Abstract
This study describes three novel erm(T)-carrying multiresistance plasmids that also harbor cadmium and copper resistance determinants. The plasmids, designated pUR1902, pUR2940, and pUR2941, were obtained from porcine and human methicillin-resistant Staphylococcus aureus (MRSA) of the clonal lineage ST398. In addition to the macrolide-lincosamide-streptogramin B (MLSB) resistance gene erm(T), all three plasmids also carry the tetracycline resistance gene tet(L). Furthermore, plasmid pUR2940 harbors the trimethoprim resistance gene dfrK and the MLSB resistance gene erm(C), while plasmids pUR1902 and pUR2941 possess the kanamycin/neomycin resistance gene aadD. Sequence analysis of approximately 18.1 kb of the erm(T)-flanking region from pUR1902, 20.0 kb from pUR2940, and 20.8 kb from pUR2941 revealed the presence of several copies of the recently described insertion sequence ISSau10, which is probably involved in the evolution of the respective plasmids. All plasmids carried a functional cadmium resistance operon with the genes cadD and cadX, in addition to the multicopper oxidase gene mco and the ATPase copper transport gene copA, which are involved in copper resistance. The comparative analysis of S. aureus RN4220 and the three S. aureus RN4220 transformants carrying plasmid pUR1902, pUR2940, or pUR2941 revealed an 8-fold increase in CdSO4 and a 2-fold increase in CuSO4 MICs. The emergence of multidrug resistance plasmids that also carry heavy metal resistance genes is alarming and requires further surveillance. The colocalization of antimicrobial resistance genes and genes that confer resistance to heavy metals may facilitate their persistence, coselection, and dissemination.
INTRODUCTION
Studies of the resistance genes present in livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) of multilocus sequence type (ST) 398 identified a certain heterogeneity of genes encoding the same resistance phenotype (1). This was particularly evident for genes conferring combined resistance to macrolides, lincosamides, and streptogramin B antibiotics (MLSB) (2–8). The major mechanism of resistance to MLSB is the methylation of the adenine at position A2058 in domain V of 23S rRNA. So far, at least four rRNA methylase genes, erm(A), erm(B), erm(C), and erm(T), have been described in LA-MRSA ST398 (1). Among them, the erm(T) gene, although initially reported in other Gram-positive bacteria, such as lactobacilli, streptococci, and enterococci (9–11), has been described for the first time in staphylococci on plasmid pKKS25 in a porcine LA-MRSA ST398 strain (12). This gene has also been detected in LA-MRSA ST398 strains from cattle (3) and food/food products of poultry origin (4). Its presence among MRSA ST398 of human origin has also been described (8). A study of erm genes in livestock manure and manure management systems found erm(T) at a high frequency in bovine and swine manure (13). Whenever erm(T) was detected in MRSA, the corresponding strains showed constitutive resistance to clindamycin, implying the presence of structural alterations in the erm(T)-associated translational attenuator (5, 8, 12).
Recent studies have also revealed the presence of the erm(T) gene among methicillin-susceptible S. aureus (MSSA) ST398 strains (usually of spa type t571) in humans who had no contact with livestock (14–18), and in some cases they were involved in serious infections in humans (19–21). In these isolates, the erm(T) genes have been shown to confer an inducible clindamycin resistance phenotype and to be located in the chromosomal DNA (15, 16). Very recently, the first erm(T)-carrying resistance plasmid, namely, pUR3912, from an MSSA isolate of human origin was described (22). This plasmid carried a functionally active cadmium resistance operon (cadDX) and represented the first erm(T)-carrying plasmid that also harbored heavy metal resistance determinants. Despite the fact that cadmium is a highly toxic metal that is neither used in agriculture nor found in the community or in the hospital sector, the presence of cadmium resistance determinants on staphylococcal plasmids has been described before (23, 24). In contrast, copper and zinc compounds are commonly used as feed supplements in livestock (25), and zinc resistance has been recently identified as part of type V SCCmec cassettes, which are commonly found in LA-MRSA ST398 (26). Thus, it has been assumed that zinc resistance plays a role in the coselection and emergence of methicillin resistance in LA-MRSA ST398 of animal origin.
This study describes novel staphylococcal multiresistance plasmids that also carry cadmium and copper resistance determinants. The aim of the present study was to determine the location and the genetic environment of the resistance genes on erm(T)-carrying multiresistance plasmids obtained from five LA-MRSA ST398 strains of porcine and human origin recently detected in Spain (5, 8).
MATERIALS AND METHODS
Bacterial strains investigated and molecular typing.
