Abstract
CMX001 is an orally available lipid acyclic nucleotide phosphonate that delivers high intracellular levels of cidofovir (CDV)-diphosphate and exhibits enhanced in vitro antiviral activity against a wide range of double-stranded DNA viruses, including cytomegalovirus (CMV). Mutations in the DNA polymerase of CMV that impart resistance to CDV also render the virus resistant to CMX001. Here, we report a novel resistance mutation that arose under the selective pressure of CMX001. The wild-type CMV strain AD169 was propagated in human foreskin fibroblasts under increasing concentrations of CMX001 over 10 months, and the resulting strain (named CMX001R) was less susceptible to CDV and CMX001 in a plaque reduction assay. Genotypic analysis of virus strain CMX001R via conventional sequencing of the genes encoding the CMV DNA polymerase (UL54) and UL97 kinase (UL97) demonstrated one mutation that changed the wild-type aspartate to glutamate at position 542 in UL54. A recombinant virus with this novel D542E mutation was generated via bacterial artificial chromosome-mediated marker transfer experiments. Subsequent phenotypic resistance analysis of the D542E mutant demonstrated reductions in susceptibility of greater than 10-fold to CMX001 and CDV, but no resistance to foscarnet (FOS) or ganciclovir (GCV). Analysis of replicative fitness showed that both strain CMX001R and the D542E mutant viruses demonstrated a smaller plaque phenotype and slower replication kinetics than their respective parent viruses. These data describe the first resistance mutation generated under the selective pressure of CMX001 and suggest that CMX001 may have a unique resistance profile associated with reduced viral replication and maintenance of sensitivity to FOS and GCV.
INTRODUCTION
Infections with human cytomegalovirus (CMV) are highly prevalent, but they are typically asymptomatic or self-limited in healthy hosts; however, they are a cause of significant morbidity and mortality in people with compromised or immature immune systems, including transplant recipients, patients with AIDS, and congenitally infected infants (1–4). Currently approved antiviral agents are often limited in their therapeutic utility due to modest antiviral activity, poor bioavailability, resistance, and/or drug toxicities (5, 6). Reports on the incidence and prevalence of resistance vary substantially depending on the patient population and how the drugs are used, but cross-resistance is a common problem with the nucleoside analogs (7, 8). For these reasons, it is important to develop new therapeutic agents for CMV with improved safety, efficacy, and resistance profiles (9).
CMX001 ({phosphonic acid, [[(S)-2-(4-amino-2-oxo-1(2H)-pyrimidinyl)-1-(hydroxymethyl)ethoxy]methyl]mono[3-(hexadecyloxy)propyl] ester}), also referred to as hexadecyloxypropyl cidofovir (HDP-CDV) in a previous publication, is an orally available lipid acyclic nucleotide phosphonate that delivers high intracellular level of CDV-diphosphate (CDV-PP) and exhibits enhanced in vitro antiviral activity against a wide range of double-stranded DNA viruses, including herpesviruses, orthopoxviruses, adenoviruses, polyomaviruses, and papillomaviruses (10, 11). CMX001 is designed for delivery into target cells where the lipid side chain is cleaved, thereby releasing CDV, which subsequently undergoes phosphorylation by intracellular kinases to the active antiviral agent, CDV-PP. CMX001 is taken up by cells much more rapidly than CDV and is associated with greater than 100-fold higher intracellular levels of CDV-PP compared to CDV (12). In CMV-infected cells, CDV-PP acts as an alternate substrate inhibitor of the UL54-encoded CMV DNA polymerase, resulting in decreased DNA synthesis and termination of chain elongation (13, 14).
The pharmacokinetic (PK) profile of CMX001 supports its clinical development due to the potential for both higher antiviral potency and lower toxicity than currently available treatment options. Due to its ability to deliver greater levels of CDV-PP intracellularly, CMX001 exhibits enhanced antiviral activity of up to 1,000-fold against CMV compared to CDV in both in vitro and in vivo models, as well as increased activity compared to ganciclovir (GCV) and foscarnet (FOS) (15–18). Data from animal studies and human trials suggests that the dose-limiting nephrotoxicity of CDV has been eliminated with CMX001 (19, 20). This has been mechanistically explained, since CMX001, unlike CDV, is not a substrate of the human organic anion transporter 1 (hOAT1) that is located in the proximal renal tubule and is primarily responsible for the toxic accumulation of CDV in the kidney (11, 19). A dose escalation PK and safety study of CMX001 in healthy adult subjects revealed no significant toxicities at doses of up to 2 mg/kg of body weight, the highest single dose tested (20).
