Abstract
Overseas travel, as a risk factor for the acquisition of infections due to antimicrobial-resistant organisms, has recently been linked to carbapenemase-producing Gram-negative bacteria. Multiresistant Klebsiella pneumoniae, Escherichia coli, and Acinetobacter baumannii strains were isolated from a wound of a Canadian patient with a recent history of hospitalization in India. This resulted in the initiation of outbreak management that included surveillance cultures. Epidemiological and molecular investigations showed that NDM-1-producing K. pneumoniae ST16 and OXA-23-producing A. baumannii ST10 strains were transmitted to 5 other patients, resulting in the colonization of 4 patients and the death of 1 patient due to septic shock caused by the OXA-23-producing A. baumannii strain. The high rate of false positivity of the screening cultures resulted in additional workloads and increased costs for infection control and clinical laboratory work. We believe that this is the first report of an infection with carbapenemase-producing Gram-negative bacteria resulting in death attributed to a patient with recent foreign hospitalization. We recommend routine rectal and wound screening for colonization with multiresistant bacteria for patients who have recently been admitted to hospitals outside Canada.
INTRODUCTION
Gram-negative bacteria, most notably Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii, are among the most important causes of serious hospital-acquired and community-onset bacterial infections in humans, and resistance to antimicrobial agents in these bacteria has become an increasingly relevant problem (1). Of special concern is the development of resistance to the carbapenems, since these agents are often the last line of effective therapy available for the treatment of infections caused by multiresistant Enterobacteriaceae, P. aeruginosa, and A. baumannii (2). Most important is the recognition of isolates that produce carbapenemases, which cause resistance to the carbapenems. Carbapenemases include class A enzymes (i.e., KPC types), class B enzymes, or the metallo-β-lactamases (MBLs [i.e., VIM, IPM, and NDM types]), and class D enzymes, or oxacillinases (i.e., OXA types) (3).
NDM enzymes were first reported for K. pneumoniae and E. coli strains recovered from a Swedish patient who had been hospitalized in New Delhi, India, previously (4). Recent reports from the Indian subcontinent (including India, Pakistan, and Bangladesh) show that the distribution of NDM β-lactamases among clinical and environmental isolates of Gram-negative bacteria is widespread in these countries (5–8). The majority of NDM-1-producing bacteria (most often E. coli and K. pneumoniae) are broadly resistant to various drug classes, leaving very limited treatment options (3). Infections with NDM-producing Enterobacteriaceae in areas where they are not endemic, such as Canada, have most often been associated with visits to, and hospitalization in, areas of endemicity, such as the Indian subcontinent (9).
A. baumannii is an important nosocomial pathogen associated with a wide range of infections, including respiratory tract, bloodstream, urinary tract, and surgical-site infections (10). The spread of A. baumannii between different patients in the hospital setting is difficult to control due to the bacterium's ability to persist in the environment. Therefore, numerous nosocomial intensive-care unit (ICU) outbreaks due to A. baumannii have been reported (10). The carbapenemases described in A. baumannii include the MBLs (i.e., VIM, IMP, SIM, and NDM types) and, more commonly, the OXA types (i.e., OXA-23-like, OXA-24-like, OXA-51-like, and OXA-58-like enzymes) (11). OXA-23-like enzymes are the most common carbapenemases described in A. baumannii and have been reported in various European, North American, South American, and Asian countries, including India, the United States, and Canada (12). A. baumannii strains that produce OXA-type β-lactamases have been responsible for a range of infectious syndromes in military personnel returning to their home countries after being injured in the conflicts in Iraq and Afghanistan (13).
During April 2012, multiresistant K. pneumoniae, E. coli, and A. baumannii strains were isolated from a wound culture from a patient in Edmonton, Canada, with a recent history of hospitalization in India. Infection control precautions and active surveillance were implemented immediately upon the isolation of Gram-negative organisms from the wound culture. However, secondary spread occurred and was associated with the death of another patient. The clinical and infection control aspects of the outbreak were recently presented at IDWeek (14). This study describes the laboratory aspects of screening for carbapenemase-producing bacteria and the molecular characterization of the bacteria involved in the outbreak.
MATERIALS AND METHODS
Index patient.
A 62-year-old female (patient A) was admitted with a postoperative wound infection to a surgical ward in a Canadian hospital in Edmonton, Alberta, Canada, on 31 March 2012. She was housed in a room shared with three other patients. She had just returned from India after being admitted to a hospital in that country for a prolonged period. Multiresistant A. baumannii (AB01), K. pneumoniae (Kp01), and E. coli (Ec01) strains were cultured from thigh tissue obtained on 1 April 2012 during surgical debridement. K. pneumoniae (Kp02) was cultured from a rectal screening swab obtained from this patient on 3 April 2012.