Five erm(T)-positive MRSA ST398 strains identified in previous studies were included (5, 8). Strains C1902, C1905, and C1906 were isolated from healthy pigs and presented the agr allotype I, spa type t011, and SCCmec IVa, while strains C2940 and C2941 came from humans with different diseases, showed agr allotype I, spa type t011, and harbored the SCCmec type V (5, 8). All strains were multiresistant (Table 1). To estimate the clonal relatedness of the strains, pulsed-field gel electrophoresis (PFGE) of total DNA after digestion with ApaI (Roche Pharma, Madrid, Spain) was performed by following the HARMONY protocol (27). ApaI fragments were separated for 20 h at 6 V/cm using pulse time ramping from 2 to 5 s (6).
Table 1.
Comparative analysis of the MICs of the original erm(T)-carrying MRSA strains, S. aureus RN4220, and S. aureus RN4220 transformants carrying the three novel erm(T)-positive plasmidsd
| Bacterial strain | Resistance genes detected for: |
MIC (μg/ml) of antimicrobial agentb |
MIC (mM) of metal |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial agentsa | Metal compounds | PEN | OXA | ERY | CLI | TIA | TET | GEN | KAN | NEO | SPE | TMP | CdSO4 | CuSO4 | ZnCl2 | |
| RN4220 | 0.03 | 0.12 | 0.5 | 0.12 | 0.5 | 0.5 | 0.25 | 4 | 0.5 | 64 | 1 | 0.015 | 8 | 1 | ||
| C1902c | blaZ, mecA, erm(T), tet(L), tet(M), aadD, aacA-aphD | cadD, cadX, mco, copA | 16 | ≥16 | ≥64 | ≥128 | 1 | 64 | 8 | ≥512 | 128 | 128 | 2 | 0.25 | 8 | 2 |
| RN4220/pUR1902 | erm(T), tet(L), aadD | cadD, cadX, mco, copA | ≤0.015 | 0.12 | ≥64 | ≥128 | 0.25 | 64 | 0.25 | 32 | 16 | 64 | 0.5 | 0.125 | 16 | 1 |
| C2940 | blaZ, mecA, erm(T), erm(C), vga(A), tet(L), tet(M), dfrK, aacA-aphD | cadD, cadX, Δmco, copA, czrC | 8 | 8 | ≥64 | ≥128 | ≥128 | 64 | 128 | ≥512 | 2 | 128 | ≥256 | 0.5 | 16 | 8 |
| RN4220/pUR2940 | erm(T), erm(C), tet(L), dfrK | cadD, cadX, Δmco, copA | ≤0.015 | 0.06 | ≥64 | ≥128 | 0.5 | 64 | 0.25 | 4 | 0.5 | 64 | ≥256 | 0.125 | 8 | 1 |
| C2941 | blaZ, mecA, erm(T), erm(C), tet(L), tet(M), aadD | cadD, cadX, mco, copA, czrC | 16 | ≥16 | ≥64 | ≥128 | 0.5 | 64 | 0.5 | 64 | 64 | ≥512 | 0.25 | 0.5 | 16 | 8 |
| RN4220/pUR2941 | erm(T), tet(L), aadD | cadD, cadX, mco, copA | ≤0.015 | 0.12 | ≥64 | ≥128 | 0.25 | 64 | 0.25 | 32 | 32 | 64 | 0.5 | 0.125 | 16 | 1 |
The antimicrobial resistance genes present in the original strains C1902, C2940, and C2941 were described in previous studies (5, 8).
PEN, penicillin; OXA, oxacillin; ERY, erythromycin; CLI, clindamycin; TIA, tiamulin; TET, tetracycline; GEN, gentamicin; KAN, kanamycin; NEO, neomycin; SPE, spectinomycin; TMP, trimethoprim.
Strains C1905 and C1906 showed the same profile to the antimicrobials and metal compounds tested.
Phenotypes considered resistant to the respective antimicrobials and those tentatively considered resistant to the respective metals are indicated by grey shading.
Isolation and transfer of erm(T)-carrying plasmids.
Plasmids were extracted and purified using a modified alkaline lysis method (12). Obtained plasmids were transformed by protoplast transformation into S. aureus RN4220 with subsequent selection on regeneration plates containing erythromycin (15 μg/ml). The presence of the erm(T) gene in the transformants was confirmed by a specific PCR (Table 2). The approximate sizes of the transformed plasmids pUR1902, pUR2940, and pUR2941 were calculated as the sum of the fragment sizes obtained after digestion of the plasmids with the restriction endonucleases EcoRI and BglII (Roche) in independent experiments.
Table 2.