In CMV, antiviral resistance to CDV maps to the DNA polymerase gene (UL54). Since CMX001 is converted intracellularly to CDV-PP, an alternate substrate inhibitor of UL54, resistance to this agent would also be expected to map to UL54. The impact of the higher intracellular CDV-PP levels achieved with CMX001 on the development of resistant CMV strains is unknown. To date, there have been no cases of treatment-emergent phenotypic resistance or mutations known to be associated with resistance in CMV antiviral treatment-naive patients receiving CMX001. The objectives of this study were to generate and characterize a de novo CMX001-resistant CMV isolate by serially passaging a wild-type laboratory strain of CMV (AD169) in the presence of increasing levels of CMX001.
MATERIALS AND METHODS
Antiviral compounds.
CMX001 was provided by Chimerix, Inc. (Durham, NC). CDV and GCV were purchased from the University of Alabama at Birmingham Hospital Pharmacy, and FOS was purchased from Sigma-Aldrich (St. Louis, MO).
Cell culture and virus strains.
Human foreskin fibroblast (HFF) cells were prepared and routinely propagated as monolayers in minimal essential medium (Mediatech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum, l-glutamine, penicillin, and gentamicin. The CMV strain AD169 was obtained from the American Type Culture Collection (Manassas, VA), the titers of the virus were determined at 2 × 105 PFU/ml, and virus was stored at −80°C.
Resistant strain selection.
The CMV strain AD169 was used to infect low-passage-number HFF cells at a low multiplicity of infection (MOI) (0.1 PFU/cell) in the presence of 0.01 μM CMX001. The culture was passaged in increasing concentrations of CMX001 up to a final concentration of 0.5 μM, which is well below the concentration needed (>30 μM) to reduce cell viability by 50% (CC50) (15). Infected monolayers were visually inspected twice a week, and the virus was harvested either when significant cytopathology was observed or 3 weeks following infection, whichever occurred first. The total passage time was 10 months, beyond which no virus could be recovered, as the concentrations of CMX001 exceeded 0.5 μM. The titer of virus resulting from passaging was determined at 3.5 × 104 PFU/ml, and the virus strain was designated CMX001R.
Plaque reduction assays.
HFF cells were plated in six-well plates and incubated at 37°C. When the cell layer reached confluence, the medium was aspirated from the wells, 0.2 ml of virus was added to each of three wells to yield 20 to 30 plaques per well, and 0.2 ml of medium was added to each of the three uninfected wells to test for drug toxicity. The plates were then incubated for 1 h with gentle rocking every 15 min after which the following drug concentration ranges were added to duplicate wells: CMX001 from 1 μM to 0.0003 μM, CDV from 100 μM to 0.03 μM, GCV from 100 μM to 0.03 μM, and FOS from 500 μM to 0.16 μM. After 8 days of incubation, cell monolayers were stained with 1% neutral red solution, the stain was aspirated, and plaques were counted using a stereomicroscope. The concentration of drug that reduced plaque formation by 50% (EC50) was determined by comparing drug-treated cultures with untreated cultures. Values from three to five independent experiments were used to calculate the mean and standard deviation values.
DNA sequencing.
The UL54 and UL97 genes were amplified from virus strain CMX001R using double-nested PCR methods in the following manner. A fragment of UL97 was amplified using the primers UL97 inner F1 (5′-TCC GCA CTT CGG TCT C-3′) and UL97 inner R1 (5′-AAC AGT TGG CGG CAG-3′) followed by a second amplification with the primers UL97 short F1 (5′-CTG AGT TCC GTC AGC A-3′) and UL97 short R1 (5′-GGT CCT CCT CGC AGA T-3′). Similarly, the UL54 open reading frame (ORF) was amplified using the primers UL54 For (For for forward) (5′-CGT AAG CTG TCA GCC TCT CA-3′) and UL54 Rev (Rev for reverse) (5′-CAG TCT CAG CAG CAT CAT CAC-3′), followed by primers UL54 Inner For (5′-CTC ACG GTC CGC TAT GTT TT-3′) and UL54 Inner Rev (5′-CGC TGT TTC TCA ACA GCA TTC-3′). All amplifications were done with High Fidelity PCR Master (Roche Applied Science, Indianapolis, IN). The resulting DNA fragments were purified via QIAquick gel extraction and PCR purification kits (Qiagen, Valencia, CA) and underwent conventional dideoxy chain termination sequencing at the University of Alabama at Birmingham Department of Genetics Core Sequencing Facility to identify any amino acid changes. DNA sequences were analyzed using Informax Vector NTI Contig Express 2003 (Invitrogen, Carlsbad, CA). Consensus DNA sequences were compared to that of the laboratory strain AD169 (GenBank accession number X17403).