Screening of patients for carbapenemase-producing bacteria.
Surveillance cultures (i.e., from rectal, wound, ostomy, and endotracheal suction specimens) were used to identify all epidemiologically linked patients who may have become colonized. This procedure was directed by the Infection Prevention and Control (IPC) team and formed part of contact tracing and unitwide prevalence screening. Those screened included inpatients in the same wards/units as the index patient and subsequent case patients. Surveillance cultures were undertaken weekly for all patients on the affected units until negative screening cultures were obtained for two consecutive weeks. Roommates and unit contacts of case patients also had surveillance specimens collected before discharge. In addition, inpatients of surgical units were screened before transfer to another service within the hospital. No environmental or health care worker screenings were performed. An incident case was defined as the isolation of any carbapenemase-producing Gram-negative organism in clinical or surveillance cultures from 1 April 2012 to 25 May 2012. Clinical specimens were obtained from body sites as directed by an attending physician for the clinical management of a patient. Active surveillance cultures were performed on stool, rectal swabs, ostomy output, wound swabs, and endotracheal suction specimens as requested by the hospital's IPC team and involved a total of 410 patients.
Screening swabs obtained from patients were placed in a Copan M40 Transystem containing Amies gel transport medium. Stool and endotracheal suction specimens were submitted in sterile containers without transport medium. The Centers for Disease Control and Prevention (CDC, Atlanta, GA) protocol was used to screen for carbapenemase-producing Gram-negative bacteria (15). The specimens were inoculated into 5 ml of Trypticase soy broth (TSB) containing a 10-μg-meropenem disk and were incubated overnight (at 35°C, in ambient air), and 100 μl was then plated onto a MacConkey agar (MAC) plate and was streaked for isolation. The MAC plate was incubated overnight (at 35°C, in ambient air) and was then examined for growth. Single colonies of Gram-negative bacilli were subcultured to a MAC plate containing a meropenem disk (10 μg). Non-meropenem-susceptible isolates underwent identification and susceptibility testing using a Vitek 2 instrument (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, MO). The identification of A. baumannii was confirmed by using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, MO) and detection of chromosomal OXA-51-like carbapenemases (16).
Susceptibility testing.
Antimicrobial susceptibility was determined with MicroScan Neg MIC panels (type 38; Siemens, Burlington, ON, Canada). The MICs of the following drugs were determined: amoxicillin-clavulanic acid (AMC), piperacillin-tazobactam (TZP), cefoxitin (FOX), ceftriaxone (CRO), ceftazidime (CAZ), aztreonam (ATM), imipenem (IPM), meropenem (MER), ertapenem (ERT), amikacin (AMK), gentamicin (GEN), tobramycin (TOB), ciprofloxacin (CIP), trimethoprim-sulfamethoxazole (SXT), and tigecycline (TIM). Additional tests for susceptibility to colistin (COL) were performed using Etest methodology according to the manufacturer's instructions (bioMérieux, Marcy l'Etoile, France). Throughout this study, results were interpreted using the 2012 CLSI criteria for broth dilution (17). The European Committee for Antimicrobial Susceptibility Testing (EUCAST) breakpoint (for K. pneumoniae and E. coli) was used for COL, and the FDA breakpoint was used for TIM.
β-Lactamase identification.
The presence of carbapenemase was determined with the modified Hodge test (MHT) (17) and the MBL Etest (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions. PCR amplification for blaCTX-M, blaKPC, blaVIM, blaIMP, blaNDM, blaOXA-48-like, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58-like was carried out on the isolates with a GeneAmp thermal cycler, model 9700 (Applied Biosystems, Norwalk, CT), using PCR conditions and primers described previously (16, 18–20). blaCTX-M, blaNDM, blaOXA-48-like, and blaOXA-23-like were sequenced using PCR conditions and primers described previously (19, 21, 22).
Plasmid-mediated quinolone resistance determinants.
The qnrA, qnrS, and qnrB genes were amplified by multiplex PCR (23). aac(6′)-Ib and qepA were amplified in a separate PCR using primers and conditions described previously (24, 25). The variant aac(6′)-Ib-cr was further identified by digestion with BstF5I (New England Biolabs, Ipswich, MA).
16S rRNA methylation.
Genes coding for 16S rRNA methylases were amplified using a multiplex PCR described by Doi and Arakawa (26).
PFGE.
The E. coli and K. pneumoniae isolates were examined for genetic relatedness by pulsed-field gel electrophoresis (PFGE) following the extraction of genomic DNA and digestion with XbaI using the standardized E. coli (O157:H7) protocol established by the CDC, Atlanta, GA (27). The A. baumannii isolates were examined for genetic relatedness by PFGE following the extraction of genomic DNA and digestion with ApaI as described previously (28). Subsequent PFGE analyses were performed on a CHEF Mapper apparatus (Bio-Rad Laboratories, Hercules, CA). PFGE banding patterns were analyzed with BioNumerics software (Applied Maths, Kortrijk, Belgium), and relatedness was calculated using the UPGMA (unweighted-pair group method using average linkages) algorithm, with the Dice coefficient for assessment of the similarity of bands. Cluster designations were based on the criteria of Tenover et al. (29).
