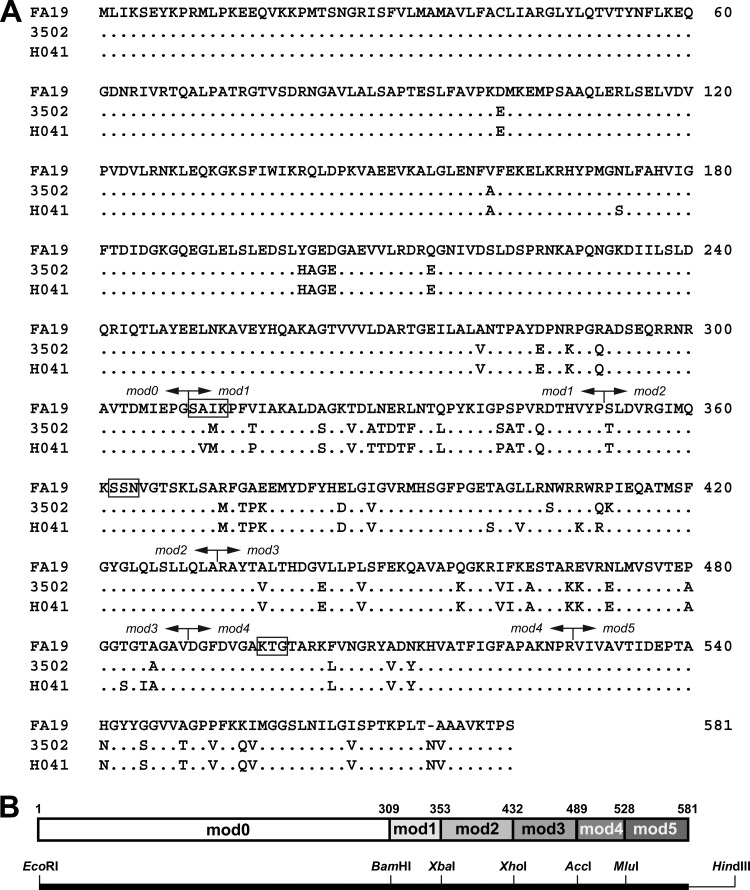
Fig 1.
Alignment of PBP2 from FA19 (wild-type), 35/02 (Cephi), and H041 (cephalosporin resistant). (A) The protein sequences from FA19, 35/02, and H041 were aligned by ClustalX (version 2.0.10 [37]). Dots represent identical amino acids, a dash represents a deletion, the active-site motifs (i.e., SxxK, SxN, and KTG) are indicated by boxes, and the boundaries of the modules (see below) are depicted by arrows. (B) Schematic showing the modules in PBP2 used to identify the regions of PBP2H041 containing amino acids that confer resistance to ceftriaxone and cefixime. The protein sequence (with numbers denoting the amino acid positions of the junctions) is shown at the top, and the corresponding DNA with the silent restriction sites is shown at the bottom.
Fig 2.
Increases in the MICs of ceftriaxone and cefixime in FA19 or FA6140 transformed with the penA alleles from either 35/02 (penA35) or H041 (penA41). (A) MICs of ceftriaxone and cefixime for the indicated penA alleles transformed into FA19; (B) MICs of ceftriaxone and cefixime for the indicated penA alleles transformed into FA6140. Values are the averages of a minimum of 3 separate determinations (usually 3 to 6) and are indicated above the bar.
We then sought to identify which of the amino acids in PBP2H041 that differ from those in PBP235/02 are responsible for conferring the large increases in cephalosporin resistance. In a previous study to define the important amino acids that increase resistance in the mosaic penA35 allele (25), we generated constructs in wild-type penA and penA35 with silent restriction sites to create six modules (called mod0 to mod5) that could be replaced by corresponding modules in the other gene (Fig. 1B). This approach allowed us to identify regions of penA35 that contained the most important amino acids for increasing resistance to ceftriaxone and cefixime, and we used a similar strategy to identify the regions containing crucial amino acids involved in resistance in penA41.
Identification of the key amino acids in penA41 that confer substantially increased resistance to expanded-spectrum cephalosporins.