Generation of recombinant viruses.
To further investigate a novel UL54 mutation identified during genotypic resistance analysis, the D542E mutation was reconstructed in the AD169 strain in the HB5 bacterial artificial chromosome (BAC) using methods similar to those previously reported (21). A plasmid containing UL54 with a kanamycin resistance marker (Kan) inserted immediately after the UL54 stop codon was constructed (UL54kan pEXP5 NT, designated pMP302) to facilitate mutagenesis. The D542E mutation was constructed by quick change mutagenesis (QuikChange II site-directed mutagenesis kit; Stratagene, La Jolla, CA) with primer UL54 D542E F (F for forward) (5′-CCG TTA CTG TCT GCA G-3′) and primer UL54 D542E R (R for reverse) (5′-CCA ATA CGG CCT CCT G-3′) and pMP302 as a template. The resulting clone, pMP547, was amplified using primers flanking the Kan marker and the UL54 locus, and the PCR product was electroporated into the SW102 recombineering strain containing the HB5 BAC and plated on selective medium containing kanamycin and chloramphenicol, and the resultant BAC was designated pMP573. In a similar manner, a wild-type UL54 Kan BAC, designated pMP556, was created from the HB5 BAC using an equivalent PCR product from pMP302 but with no engineered mutations.
HindIII restriction digested products of all BACs were performed to ensure that no large rearrangements had occurred. UL54 ORFs from each BAC were sequenced to confirm that they had only the engineered mutation. As intended, pMP573 contained only the D542E mutation, while pMP556 contained no UL54 mutations.
Recombinant mutant viruses were reconstituted from pMP573 and pMP556 BACs via transfection into HFF cells and were designated RC573 and RC556, respectively. These recombinant viruses underwent plaque reduction assay as described above to determine EC50s against CMX001 and control compounds. The UL54 Kan wild-type recombinant virus (RC556) was used as a control for phenotypic studies of RC573.
Replication kinetics.
Monolayers of HFF cells in duplicate 12-well plates were infected at an MOI of 0.01 PFU/cell with virus strain CMX001R or the D542E mutant (RC573), and the comparator viruses AD169 and UL54 Kan (RC556), respectively. Two hours and 4, 5, 6, 8, 10, 12, and 14 days following infection, the duplicate plates were frozen at −80°C for subsequent evaluation by real-time PCR. Total DNA was extracted by lysing the cells in a buffer containing 10 mM Tris (pH 7.4), 1 mM EDTA, and 1% SDS and incubating with 100 μg of proteinase K for 60 min at 37°C. DNA samples were purified using the QIAquick PCR purification kit according to the manufacturer's protocol. Genome copy number was determined on two independent cultures at each time point using primers 5′-AGG TCT TCA AGG AAC TCA GCA AGA-3′ and 5′-CGG CAA TCG GTT TGT TGT AAA-3′, with the probe 5′-(6-carboxyfluorescein [6FAM])-CCG TCA GCC ATT CTC TCG GC 6-carboxytetramethylrhodamine (TAMRA)-3′, and the plasmid pMP217 to provide absolute quantification. Data shown represent the average of the log of the genome copy number determined from two independent cultures at each point in time.
RESULTS
Selection of CMX001-resistant virus.
The generation of CMV strains resistant to CDV is typically slow, and this proved to be the case for CMX001 as well. Serial passage over a period of approximately 10 months was required to generate a viral isolate that replicated in the presence of 0.5 μM CMX001. This isolate, designated CMX001R, was evaluated for susceptibility to CMX001 and other relevant compounds (Table 1). The resistant isolate exhibited an EC50 for CMX001 that was 17-fold higher than that of the parent virus. This isolate was also resistant to CDV as expected, yet it remained fully susceptible to GCV and FOS.
Table 1.