MLST.
Multilocus sequence typing (MLST) was performed on the K. pneumoniae isolates by using seven conserved housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, tonB) as described previously (30). MLST was performed on the E. coli isolate by using seven conserved housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, recA). A detailed protocol of the MLST procedure for E. coli, including allelic type and sequence type (ST) assignment methods, is available at MLST Databases at the ERI, University College Cork (http://mlst.ucc.ie/mlst/dbs/Ecoli). MLST was performed on the A. baumannii isolates by using seven conserved housekeeping genes (cpn60, fusA, gltA, pyrG, recA, rplB, rpoB). A detailed protocol of the MLST procedure for A. baumannii, including allelic type and ST assignment methods, is available at the Institut Pasteur website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/primers_Abaumannii.html).
RESULTS
Summary of the nosocomial outbreak.
The details of the clinical presentations, nosocomial outbreak, and infection control procedures were recently presented at IDWeek, San Diego, CA (14). Outbreak management was initiated immediately upon the identification of multiresistant Gram-negative bacteria in the index patient (patient A).
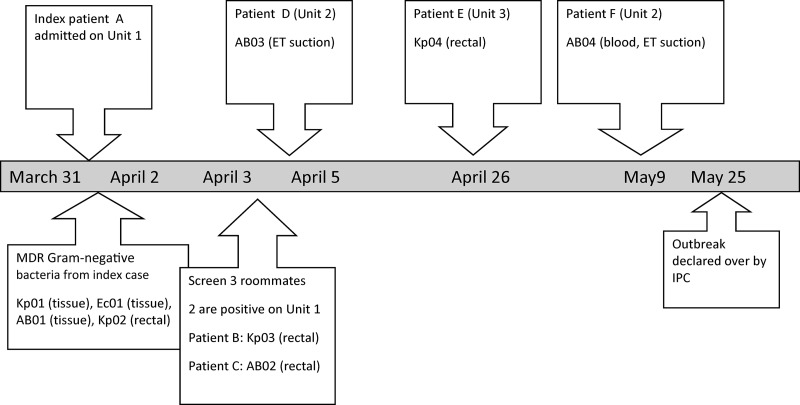
The details of the outbreak and the chronological summary of events are given in Fig. 1 and Table 1. In brief, the index patient underwent surgery in India for a fractured femur that subsequently became infected and did not respond to treatment. Eventually, she discharged herself and flew back to Edmonton, Canada, where she was admitted to the surgical ward (unit 1), sharing a room with 3 other patients, on 31 March 2012. Between 3 April and 9 May 2012, several multiresistant Gram-negative bacteria were cultured from 6 different patients in 3 different units. On 3 April, K. pneumoniae (Kp03) was cultured from a rectal swab from patient B, and A. baumannii (AB02) was cultured from a rectal swab from patient C. Patients B and C were roommates of patient A in unit 1.
Fig 1.
Timeline of events in the nosocomial outbreak with multidrug-resistant (MDR) Gram-negative bacteria. ET, endotracheal.
Table 1.
Clinical characteristics of the patients involved in the outbreak
| Patient | Age (yr) | Sex | Admitting diagnosis | Organism(s) isolated | Date of culture | Unit | Specimena |
|---|---|---|---|---|---|---|---|
| A | 62 | F | Femoral fracture with deep postoperative infection | K. pneumoniae (Kp01) | 1 Apr. | 1 | Tissue |
| E. coli (Ec01) | 1 Apr. | Tissue | |||||
| A. baumannii (AB01) | 1 Apr. | Tissue | |||||
| K. pneumoniae (Kp02) | 3 Apr. | Rectal | |||||
| B | 83 | F | Ankle fracture requiring operative fixation | K. pneumoniae (Kp03) | 3 Apr. | 1 | Rectal |
| C | 69 | F | Infected thigh requiring incision and drainage | A. baumannii (AB02) | 19 Apr. | 1 | Rectal |
| D | 58 | M | Intra-abdominal sepsis due to perforated viscus | A. baumannii (AB03) | 4 Apr. | 2 | ET suction |
| E | 80 | M | Ischemic bowel due to small-bowel obstruction | K. pneumoniae (Kp04) | 26 Apr | 3 | Rectal |
| F | 74 | M | Segmental resection for pulmonary nodule | A. baumannii (AB04) | 8 May | 2 | ET suction, blood |
ET, endotracheal.