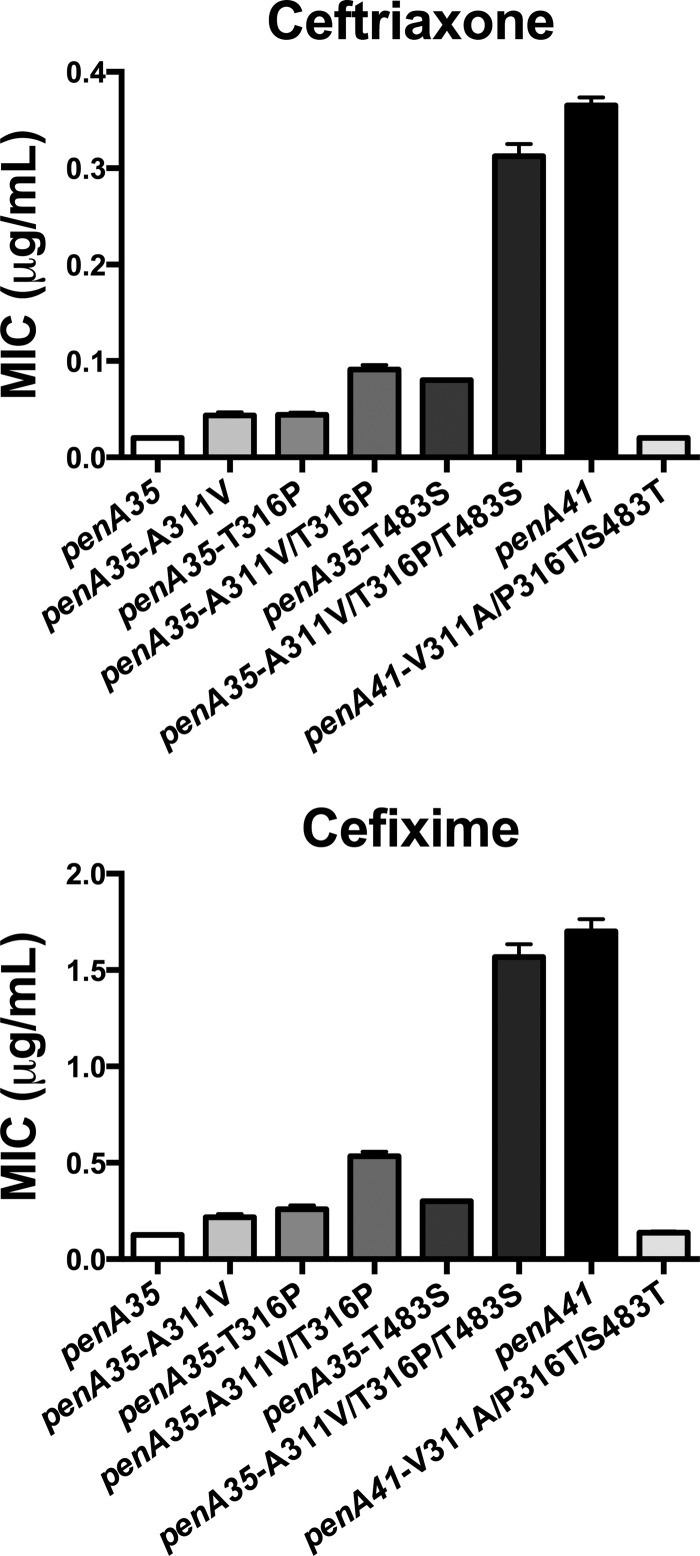
Of the 12 mutations in the penicillin-binding domain of PBP2, 4 are in mod1, 6 are in mod2, and 2 are in mod3 (we have shown previously [25] that mutations in mod0 comprising the first 300 amino acids have no effect on the MIC, and thus, we did not examine the lone mutation in this region). We first focused on mod1 and mod2 by transferring mod1, mod2, or mod1 and mod2 together from H041 into penA35, transforming the resulting constructs into FA19, and selecting for increased cefixime resistance. Transfer of mod1 and of mod1 and mod2 together, but not mod2 alone, increased the MIC of cefixime. Mutations within mod2 were therefore discounted, and we focused on the mutations in mod1. The 4 amino acid mutations residing in mod1 of PBP2H041 that differ from PBP235/02 are A311V, T316P, A328T, and S341P (Fig. 1A). These mutations were introduced individually into penA35, the constructs were transformed into FA19, and resistant colonies were selected with cefixime concentrations just above the MIC for FA19 penA35. Of these, only constructs with an A311V or T316P mutation gave rise to colonies, with each mutation conferring about the same increase in MICs of the expanded-spectrum cephalosporins (Fig. 3). We also constructed a strain containing PBP235/02 with an A311V/T316P double mutation, and the two mutations were additive, with an MIC ∼2-fold higher than that with either individual mutation alone. However, these two mutations together conferred only 30% of the cephalosporin resistance contributed by penA41, indicating that other mutations present in penA41 are also important for increasing resistance (Fig. 3).