Susceptibility of CMV strain CMX001R and a recombinant UL54 D542E mutant virus
| Antiviral agent | EC50 ± SD (μM)a |
Fold increaseb | EC50 ± SD (μM)a |
Fold increaseb | ||
|---|---|---|---|---|---|---|
| CMX001R | AD169 | D542E mutant (RC573) | Wild-type UL54 Kan (RC556) | |||
| CDV | 8.4 ± 5.5 | 1.1 ± 0.78 | 7.6c | 2.8 ± 0.62 | 0.23 ± 0.10 | 12c |
| CMX001 | 0.017 ± 0.003d | 0.001 ± 0.0005d | 17c | 0.026 ± 0.007 | 0.0008 ± 0.0007 | 32c |
| FOS | 59 ± 12d | 90 ± 21d | 0.66 | 34 ± 36 | 20 ± 15 | 1.7 |
| GCV | 4.3 ± 2.0d | 3.5 ± 0.55d | 1.2 | 9.6 ± 5.2e | 6.5 ± 8.7 | 1.5 |
Values represent the mean ± standard deviation values from four independent experiments unless otherwise noted.
The relative resistance compared to the parent virus is shown as the fold increase in the EC50.
Significantly different (P < 0.05).
Values (means ± standard deviations) for three separate experiments.
Values (means ± standard deviations) for five separate experiments.
Genotypic resistance analysis.
To identify the molecular determinants of resistance in virus strain CMX001R, the genes encoding the CMV UL97 kinase and DNA polymerase were sequenced. No mutations were observed in UL97, but a single nucleotide mutation leading to a change in the inferred amino acid sequence (D542E) was identified in UL54. The D542E mutation is interesting, because it has not been previously reported, is located within the conserved UL54 δC/exonuclease III (ExoIII) domain (7), and is proximal to a known mutation (L545S) that confers resistance to both CDV and GCV (22). Thus, it was considered highly probable that the UL54 D542E mutation would be sufficient to confer reduced susceptibility to CDV and CMX001.
Recombinant phenotyping of the novel mutation.
To confirm these data, the D542E mutation was reconstructed in plasmid pMP547, which was used to transfer the mutation to the HB5 BAC using a kanamycin selectable marker, and the resulting BAC was designated pMP573. DNA sequencing of the UL54 ORF of pMP573 confirmed the presence of the engineered D542E mutation and no other mutations. This BAC was then used to regenerate a recombinant CMV strain carrying the UL54 D542E mutation. This recombinant virus was also sequenced to confirm that D542E was the only change in the UL54 ORF. This D542E mutant strain was designated RC573. Because RC573 carries the Kan selective marker near UL54, another recombinant virus with the Kan marker in the same locus but without the D542E mutation in UL54 was engineered from the same HB5 parent BAC (designated RC556) and used as a wild-type control for phenotypic studies of the recombinant D542E mutant virus.
In a series of plaque reduction assays, the D542E mutant virus, RC573, recapitulated the resistance phenotype of isolate CMX001R, as shown in Table 1, with reduced susceptibility to CDV and CMX001, but no resistance to GCV or FOS. EC50s for the D542E mutant strain demonstrated reductions in susceptibility of greater than 30-fold to CMX001 and greater than 10-fold to CDV compared to the parent wild-type UL54 Kan isolate, RC556.
Assessment of replication kinetics.