A. baumannii (AB03) was cultured from an endotracheal specimen collected on 4 April from patient D in unit 2. Patient D had no symptoms or signs of pulmonary infection. Patient D stayed briefly in the same unit as patient A and was then transferred to unit 2. K. pneumoniae (Kp04) was cultured from a rectal swab collected on 26 April from patient E in unit 3. Patient E was briefly admitted to unit 2. A. baumannii (AB04) was cultured from an endotracheal specimen and blood collected on 8 May from patient F in unit 2. Patient F developed septic shock and organ failure, was transferred to the intensive-care unit, and died a few hours later. None of patients B, C, D, E, and F had a history of hospitalization in a foreign country or of travel to a foreign country in the preceding 12 months.
Active surveillance cultures.
Over a period of 55 days, the clinical microbiology laboratory received 442 screening specimens from 410 patients. The true-positive rate (i.e., the proportion of carbapenemase producers) was 1.1% (5/442). The 5 true-positive specimens were from patients B through F. Sixty-three percent of specimens (278/442) had no growth, while 35.7% (158/442) grew organisms that subsequently proved not to be resistant to the carbapenems. One specimen grew carbapenem-resistant Enterobacter cloacae that tested negative for carbapenemases by phenotypic and genotypic tests. The subsequent growth from the TSB containing a 10-μg-meropenem disk resulted in 246 different isolates that required further workup; 5 were true positives, and 241 included the following bacteria: 203 Enterobacteriaceae that subsequently tested susceptible to MER by disk diffusion, 21 Pseudomonas aeruginosa isolates that tested susceptible to MER, 3 P. aeruginosa isolates intermediate to MER, 3 P. aeruginosa isolates that tested resistant to MER but were negative for carbapenemases, 1 Pseudomonas fluorescens isolate, 4 Stenotrophomonas maltophilia isolates, 2 yeast isolates, and 4 Enterococcus spp. This part of the investigation required an additional 120 technologist hours, with an estimated laboratory cost of Can$7,000.
Susceptibility testing and β-lactamases.
Table 2 shows the results of susceptibility and molecular tests for carbapenemase-producing bacteria. Kp01 was susceptible to AMK (MIC, 16 μg/ml), GEN (MIC, 2 μg/ml), TIM (MIC, 2 μg/ml), and COL (MIC, 0.25 μg/ml); Kp02, Kp03, and Kpn04 were susceptible only to COL (MIC, 0.5 μg/ml); Ec01 was susceptible to TIM (MIC, 1 μg/ml) and COL (MIC, 0.12 μg/ml); AB01, AB02, AB03, and AB04 were susceptible to COL (0.12 μg/ml). The TIM MIC for Kp02, Kp03, and Kp04 was 4 μg/ml.
Table 2.
Susceptibilities and molecular characteristics of carbapenemase-producing bacteria
| Isolate no. | Organism | Susceptibilitya (MIC [μg/ml]) |
Phenotypic test result |
β-Lactamases | PFGE patternb | MLST | PMQRc determinant(s) | 16S rRNA methylase | ||
|---|---|---|---|---|---|---|---|---|---|---|
| NS | S | MHT | MBL Etest | |||||||
| Kp01 | K. pneumoniae | TZP (>64/4), FOX (>32), CRO (>32), CAZ (>16), ATM (>16), IPM (>8), MER (>8), ERT (>4), TOB (>8), CIP (>2), SXT (>2/38) | AMK (16), GEN (2), TIM (2), COL (0.25) | Positive | Positive | NDM-1, CTX-M-15, OXA-181 | A | ST972 | qnrS, aac6′-Ib-cr | Negative |
| Kp02 | K. pneumoniae | TZP (>64/4), FOX (>32), CRO (>32), CAZ (>16), ATM (>16), IPM (>8), MER (>8), ERT (>4), GEN (>8), TOB (>8), AMK (>32), CIP (>2), SXT (>2/38), TIM (4) | COL (0.5) | Negative | Negative | NDM-1, CTX-M-15 | B | ST16 | qnrB, aac6′-Ib-cr | Negative |
| Kp03 | K. pneumoniae | TZP (>64/4), FOX (>32), CRO (>32), CAZ (>16), ATM (>16), IPM (>8), MER (>8), ERT (>4), GEN (>8), TOB (>8), AMK (>32), CIP (>2), SXT (>2/38), TIM (4) | COL (0.5) | Negative | Negative | NDM-1, CTX-M-15 | B | ST16 | qnrB, aac6′-Ib-cr | Negative |