Fig 3.
Increases in the MICs of ceftriaxone and cefixime conferred by penA35 containing mutations from penA41. The indicated mutations were incorporated into the penA35 allele, the resulting DNA constructs were used to transform FA19 to increased cefixime resistance, and the penA sequences of individual transformants were confirmed by PCR amplification and sequencing. The MICs of the two expanded-spectrum cephalosporins were determined by agar dilution on GCB plates containing <2-fold increases in the concentrations of the two antibiotics for a minimum of 2 independent transformants (usually 3 to 4).
We next examined the last remaining mutations in penA41, T483S and T485I, that are present in mod3. These mutations, both individually and together, were introduced into both penA35 and penA35 encoding both the A311V and V316P codon mutations (penA35-A311V/V316P), the resulting plasmids were transformed into FA19, and transformants with MICs of cefixime above those containing the parental genes were selected. Colonies were obtained with both the T483S single mutation and the T483S/T485I double mutation but not with the T485I single mutation, and initial experiments indicated that the MICs of the transformants with just the T483S mutation were no different than those with both mutations, so we focused solely on T483S. Transformants carrying penA35-T483S had an ∼2-fold increase in the MIC of ceftriaxone compared to either the penA35-A311V or -V316P mutant alone and about the same MIC as both of these mutations together, whereas the MIC of cefixime was about the same as that for transformants containing the two individual mod1 mutations (Fig. 3). However, when T483S was incorporated into the penA35-A311V/V316P double mutant, the MICs of both ceftriaxone and cefixime increased to essentially the same levels as those for FA19 harboring penA41, i.e., nearly 15-fold higher than those for FA19 harboring penA35.
To confirm that these three amino acid changes (A311V, V316P, and T483S) were the key mutations in penA41, we reverted the three residues in penA41 back to the amino acids found in penA35 and transformed the construct into FA19. Transformants of the reverted penA41 allele had MICs of both ceftriaxone and cefixime that were identical to those for FA19 harboring penA35 (Fig. 3). These data clearly show that only three codon alterations, A311V, V316P, and T483S, are responsible for the substantially increased cephalosporin resistance of penA41 over penA35 (Fig. 3).
Acylation rates of PBP2 variants with penicillin G, ceftriaxone, and cefixime.
The second-order acylation rate constants for key mutants (Table 1) were determined for penicillin G, ceftriaxone, and cefixime, as described in Materials and Methods. PBP2 from H041 showed a remarkable ∼11,000-fold decrease in the k2/Ks value for cefixime and a 2,300-fold decrease for ceftriaxone compared to wild-type PBP2, highlighting the effectiveness of active-site remodeling in lowering the acylation rates of the expanded-spectrum cephalosporins without completely ablating essential transpeptidase activity. The k2/Ks values for the A311V and T316P (double), T483S (single), and A311V, T316P, and T483S (triple) PBP235/02 mutants generally followed a pattern consistent with the cephalosporin MICs, although it is difficult to derive an exact relationship between the MICs and acylation rates.
Table 1.