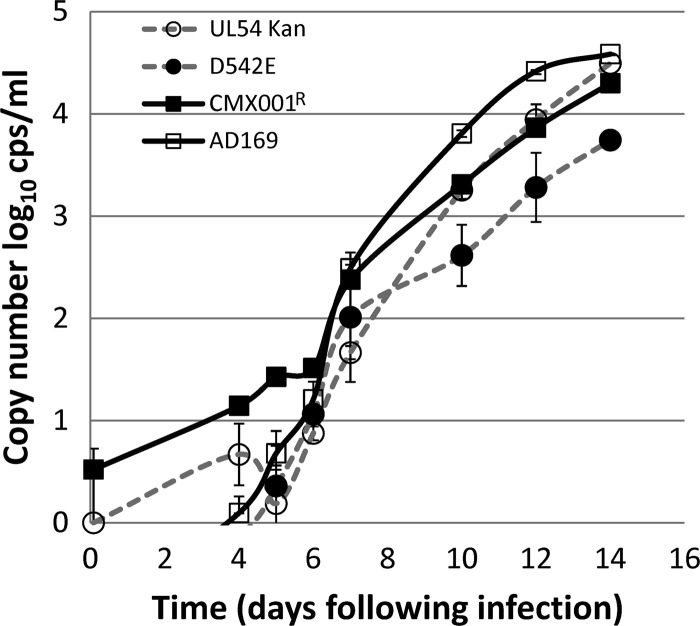
Both the isolate CMX001R and D542E mutant isolates exhibited a small-plaque phenotype that is an indicator of impaired replication kinetics in some CMV strains. Further investigations were carried out to evaluate the replicative fitness of each strain in comparison to its respective parent virus (Fig. 1). Cells were infected at a low MOI and were evaluated by real-time PCR from 2 h to 14 days to assess the replication kinetics of the viruses. For the first several days, few differences in replication were observed, but by day 10 following infection, cells infected with the virus strain CMX001R exhibited reduced levels of progeny DNA compared to the AD169 progenitor. The levels of virus strain CMX001R DNA were less than those of the parent virus on days 10, 12, and 14 and were statistically significant (P < 0.002 by paired Student's t test). Likewise, the recombinant virus with the D542E mutation produced lower levels of DNA starting at day 10 after infection compared to the comparator virus RC556 (wild-type UL54 Kan), which was also statistically significant (P < 0.001). Cells infected with either strain CMX001R or D542E mutant strain contained approximately 0.5 log10 copies/ml lower levels of progeny DNA compared to the comparator viruses on days 10, 12, and 14. These data indicate that the UL54 D542E mutation is sufficient to reduce the replicative fitness of viruses that carry this lesion, resulting in slower replication kinetics that are consistent with the small-plaque phenotype observed for both viruses containing this mutation.
Fig 1.
Replication kinetics. HFF cells infected at an MOI of 0.01 PFU/cell were harvested at the times indicated on the x axis. The average DNA copy number of two replicate cultures is plotted with standard deviation values indicated by the error bars. Virus strain CMX001R replicated more slowly than the AD169 strain from which it was derived. Similarly, the recombinant virus (RC573) containing the D542E mutation also replicated more slowly than did the wild-type UL54 Kan (RC556) comparator virus.
DISCUSSION
CMX001 has the potential to improve upon the efficacy and toxicity profiles of current CMV antiviral agents. Initial human studies indicate that it avoids the toxicities of myelosuppression and renal injury that often limit the use of GCV, CDV, and FOS (20). To date, emergence of resistant CMV has not been observed in clinical studies of CMX001 where prior anti-CMV therapy was limited or absent, limiting our knowledge of de novo resistance to CMX001 and the theoretical associated cross-resistance.
Here, we report the generation of a CMX001-resistant CMV strain under prolonged selective pressure in cell culture. Genotypic resistance analysis identified the presence of a single UL54 mutation (D542E), and this mutation was transferred to a control laboratory strain of CMV in order to assess antiviral susceptibility to CMX001 and cross-resistance to other CMV antivirals. These studies indicate that the D542E mutation in UL54 is sufficient to confer resistance to CMX001 and CDV, but it does not appear to confer GCV or FOS cross-resistance.
D542E has not been previously identified in clinical or laboratory strains of resistant CMV and is the first resistance mutation described as emerging de novo under CMX001 selective pressure, either in vivo or in vitro. It is located in the δC/ExoIII functional region of UL54, which is a conserved region found within the 3′-5′ exonuclease domain. This exonuclease domain is highly associated with amino acid changes known to confer GCV and CDV cross-resistance, as well as some FOS resistance (7). Since a majority of CDV resistance-associated mutations also confer cross-resistance to GCV and/or FOS, it is somewhat surprising that D542E does not. Taken together, the fact that D542E was not a previously known CDV-resistant mutation and that it varies from the typical cross-resistance profile of mutations in the exonuclease domain indicate that the antiviral pressure of CMX001 may drive the selection of resistance mutations that are distinct from those generated by other antiviral agents. This may be attributable to the substantially higher intracellular levels of the active antiviral achieved with CMX001 compared to CDV, which results from the lipid-facilitated cellular uptake of CMX001. A similar situation in which differing levels of antiviral drug exposure leads to distinct genotypic profiles has been described for the HIV protease inhibitor, fosamprenavir, where I54L/M is the predominant resistance mutation in regimens with low trough plasma drug concentrations, whereas a different mutation (I50V) predominates in those with a high trough plasma drug concentration (23).