| Kp04 | K. pneumoniae | TZP (>64/4), FOX (>32), CRO (>32), CAZ (>16), ATM (>16), IPM (>8), MER (>8), ERT (>4), GEN (>8), TOB (>8), AMK (>32), CIP (>2), SXT (>2/38), TIM (4) | COL (0.5) | Negative | Negative | NDM-1, CTX-M-15 | B | ST16 | qnrB, aac6′-Ib-cr | Negative |
| Ec01 | E. coli | TZP (>64/4), FOX (>32), CRO (>32), CAZ (>16), ATM (>16), IPM (>8), MER (>8), ERT (>4), GEN (>8), TOB (>8), AMK (>32), CIP (>2), SXT (>2/38) | TIM (1), COL (0.12) | Negative | Positive | NDM-1, CTX-M-15 | NP | ST410 | aac6′-Ib-cr | Negative |
| AB01 | A. baumannii | TZP (>64/4), CAZ (>16), ATM (>16), IPM (>8), MER (>8), AMK (>32), GEN (>8), TOB (>8), CIP (>2), SXT (>2/38) | COL (0.12) | Positive | Positive | OXA-23, OXA-51 | C | ST10 | aac6′-Ib-cr | armA |
| AB02 | A. baumannii | TZP (>64/4), CAZ (>16), ATM (>16), IPM (>8), MER (>8), AMK (>32), GEN (>8), TOB (>8), CIP (>2), SXT (>2/38) | COL (0.12) | Positive | Positive | OXA-23, OXA-51 | C | ST10 | aac6′-Ib-cr | armA |
| AB03 | A. baumannii | TZP (>64/4), CAZ (>16), ATM (>16), IPM (>8), MER (>8), AMK (>32), GEN (>8), TOB (>8), CIP (>2), SXT (>2/38) | COL (0.12) | Positive | Positive | OXA-23, OXA-51 | C | ST10 | aac6′-Ib-cr | armA |
| AB04 | A. baumannii | TZP (>64/4), CAZ (>16), ATM (>16), IPM (>8), MER (>8), AMK (>32), GEN (>8), TOB (>8), CIP (>2), SXT (>2/38) | COL (0.12) | Positive | Positive | OXA-23, OXA-51 | C | ST10 | aac6′-Ib-cr | armA |
NS, nonsusceptible (resistant or intermediate); S, sensitive; TZP, piperacillin-tazobactam; FOX, cefoxitin; CRO, ceftriaxone; CAZ, ceftazidime; ATM, aztreonam; IPM, imipenem; MER, meropenem; ERT, ertapenem; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; TIM, tigecycline; COL, colistin.
NP, not performed.
PMQR, plasmid-mediated quinolone resistance.
Kp01 tested positive by the modified Hodge test and the MBL Etest; Kp02, Kp03, and Kp04 were negative by the modified Hodge test and the MBL Etest; Ec01 was negative by the modified Hodge test and positive by the MBL Etest. AB01, AB02, AB03, and AB04 tested positive by the MBL Etest and the modified Hodge test using IPM as the substrate. PCR results showed that Kp01 was positive for blaNDM, blaCTX-M, and blaOXA-48-like, and sequencing identified the blaNDM enzyme as NDM-1, the blaCTX-M enzyme as CTX-M-15, and the blaOXA-48-like enzyme as OXA-181. Kp02, Kp03, Kp04, and Ec01 were positive for blaNDM and blaCTX-M, and sequencing identified the blaNDM enzyme as NDM-1 and the blaCTX-M enzyme as CTX-M-15. AB01, AB02, AB03, and AB04 were positive for blaOXA-23-like and blaOXA-51, and sequencing identified the blaOXA-23-like enzyme as OXA-23. The A. baumannii isolates were negative for MBLs by use of a different multiplex PCR (31).
Molecular characterization and typing.
PFGE showed that Kp02 was indistinguishable from Kp03 and Kp04 but was not related to Kp01. MLST identified Kp02, Kp03, and Kp04 as ST16, while Kp01 belonged to ST972 (Table 2). PFGE showed that AB01, AB02, AB03, and AB04 were indistinguishable from each other, and MLST identified them as ST10 (Table 2). Ec01 belonged to ST410. The presence of the different plasmid-mediated quinolone resistance determinants and 16S rRNA methylation is shown in Table 2.
DISCUSSION
Easy access to air and ground transportation is making it possible for people to travel to different countries and continents in a matter of hours or days, whether as tourists (medical, business, study, or recreational), immigrants, refugees, asylum seekers, or migrant workers (9). Overseas travel, as a risk factor for the acquisition of infections due to antimicrobial-resistant organisms, has recently been linked to infections due to CTX-M-producing E. coli strains or to various carbapenemase-producing Gram-negative bacteria (9).