Acylation rate constants for PBP2 variantsb
| Protein | Mean acylation rate constant (M−1 s−1) ± SD |
||
|---|---|---|---|
| Ceftriaxone | Cefixime | Penicillin G | |
| PBP2WTa | 1,710,000 ± 90,000 | 1,480,000 ± 22,000 | 75,700 ± 2,300 |
| PBP235/02a | 11,300 ± 400 | 7,170 ± 300 | 510 ± 90 |
| PBP2H041 | 741 ± 28 | 135 ± 21 | 55 ± 14 |
| PBP235/02-A311V/V316P | 19,900 ± 1,200 | 2,480 ± 210 | 890 ± 40 |
| PBP235/02-T483S | 1,730 ± 120 | 710 ± 32 | 62 ± 8 |
| PBP235/02-A311V/V316P/T483S | 1,230 ± 28 | 193 ± 10 | 88 ± 8 |
| PBP2WT-A311V/V316P/T483S | 1,300,000 ± 110,000 | 660,000 ± 41,000 | 43,000 ± 2,000 |
Values were reported previously by Tomberg et al. (25).
The acylation rate constants were derived from kinetic measurements of the formation of the acyl-enzyme complex, as described in Materials and Methods. The rates were determined directly with [14C]penicillin G and by the competition method with ceftriaxone and cefixime. Values were derived from a minimum of 3 (usually 3 to 7) separate determinations. PBP2WT, PBP2 from strain FA19; PBP235/02, PBP2 from strain 35/02; PBP2H041, PBP2 from strain H041.
We also purified wild-type PBP2 harboring the three mutations and determined the k2/Ks for penicillin G, ceftriaxone, and cefixime. In the wild-type background, these mutations had a minimal effect (less than a 2-fold decrease) on the acylation rate constants of all three antibiotics relative to wild-type PBP2, but in the PBP235/02 background, they decreased the acylation rate constants by 6- to 35-fold compared to PBP235/02 (Table 1). Thus, the capacity of the three mutations to markedly alter the acylation rates of the antibiotics is dependent on other mutations present in mosaic PBP2 variants. These results are consistent with our previous study demonstrating the complex interdependency of mutations in the mosaic PBP2 background required to alter the acylation rate constants of β-lactam antibiotics (25).
DISCUSSION
The emergence of H041, which is essentially untreatable with expanded-spectrum cephalosporins and most other antimicrobials, is a wake-up call that the postantibiotic era for treatment of N. gonorrhoeae infections may be imminent (3, 24). The spread of this high-level ceftriaxone-resistant strain or other ceftriaxone-resistant strains such as those recently identified in France (29) and Spain (34) would be a public health disaster. Part of the response to this threat is to identify the amino acids that result in cephalosporin resistance in order to carry out more effective surveillance of the spread of cephalosporin-resistant strains, to understand the mechanisms underlying this resistance, and to develop genetic testing methods for cephalosporin resistance in the future.
In this study, we have identified three amino acid alterations in PBP2 from H041, A311V, V316P, and T483S, that together are responsible for conferring high-level resistance to expanded-spectrum cephalosporins above that conferred by mosaic penA alleles from strains such as 35/02 (16, 22, 25). These mutations, when incorporated into the mosaic penA35 allele, increase the MICs of ceftriaxone and cefixime for a wild-type recipient strain by 15-fold (to essentially the same level as the penA41 allele itself) over the same strain harboring the traditional penA35 mosaic XXIX allele (Fig. 3). The capacity of these mutations to decrease the acylation rates of the expanded-spectrum cephalosporins for PBP2 (and thereby increase the MICs), however, depends on the presence of other mutations in the mosaic PBP2 background, as the three mutations have very little effect when incorporated into wild-type PBP2 (Table 1). These data highlight the ability of a small number of mutations to confer very large increases in resistance when incorporated into a mosaic penA allele and also reflect the complex interactions between the mutations within a given PBP2 background.
One of the major hurdles inherent in the evolution of cephalosporin-resistant penA alleles is to remodel the active site of PBP2 to cause a decrease in acylation rates with β-lactam antibiotics while retaining essential transpeptidase (TPase) activity. Because β-lactam antibiotics are substrate analogs, these changes must be subtle and, by extension, must affect the reaction steps that are most important for rapid acylation by β-lactams but less important for transpeptidation. Unfortunately, it is not possible to quantify TPase activity of this class (class B) of PBPs in vitro, and the only assessment of transpeptidase activity is a qualitative determination of whether the encoded PBP2 variant supports growth of gonococci. Moreover, since mutations that lower acylation with β-lactam antibiotics may also lower TPase activity of PBP2, it is unknown how much TPase activity can be lost while still allowing normal growth. We know only that such mutations do not cripple TPase function and that whatever activity is retained is sufficient for growth of H041.