The location of codon position 542 within the CMV DNA polymerase molecule may give insight into the mechanism of resistance for these resistant isolates. Although studies of another resistance mutation (K513N) within the δC/ExoIII region demonstrated that it severely restricted the exonuclease function of UL54 (22), 3-dimensional modeling of the CMV DNA polymerase enzyme suggests that some mutations within this region may actually enhance the proofreading activity of the exonuclease, thereby facilitating the excision of incorporated nucleotide analogs (24). It is theorized that more efficient excision of incorporated nucleoside-based antivirals from the elongating viral DNA chain produces resistance to that antiviral.
Compared to mutations in UL97, UL54 mutations are more likely to be associated with replicative deficits that reduce the growth rate of affected viruses (22, 25, 26). In our studies, the CMX001-resistant CMV strain generated in cell culture (strain CMX001R) and the recombinant D542E mutant virus (strain RC573) both exhibited small-plaque phenotypes that are often indicative of compromised viral growth kinetics (27). Replicative fitness assays confirmed our observation by showing that virus strains CMX001R and RC573 both demonstrated modest growth attenuation compared to their parent viruses. Not surprisingly, the UL54 Kan strain (RC566) from which RC573 was derived showed a slight decrease in replicative fitness compared to unmodified AD169, likely due to the Kan insert. Increased variation in plaque reduction assays was also noted with both virus strains CMX001R and RC573, and we hypothesize that this is also associated with the D542E mutation. A growth deficiency of this magnitude would be predicted to lead to a minor replicative disadvantage in a mixed virus population, but it could allow for clonal expansion of the resistant population under selective pressure from an antiviral agent.
In conclusion, we have generated the first de novo CMX001-resistant CMV strain under prolonged in vitro selective pressure, identified a novel UL54 mutation (D542E), and demonstrated with recombinant phenotyping techniques that this mutation confers resistance to CMX001 and CDV, but not to GCV or FOS. No other UL54 mutations and no UL97 mutations were found. As is common with UL54 resistance mutations, D542E is associated with reduced viral growth in cell culture. Clinical trials of CMX001 in high-risk patient populations should continue surveillance aimed at defining the potential for emerging CMX001 resistance, noting that it may be associated with previously unrecognized UL54 mutations.
ACKNOWLEDGMENTS
These studies were funded in whole or in part with federal funds from National Institutes of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contracts N01-AI-30049 and HHSN2722011000010C.
Footnotes
Published ahead of print 6 May 2013
REFERENCES
- 1. Torres-Madriz G, Boucher HW. 2008. Immunocompromised hosts: perspectives in the treatment and prophylaxis of cytomegalovirus disease in solid-organ transplant recipients. Clin. Infect. Dis. 47:702–711 [DOI] [PubMed] [Google Scholar]
- 2. Tuthill M, Chen F, Paston S, De La Pena H, Rusakiewicz S, Madrigal A. 2009. The prevention and treatment of cytomegalovirus infection in haematopoietic stem cell transplantation. Cancer Immunol. Immunother. 58:1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kedhar SR, Jabs DA. 2007. Cytomegalovirus retinitis in the era of highly active antiretroviral therapy. Herpes 14:66–71 [PubMed] [Google Scholar]
- 4. Dollard SC, Grosse SD, Ross DS. 2007. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev. Med. Virol. 17:355–363 [DOI] [PubMed] [Google Scholar]
- 5. Andrei G, De Clercq E, Snoeck R. 2009. Drug targets in cytomegalovirus infection. Infect. Disord. Drug Targets 9:201–222 [DOI] [PubMed] [Google Scholar]
- 6. Price NB, Prichard MN. 2011. Progress in the development of new therapies for herpesvirus infections. Curr. Opin. Virol. 1:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lurain NS, Chou S. 2010. Antiviral drug resistance of human cytomegalovirus. Clin. Microbiol. Rev. 23:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. James SH, Prichard MN. 2011. The genetic basis of human cytomegalovirus resistance and current trends in antiviral resistance analysis. Infect. Disord. Drug Targets. 