Kohlenberg and colleagues described an outbreak of OXA-23-producing A. baumannii over a 10-month period at a German hospital that involved 32 patients in 5 intensive-care units and 2 regular wards (32). The index patient had been transferred to the German hospital by an ambulance flight from Thailand, where he had been hospitalized after a motor vehicle accident. Hrabak and colleagues reported an NDM-1-producing A. baumannii isolate from a patient transferred from a hospital in Egypt to the Czech Republic (33). A second A. baumannii isolate that produced NDM-1 was recovered 6 days later from the airways of another patient sharing the same room. That patient died due to respiratory failure; however, it was not clear if his death was due to the A. baumannii infection. MLST showed that both isolates belonged to ST1, which represents the epidemiologically successful European clone I. A report from the CDC described the spread of NDM-producing K. pneumoniae from an index patient to 2 other patients (34). One month prior to being admitted to a hospital in Rhode Island, the index patient had been hospitalized in Vietnam. Lowe and colleagues from Toronto, Canada, analyzed a cluster of patients infected or colonized with NDM-producing K. pneumoniae over a 15-month period and identified 2 index patients who were responsible for the transfer of 2 different clones to 7 contacts. One of the index patients had previously received medical care in India (35).
We describe the characteristics of carbapenemase-producing Enterobacteriaceae and A. baumannii strains that were linked to an index patient who had been hospitalized in India recently when she injured her leg while on vacation. Epidemiological and molecular investigations showed that NDM-1-producing K. pneumoniae ST16 and OXA-23-producing A. baumannii ST10 were transmitted to 5 other patients, even after infection control precautions and active surveillance had been implemented. One of these patients died due to septic shock caused by the same OXA-23-producing A. baumannii strain. We believe this is the first report to describe infection with multiresistant Gram-negative bacteria resulting in death attributed to a patient with recent foreign hospitalization.
Of special interest was the fact that the index patient was colonized/infected with 2 different isolates of NDM-producing K. pneumoniae (belonging to ST16 and ST972). However, only the ST16 isolate was transmitted to other patients. K. pneumoniae ST16 was first described in Paris during the early 2000s (30) and had been responsible for nosocomial outbreaks of CTX-M-15-producing K. pneumoniae in Sweden (36) and Denmark (37) during the mid- to late 2000s. This sequence type had also been reported with KPC-2 from Brazil (38) and with OXA-48 from Spain (39). A. baumannii ST10 with acinetobacter-derived cephalosporinase (ADC) from companion animals and horses in Switzerland had been described previously (40).
Active surveillance screening of patients played an important role in managing and eventually controlling this outbreak. We chose to use the CDC screening procedure for carbapenemase-producing Gram-negative bacteria because of its cost-effectiveness and the fact that all supplies were readily available in our laboratory (15). The sensitivity of this method compares well to that of CHROMagar chromogenic media for the detection of carbapenemase-producing Gram-negative bacteria (41). However, we found the CDC screening method labor-intensive, with high breakthrough growth (i.e., 35.7%) and slow turnaround (a minimum of 2 days for negative specimens and longer for positive specimens). Isolates from surveillance cultures were reported to Infection Control on day 1 (including those that subsequently tested negative for carbapenemases). The high rate of false positivity of the screening cultures resulted in additional workloads, closure of hospital beds and wards, and increased costs for infection control and the clinical laboratory. Collaboration and good communication between the Infection Prevention and Control team and the clinical laboratories were crucial to the successful investigation and control of this outbreak.
The phenotypic confirmation tests, such as the MBL Etest and the modified Hodge test (MHT), performed poorly during our investigation. Previous evaluations of the MHT have shown that this method reliably detects KPC- and OXA-48-producing isolates (42). Unfortunately, the MHT performs poorly in the detection of MBL-producing isolates (3). This would explain why Kp01 tested positive by the MHT while the other K. pneumoniae isolates tested negative. Interestingly, the OXA-23-producing A. baumannii isolate tested positive by the MBL Etest, a result attributed to the destabilization of OXA dimers by EDTA and/or to the antimicrobial activity of EDTA against certain isolates of A. baumannii (43, 44). We believe that the presence of carbapenemases among Gram-negative bacteria is an infection control emergency and that the detection of these bacteria in clinical laboratories is a critical step required for the appropriate management of patients and for infection prevention and control efforts. We urgently need rapid, cost-effective phenotypic methods, such as the Carba NP test, that will reliably detect carbapenemases in all Gram-negative bacteria (45).
The European Centre for Disease Prevention and Control has developed a communication platform tool dedicated to antimicrobial resistance (AMR) in health care-associated infections (HAI), which is referred to as the Epidemic Intelligence Information System (EPIS) AMR-HAI. EPIS AMR-HAI allows experts in national risk assessment bodies within the European Union to rapidly and securely exchange information related to microorganisms with emerging antimicrobial resistance that have a potential impact in the European Union (9). We recommend that similar communication platforms be established in different provinces and states on the North American continent to ensure the timely communication of emerging antimicrobial resistance mechanisms among public health officials, infection control practitioners, and clinical microbiologists.