It is also worth noting that although we have focused primarily on resistance to ceftriaxone and cefixime, our acylation data indicate that mutations present in PBP2 from H041 markedly decrease the k2/Ks values for penicillin G as well as for the two cephalosporins (Table 1). This indicates that the mutations are not simply discriminating against the different structures of the cephalosporins relative to penicillin but are probably affecting acylation in general. Hence, even as clinical strains drive toward cephalosporin resistance, it is unlikely that penicillin will ever again become effective against gonorrhea, as the mutations introduced into the mosaic alleles that decrease acylation by cephalosporins also decrease acylation by penicillins.
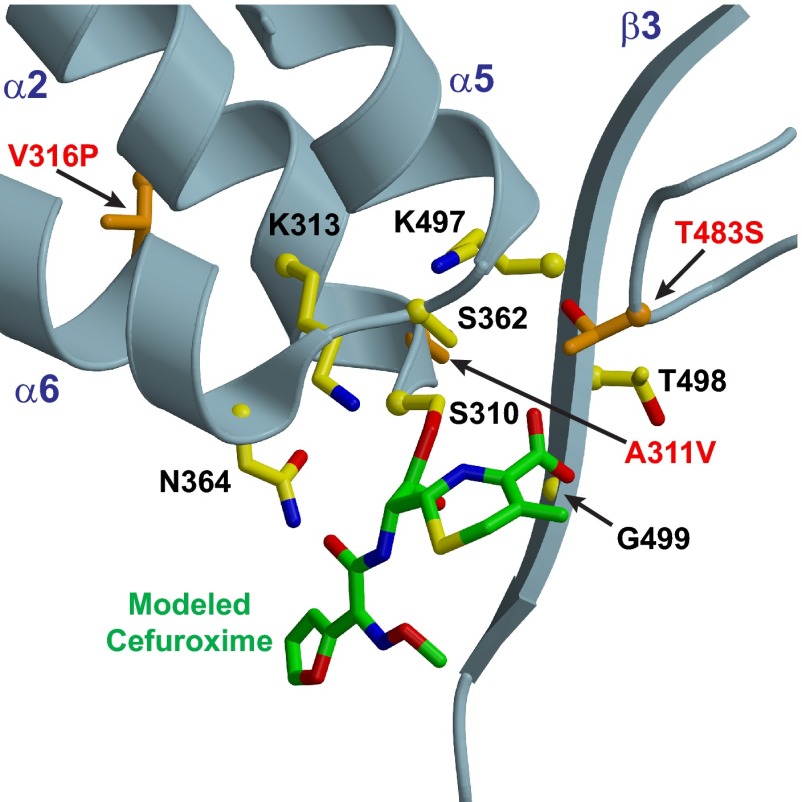
Now that the mutations in penA41 that confer the large increase in cephalosporin resistance to recipient strains above and beyond that already conferred by the mosaic penA35 allele have been identified, it is important to define how these three amino acid changes alter the structure of PBP2. In lieu of a crystal structure of PBP2H041, we can examine the positions of these amino acid alterations in the crystal structure of wild-type PBP2 (31) to infer how these mutations might affect acylation by β-lactam antibiotics. The locations of the three mutations in the wild-type structure are shown in Fig. 4. Two of these mutations (A311V and V316P) are present on the α2 helix either within or just downstream of the SxxK active-site motif containing the Ser nucleophile (Ser310) that attacks the β-lactam–d-Ala-d-Ala bond, whereas the T483S mutation is on a loop connecting α10 with β3 (the latter of which contains the KTG active-site motif). These mutations are discussed in turn below.
Fig 4.