11:504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prichard MN, Kern ER. 2011. The search for new therapies for human cytomegalovirus infections. Virus Res. 157:212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beadle JR, Hartline C, Aldern KA, Rodriguez N, Harden E, Kern ER, Hostetler KY. 2002. Alkoxyalkyl esters of cidofovir and cyclic cidofovir exhibit multiple-log enhancement of antiviral activity against cytomegalovirus and herpesvirus replication in vitro. Antimicrob. Agents Chemother. 46:2381–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hostetler KY. 2009. Alkoxyalkyl prodrugs of acyclic nucleoside phosphonates enhance oral antiviral activity and reduce toxicity: current state of the art. Antiviral Res. 82:A84–A98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aldern KA, Ciesla SL, Winegarden KL, Hostetler KY. 2003. Increased antiviral activity of 1-O-hexadecyloxypropyl-[2-14C]cidofovir in MRC-5 human lung fibroblasts is explained by unique cellular uptake and metabolism. Mol. Pharmacol. 63:678–681 [DOI] [PubMed] [Google Scholar]
- 13. Xiong X, Smith JL, Kim C, Huang ES, Chen MS. 1996. Kinetic analysis of the interaction of cidofovir diphosphate with human cytomegalovirus DNA polymerase. Biochem. Pharmacol. 51:1563–1567 [DOI] [PubMed] [Google Scholar]
- 14. Xiong X, Smith JL, Chen MS. 1997. Effect of incorporation of cidofovir into DNA by human cytomegalovirus DNA polymerase on DNA elongation. Antimicrob. Agents Chemother. 41:594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Williams-Aziz SL, Hartline CB, Harden EA, Daily SL, Prichard MN, Kushner NL, Beadle JR, Wan WB, Hostetler KY, Kern ER. 2005. Comparative activities of lipid esters of cidofovir and cyclic cidofovir against replication of herpesviruses in vitro. Antimicrob. Agents Chemother. 49:3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wan WB, Beadle JR, Hartline C, Kern ER, Ciesla SL, Valiaeva N, Hostetler KY. 2005. Comparison of the antiviral activities of alkoxyalkyl and alkyl esters of cidofovir against human and murine cytomegalovirus replication in vitro. Antimicrob. Agents Chemother. 49:656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bidanset DJ, Beadle JR, Wan WB, Hostetler KY, Kern ER. 2004. Oral activity of ether lipid ester prodrugs of cidofovir against experimental human cytomegalovirus infection. J. Infect. Dis. 190:499–503 [DOI] [PubMed] [Google Scholar]
- 18. Kern ER, Collins DJ, Wan WB, Beadle JR, Hostetler KY, Quenelle DC. 2004. Oral treatment of murine cytomegalovirus infections with ether lipid esters of cidofovir. Antimicrob. Agents Chemother. 48:3516–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ciesla SL, Trahan J, Wan WB, Beadle JR, Aldern KA, Painter GR, Hostetler KY. 2003. Esterification of cidofovir with alkoxyalkanols increases oral bioavailability and diminishes drug accumulation in kidney. Antiviral Res. 59:163–171 [DOI] [PubMed] [Google Scholar]
- 20. Painter W, Robertson A, Trost LC, Godkin S, Lampert B, Painter G. 2012. First pharmacokinetic and safety study in humans of the novel lipid antiviral conjugate CMX001, a broad-spectrum oral drug active against double-stranded DNA viruses. Antimicrob. Agents Chemother. 56:2726–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD, Kern ER. 2008. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J. Virol. 82:5054–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cihlar T, Fuller MD, Mulato AS, Cherrington JM. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382–393 [DOI] [PubMed] [Google Scholar]
- 23. Paulsen D, Elston R, Snowden W, Tisdale M, Ross L. 2003. Differentiation of genotypic resistance profiles for amprenavir and lopinavir, a valuable aid for choice of therapy in protease inhibitor-experienced HIV-1-infected subjects. J. Antimicrob. Chemother. 52:319–323 [DOI] [PubMed] [Google Scholar]
- 24. Shi R, Azzi A, Gilbert C, Boivin G, Lin SX. 2006. Three-dimensional modeling of cytomegalovirus DNA polymerase and preliminary analysis of drug resistance. Proteins 64:301–307 [DOI] [PubMed] [Google Scholar]
- 25. Chou S, Lurain NS, Thompson KD, Miner RC, Drew WL. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32–39 [DOI] [PubMed] [Google Scholar]
- 26. Chou S, Marousek GI, Van Wechel LC, Li S, Weinberg A. 2007. Growth and drug resistance phenotypes resulting from cytomegalovirus DNA polymerase region III mutations observed in clinical specimens. Antimicrob. Agents Chemother. 51:4160–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prichard MN, Gao N, Jairath S, Mulamba G, Krosky P, Coen DM, Parker BO, Pari GS. 1999. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 73:5663–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]