In order to prevent the introduction and spread of multiresistant bacteria from travelers returning to their home countries, it is essential to rapidly identify patients colonized or infected by these bacteria (9, 46). We recommend that patients recently admitted to hospitals in areas of endemicity be screened, by culture of rectal and wound specimens using CHROMagar, for colonization with multiresistant bacteria. We further recommend careful attention to routine infection control measures (i.e., hand hygiene, cleaning of shared equipment, effective environmental cleaning) with consideration of preemptive contact isolation for patients with a recent history (i.e., 6 months) of foreign hospitalization. Such measures will help to prevent nosocomial outbreaks with multiresistant Gram-negative bacteria, which will ultimately save lives.
ACKNOWLEDGMENTS
J.D.D.P. previously received research funds from Merck and Astra Zeneca. All other authors have nothing to declare.
This work was supported by a research grant (73-6350) from Calgary Laboratory Services.
We thank Sharla Manca and Amber-Leah Wolfe for their assistance during the outbreak investigation.
Footnotes
Published ahead of print 22 April 2013
REFERENCES
- 1. Paterson DL. 2006. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control 34:S20–S28, S64–S73 [DOI] [PubMed] [Google Scholar]
- 2. Livermore DM, Woodford N. 2006. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 14:413–420 [DOI] [PubMed] [Google Scholar]
- 3. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob. Agents Chemother. 55:1274–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castanheira M, Mendes RE, Woosley LN, Jones RN. 2011. Trends in carbapenemase-producing Escherichia coli and Klebsiella spp. from Europe and the Americas: report from the SENTRY Antimicrobial Surveillance Programme (2007–09). J. Antimicrob. Chemother. 66:1409–1411 [DOI] [PubMed] [Google Scholar]
- 7. Lascols C, Hackel M, Marshall SH, Hujer AM, Bouchillon S, Badal R, Hoban D, Bonomo RA. 2011. Increasing prevalence and dissemination of NDM-1 metallo-beta-lactamase in India: data from the SMART study (2009). J. Antimicrob. Chemother. 66:1992–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 9. van der Bij AK, Pitout JD. 2012. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J. Antimicrob. Chemother. 67:2090–2100 [DOI] [PubMed] [Google Scholar]
- 10. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poirel L, Nordmann P. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826–836 [DOI] [PubMed] [Google Scholar]
- 12. Durante-Mangoni E, Zarrilli R. 2011. Global spread of drug-resistant Acinetobacter baumannii: molecular epidemiology and management of antimicrobial resistance. Future Microbiol. 6:407–422 [DOI] [PubMed] [Google Scholar]
- 13. Dallo SF, Weitao T. 2010. Insights into acinetobacter war-wound infections, biofilms, and control. Adv. Skin Wound Care 23:169–174 [DOI] [PubMed] [Google Scholar]
- 14. Chandran A, Wolfe AL, Manca S, Ahmed-Bentley J, Pitout JD, Barclay J, Joffe AM. 2012. Investigation of multiple multidrug-resistant Gram-negative bacilli outbreak in a Canadian hospital (or Help!! We have CRE!!), abstr. 1402. IDWeek 2012. https://idsa.confex.com/idsa/2012/webprogram/Paper35866.html
- 15. Centers for Disease Control and Prevention 2012. 2012 CRE toolkit—guidance for the control of carbapenem-resistant Enterobacteriaceae (CRE). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/hai/organisms/cre/cre-toolkit/ [Google Scholar]
- 16. Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353 [DOI] [PubMed] [Google Scholar]
- 17. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement. M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 18. Pitout JD, Church DL, Gregson DB, Chow BL, McCracken M, Mulvey MR, Laupland KB. 2007. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15-producing isolates. Antimicrob. Agents Chemother. 51:1281–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peirano G, Pillai DR, Pitondo-Silva A, Richardson D, Pitout JD. 2011. The characteristics of NDM-producing Klebsiella pneumoniae from Canada. Diagn. Microbiol. Infect. Dis. 71:106–109 [DOI] [PubMed] [Google Scholar]
- 20. Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. 2012. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 50:3877–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Afzal-Shah M, Woodford N, Livermore DM. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D beta-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67:1597–1606 [DOI] [PubMed] [Google Scholar]
- 23. Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83–88 [DOI] [PubMed] [Google Scholar]