Structure of PBP2 with modeled cefuroxime showing the locations of the three mutations from PBP2H041 that confer essentially all of the increased resistance above that of PBP235/02. The A311V and V316P mutations are located on the opposite side of the α2 helix containing the active-site nucleophile Ser310. These mutations may perturb the position of Ser310, thus increasing the activation energy of the transition state during acylation and lowering the acylation rate. The T483S mutation is on a loop preceding the β3 strand containing the KTG motif (pictured) and is ideally located to interact with the carboxylate from cefuroxime. Cefuroxime was modeled into the active site based on superimposition of PBP2 with PBP2X from Streptococcus pneumoniae (38).
A311V/V316P.
The A311V and V316P mutations on α2 are in the same location as two of the three mutations that we previously investigated in mosaic penA alleles (25). The data from that study showed that I312M and V316T were important for the intermediate-level cephalosporin resistance phenotype conferred by the penA35 allele, as reversion of these two mutations to the wild type reduced the MICs of ceftriaxone and cefixime by ∼4-fold. Along with the I312M mutation, PBP2 from H041 contains a new mutation at Ala311 (A311V) and a different mutation at Val316, V316P (Fig. 4).
As shown in Fig. 4, Ala311 and Val316 are present on the same α2 helix that contains the serine nucleophile Ser310, with Ala311 being immediately adjacent. Changes in the hydrophobic packing of α2 conferred by the mutations could change the dynamics of transition state formation and lead to decreases in k2/Ks, the acylation rate constant. Moreover, the differences between the amino acids at residue 316 between penA35 and penA41, with the latter having a proline instead of threonine, could have a significant impact on the structure of α2. Prolines are known to promote helix kinking, and so it will be of particular interest to determine the structure of PBP2 from H041 to see if this mutation has any impact on the helical structure of α2.
T483S.
The T483S mutation is unique to H041 penA and in a region that is not highly divergent in mosaic alleles. While the T483S mutation is very conservative, the loss of the methyl group of Thr has a large impact on acylation (Fig. 3 and Table 1). Thr483 is on a loop preceding the β3 strand containing the KTG motif and is situated near the active site. Modeling of the cephalosporin cefuroxime into the active site of PBP2 (no acylated structure of PBP2 is available) reveals that the hydroxyl group of Thr483 is very close to the carboxyl group of cefuroxime (Fig. 4). Alteration of this interaction may increase the activation energy for transition state formation and thus lead to decreases in the acylation rate (k2), or alternatively, Thr483 may be important in binding (Ks), and its mutation to Ser increases Ks and thus lowers the second-order acylation rate (k2/Ks).
In conclusion, the first strain (H041) with high-level resistance to ceftriaxone (MIC = 2 to 4 μg/ml) and resistance to most other antimicrobials was isolated recently in Japan (24). We have identified the 3 amino acid mutations in penA41 that confer essentially all of the increased resistance to expanded-spectrum cephalosporins over that conferred by a common mosaic penA allele (e.g., penA35). Two of these amino acid alterations (A311V and V316P) are in a region of PBP2 already known to harbor resistance-changing mutations, but the third (T483S) is in a novel location, where it may interact with the carboxylate moiety of β-lactam antibiotics. The effect of these mutations on peptidoglycan synthesis, in vitro growth, and biological fitness in vivo remain important questions to address in order to assess the propensity of strains harboring this allele to cause disease outbreaks. In addition to the large decrease in the acylation rate, PBP2H041 may also have a marked reduction in transpeptidase activity, thereby causing a decrease in fitness. However, H041 and similar strains might also contain compensatory mutations that mitigate the fitness cost, which are frequently obtained in the laboratory (35, 36) and are likely occur in nature as well. Studies to address these questions are currently in progress.
ACKNOWLEDGMENTS
This work was supported by grants AI36901 (to R.A.N.) and GM66861 (to C.D.) from the National Institutes of Health and by a grant from the Orebro University Hospital Research Foundation, Orebro, Sweden (to M.U.).
Footnotes
Published ahead of print 15 April 2013
REFERENCES
- 1. World Health Organization 2008. Global incidence and prevalence of selected curable sexually transmitted infections—2008. World Health Organization, Geneva, Switzerland: http://www.who.int/reproductivehealth/publications/rtis/2008_STI_estimates.pdf [Google Scholar]
- 2. Unemo M, Shafer WM. 2011. Antibiotic resistance in Neisseria gonorrhoeae: origin, evolution, and lessons learned for the future. Ann. N. Y. Acad. Sci. 1230:E19–E28. 10.1111/j.1749-6632.2011.06215.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Unemo M, Nicholas RA. 2012. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol. 7:1401–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phillips I. 1976. β-Lactamase producing penicillin-resistant gonococcus. Lancet ii:656–657 [DOI] [PubMed] [Google Scholar]
- 5. CDC 2007. Update to CDC's sexually transmitted diseases treatment guidelines, 2006: fluoroquinolones no longer recommended for treatment of gonococcal infections. MMWR Morb. Mortal. Wkly. Rep. 56:332–336 [PubMed] [Google Scholar]
- 6. Golparian D, Hellmark B, Fredlund H, Unemo M. 2010. Emergence, spread and characteristics of Neisseria gonorrhoeae isolates with in vitro decreased susceptibility and resistance to extended-spectrum cephalosporins in Sweden. Sex. Transm. Infect. 86:454–460 [DOI] [PubMed] [Google Scholar]
- 7. Allen VG, Mitterni L, Seah C, Rebbapragada A, Martin IE, Lee C, Siebert H, Towns L, Melano RG, Low DE. 2013. Neisseria gonorrhoeae treatment failure and susceptibility to cefixime in Toronto, Canada. JAMA 309:163–170 [DOI] [PubMed] [Google Scholar]
- 8. CDC 2012. Update to CDC's sexually transmitted diseases treatment guidelines, 2010: oral cephalosporins no longer a recommended treatment for gonococcal infections. MMWR Morb. Mortal. Wkly. Rep. 61:590–594 [PubMed] [Google Scholar]
- 9. Bignell C, Unemo M. 2012. European guideline on the diagnosis and treatment of gonorrhoea in adults. Int. J. STD AIDS, in press [DOI] [PubMed] [Google Scholar]
- 10. Hagman KE, Pan W, Spratt BG, Balthazar JT, Judd RC, Shafer WM. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611–622 [DOI] [PubMed] [Google Scholar]
- 11. Veal WL, Nicholas RA, Shafer WM. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184:5619–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gill MJ, Simjee S, Al-Hattawi K, Robertson BD, Easmon CS, Ison CA. 1998. Gonococcal resistance to β-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob. Agents Chemother. 42:2799–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olesky M, Hobbs M, Nicholas RA. 2002. Identification and analysis of amino acid mutations in porin IB that mediate intermediate-level resistance to penicillin and tetracycline in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:2811–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Olesky M, Zhao S, Rosenberg RL, Nicholas RA. 2006. Porin-mediated antibiotic resistance in Neisseria gonorrhoeae: ion, solute, and antibiotic permeation through PIB proteins with penB mutations. J. Bacteriol. 188:2300–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ropp PA, Hu M, Olesky M, Nicholas RA. 2002. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao S, Duncan M, Tomberg J, Davies C, Unemo M, Nicholas RA. 2009. Genetics of chromosomally mediated intermediate resistance to ceftriaxone and cefixime in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 53:3744–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spratt BG. 1988. Hybrid penicillin-binding proteins in penicillin-resistant strains of Neisseria gonorrhoeae. Nature 332:173–176 [DOI] [PubMed] [Google Scholar]
- 18. Ameyama S, Onodera S, Takahata M, Minami S, Maki N, Endo K, Goto H, Suzuki H, Oishi Y. 2002. Mosaic-like structure of penicillin-binding protein 2 gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob. Agents Chemother. 46:3744–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito M, Deguchi T, Mizutani KS, Yasuda M, Yokoi S, Ito S, Takahashi Y, Ishihara S, Kawamura Y, Ezaki T. 2005. Emergence and spread of Neisseria gonorrhoeae clinical isolates harboring mosaic-like structure of penicillin-binding protein 2 in central Japan. Antimicrob. Agents Chemother. 49:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanaka M, Nakayama H, Tunoe H, Egashira T, Kanayama A, Saika T, Kobayashi I, Naito S. 2002. A remarkable reduction in the susceptibility of Neisseria gonorrhoeae isolates to cephems and the selection of antibiotic regimens for the single-dose treatment of gonococcal infection in Japan. J. Infect. Chemother. 8:81–86 [DOI] [PubMed] [Google Scholar]