- 25. Yamane K, Wachino J, Suzuki S, Arakawa Y. 2008. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 52:1564–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Doi Y, Arakawa Y. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88–94 [DOI] [PubMed] [Google Scholar]
- 27. Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pitout JD, Chow BL, Gregson DB, Laupland KB, Elsayed S, Church DL. 2007. Molecular epidemiology of metallo-beta-lactamase-producing Pseudomonas aeruginosa in the Calgary Health Region: emergence of VIM-2-producing isolates. J. Clin. Microbiol. 45:294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70:119–123 [DOI] [PubMed] [Google Scholar]
- 32. Kohlenberg A, Brummer S, Higgins PG, Sohr D, Piening BC, de Grahl C, Halle E, Ruden H, Seifert H. 2009. Outbreak of carbapenem-resistant Acinetobacter baumannii carrying the carbapenemase OXA-23 in a German university medical centre. J. Med. Microbiol. 58:1499–1507 [DOI] [PubMed] [Google Scholar]
- 33. Hrabak J, Stolbova M, Studentova V, Fridrichova M, Chudackova E, Zemlickova H. 16 February 2012. NDM-1 producing Acinetobacter baumannii isolated from a patient repatriated to the Czech Republic from Egypt, July 2011. Euro Surveill. 17(7):pii=20085. http://www.eurosurveillance.org/ViewArticle.aspx?Articleid=20085 [PubMed] [Google Scholar]
- 34. Centers for Disease Control and Prevention 2012. Carbapenem-resistant Enterobacteriaceae containing New Delhi metallo-beta-lactamase in two patients—Rhode Island, March 2012. MMWR Morb. Mortal. Wkly. Rep. 61(24):446–448 [PubMed] [Google Scholar]
- 35. Lowe CF, Kus JV, Salt N, Callery S, Louie L, Khan MA, Vearncombe M, Simor AE. 2013. Nosocomial transmission of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Toronto, Canada. Infect. Control Hosp. Epidemiol. 34:49–55 [DOI] [PubMed] [Google Scholar]
- 36. Lytsy B, Sandegren L, Tano E, Torell E, Andersson DI, Melhus A. 2008. The first major extended-spectrum beta-lactamase outbreak in Scandinavia was caused by clonal spread of a multiresistant Klebsiella pneumoniae producing CTX-M-15. APMIS 116:302–308 [DOI] [PubMed] [Google Scholar]
- 37. Nielsen JB, Skov MN, Jorgensen RL, Heltberg O, Hansen DS, Schonning K. 2011. Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur. J. Clin. Microbiol. Infect. Dis. 30:773–778 [DOI] [PubMed] [Google Scholar]
- 38. Seki LM, Pereira PS, de Souza MDPAH, Conceicao MDS, Marques EA, Porto CO, Colnago EML, Alves CDFM, Gomes D, Assef APDAC, Samuelsen O, Asensi MD. 2011. Molecular epidemiology of KPC-2-producing Klebsiella pneumoniae isolates in Brazil: the predominance of sequence type 437. Diagn. Microbiol. Infect. Dis. 70:274–277 [DOI] [PubMed] [Google Scholar]
- 39. Oteo J, Hernandez JM, Espasa M, Fleites A, Saez D, Bautista V, Perez-Vazquez M, Fernandez-Garcia MD, Delgado-Iribarren A, Sanchez-Romero I, Garcia-Picazo L, Miguel MD, Solis S, Aznar E, Trujillo G, Mediavilla C, Fontanals D, Rojo S, Vindel A, Campos J. 2013. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J. Antimicrob. Chemother. 68:317–321 [DOI] [PubMed] [Google Scholar]
- 40. Endimiani A, Hujer KM, Hujer AM, Bertschy I, Rossano A, Koch C, Gerber V, Francey T, Bonomo RA, Perreten V. 2011. Acinetobacter baumannii isolates from pets and horses in Switzerland: molecular characterization and clinical data. J. Antimicrob. Chemother. 66:2248–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vrioni G, Daniil I, Voulgari E, Ranellou K, Koumaki V, Ghirardi S, Kimouli M, Zambardi G, Tsakris A. 2012. Comparative evaluation of a prototype chromogenic medium (ChromID CARBA) for detecting carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J. Clin. Microbiol. 50:1841–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 50:477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bonnin RA, Naas T, Poirel L, Nordmann P. 2012. Phenotypic, biochemical, and molecular techniques for detection of metallo-beta-lactamase NDM in Acinetobacter baumannii. J. Clin. Microbiol. 50:1419–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Segal H, Elisha BG. 2005. Use of Etest MBL strips for the detection of carbapenemases in Acinetobacter baumannii. J. Antimicrob. Chemother. 56:598. 10.1093/jac/dki265 [DOI] [PubMed] [Google Scholar]
- 45. Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother. 56:6437–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rogers BA, Aminzadeh Z, Hayashi Y, Paterson DL. 2011. Country-to-country transfer of patients and the risk of multi-resistant bacterial infection. Clin. Infect. Dis. 53:49–56 [DOI] [PubMed] [Google Scholar]