- 21. Wang SA, Lee MV, O'Connor N, Iverson CJ, Ohye RG, Whiticar PM, Hale JA, Trees DL, Knapp JS, Effler PV, Weinstock HS. 2003. Multidrug-resistant Neisseria gonorrhoeae with decreased susceptibility to cefixime—Hawaii, 2001. Clin. Infect. Dis. 37:849–852 [DOI] [PubMed] [Google Scholar]
- 22. Lindberg R, Fredlund H, Nicholas RA, Unemo M. 2007. Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime and ceftriaxone: association with genetic polymorphisms in penA, mtrR, porB1b, and ponA. Antimicrob. Agents Chemother. 51:2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ohnishi M, Saika T, Hoshina S, Iwasaku K, Nakayama S, Watanabe H, Kitawaki J. 2011. Ceftriaxone-resistant Neisseria gonorrhoeae, Japan. Emerg. Infect. Dis. 17:148–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, Iwasaku K, Nakayama SI, Kitawaki J, Unemo M. 2011. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea? Detailed characterization of the first high-level ceftriaxone resistant strain. Antimicrob. Agents Chemother. 55:3538–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tomberg J, Unemo M, Davies C, Nicholas RA. 2010. Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry 49:8062–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maness MJ, Sparling PF. 1973. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128:321–330 [DOI] [PubMed] [Google Scholar]
- 27. Danielsson D, Faruki H, Dyer D, Sparling PF. 1986. Recombination near the antibiotic resistance locus penB results in antigenic variation of gonococcal outer membrane protein I. Infect. Immun. 52:529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kellogg DS, Peacock WL, Deacon WE, Browh L, Perkle CI. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to colonial variation. J. Bacteriol. 85:1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Unemo M, Golparian D, Nicholas R, Ohnishi M, Gallay A, Sednaoui P. 2012. High-level cefixime- and ceftriaxone-resistant N. gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob. Agents Chemother. 56:1273–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 31. Powell AJ, Tomberg J, Deacon AM, Nicholas RA, Davies C. 2009. Crystal structures of penicillin-binding protein 2 from penicillin-susceptible and -resistant strains of Neisseria gonorrhoeae reveal an unexpectedly subtle mechanism for antibiotic resistance. J. Biol. Chem. 284:1202–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frere JM, Nguyen-Disteche M, Coyette J, Joris B. 1992. Mode of action: interaction with the penicillin binding proteins, p 148–196 In Page MI. (ed), The chemistry of β-lactams. Chapman & Hall, Glasgow, United Kingdom [Google Scholar]
- 33. Stefanova ME, Tomberg J, Olesky M, Holtje JV, Gutheil WG, Nicholas RA. 2003. Neisseria gonorrhoeae penicillin-binding protein 3 exhibits exceptionally high carboxypeptidase and β-lactam binding activities. Biochemistry 42:14614–14625 [DOI] [PubMed] [Google Scholar]
- 34. Camara J, Serra J, Ayats J, Bastida T, Carnicer-Pont D, Andreu A, Ardanuy C. 2012. Molecular characterization of two high-level ceftriaxone-resistant Neisseria gonorrhoeae isolates detected in Catalonia, Spain. J. Antimicrob. Chemother. 67:1858–1860 [DOI] [PubMed] [Google Scholar]
- 35. Fermer C, Swedberg G. 1997. Adaptation to sulfonamide resistance in Neisseria meningitidis may have required compensatory changes to retain enzyme function: kinetic analysis of dihydropteroate synthases from N. meningitidis expressed in a knockout mutant of Escherichia coli. J. Bacteriol. 179:831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kunz AN, Begum AA, Wu H, D'Ambrozio JA, Robinson JM, Shafer WM, Bash MC, Jerse AE. 2012. Impact of fluoroquinolone resistance mutations on gonococcal fitness and in vivo selection for compensatory mutations. J. Infect. Dis. 205:1821–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 38. Gordon E, Mouz N, Duee E, Dideberg O. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477–485 [DOI] [PubMed] [Google Scholar]