Primers and PCR conditions employeda
| Gene or region amplified | Primer designation | Primer sequence (5′→3′) | Nucleotide position in published sequence | Amplicon size (bp) | Reference | GenBank accession nos.b |
|---|---|---|---|---|---|---|
| erm(T)c | ermT_fw | ATTGGTTCAGGGAAAGGTCA | 109–128 in erm(T) | 536 | 3 | HF583290, HF583291, HF583292 |
| ermT_rv | GCTTGATAAAATTGGTTTTTGGA | 622–644 in erm(T) | ||||
| cadD | cadD-fw2 | TGCTAGAGCAAAGACTAGGAAAGA | 93–116 in cadD (81–104 in cadD of pUR3912) | 460 | This study | HF583290, HF583291, HF583292, HE805623 |
| cadD-rv | AGCCATAATCCAACGACCAA | 533–552 in cadD (521–540 in cadD of pUR3912) | ||||
| cadX | cadX-fw | TGCTTGTGATGTGATCTGTGT | 15–35 in cadX | 213 | This study | HF583290, HF583291, HF583292, HE805623 |
| cadX-rv | TGATGTGAAGTTGAAGCAACAC | 206–227 in cadX | ||||
| Promoter cadDX | Pr_cadDX-fw | CTGACGATGCCAGGAAACTT | 438–457 upstream of cadD | 579, 419d | This study | HF583290, HF583291, HF583292, HE805623 |
| Pr_cadDX(pUR3912)-fw | AGTAAGGGTGCAGTGCCAAT | 290–309 upstream of cadD of pUR3912 | ||||
| Pr_cadDX-rv2 | CGATATTCTTTCCTAGTCTTTGCTC | 98–122 in cadD (86–110 in cadD of pUR3912) | ||||
| copA | copA-fw | CATGCTTTAGGCTTGGCAAT | 931–950 in copA | 662 | This study | HF583290, HF583291, HF583292 |
| copA-rv | TCTTCTGGCATGAGTTGTGC | 1573–1592 in copA | ||||
| mco | mco-fw | TCCCTCCCCAAATACAGCTA | 537–556 in mco | 699 | This study | HF583290, HF583291, HF583292 |
| mco-rv | GTTCCGTGGATATGGAATGG | 1216–1235 in mco | ||||
| czrC | czrC-fw | TAGCCACGATCATAGTCATG | 48–67 in czrC | 655 | 26 | JCSC6944 |
| czrC-rv | ATCCTTGTTTTCCTTAGTGACTT | 680–702 in crzC |
Primers and PCR conditions employed in this study to detect genetic determinants for resistance to the different metal compounds detected in the three novel plasmids described in the study and those detected in the recently described pUR3912, as well as those for the MLSB resistance gene erm(T). The PCRs were performed using BioTaq DNA polymerase (Bioline; Cultek, Madrid, Spain) and the following conditions: initial cycle of 3 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 56°C [45°C for the erm(T) gene], and 1 min at 72°C, and with a final step of 5 min at 72°C.
Published sequences from which primers were designed and/or coordinates have been established.
It should be noted that this pair of primers could not properly amplify the erm(T) gene of a set of erm(T)-positive MSSA isolates of the sublineage ST398, which seems to be associated with humans.
Amplicon size obtained using the combination of Pr_cadDX(pUR3912)-fw and Pr_cadDX-rv2.
Cloning of erm(T) and flanking regions from plasmids pUR1902, pUR2940, and pUR2941.
Plasmids were digested with EcoRI (for pUR1902 and pUR2941) and by EcoRI and BglII (for pUR2940). The corresponding fragments were cloned into the plasmid vector pBluescript II SK(+) (Stratagene, Amsterdam, The Netherlands), and recombinant plasmids were transformed into Escherichia coli JM101. The cloned fragments of interest were sequenced by primer walking on both strands, starting with M13 universal and reverse primers (22). Linkage between sequenced fragments as well as determination of DNA regions that could not be cloned in repeated experiments were performed by PCR mapping. For this, primers were designed from the sequences of already known segments deposited in the GenBank database and the amplicons were sequenced.
Antimicrobial susceptibility testing.
MICs for the antimicrobial agents listed in Table 1 were determined for the original strains and their S. aureus RN4220 transformants by broth microdilution (28) using custom-made microtiter plates (MCS Diagnostics, Swalmen, The Netherlands). S. aureus ATCC 29213 served as a quality control strain.
MIC determinations for cadmium, copper, and zinc compounds and testing of the resistance genes involved.
The MIC for cadmium sulfate (CdSO4; Panreac, Barcelona, Spain) was determined by agar dilution in three independent assays on Mueller-Hinton (MH; Becton Dickinson, Madrid, Spain) agar plates, while the MICs for copper sulfate and zinc chloride (CuSO4 and ZnCl2; Scharlau, Barcelona, Spain) were likewise determined on cation-adjusted Mueller-Hinton II (MH-II; Becton Dickinson, Madrid, Spain) agar with the pH of the medium adjusted to 5.5 for ZnCl2 or to 7.4 for CuSO4 (25, 26). For this, the original strains C1902, C1905, C1906, C2940, and C2941, the recipient strain S. aureus RN4220, and also the isogenic S. aureus RN4220 transformants were used. Concentration ranges for CdSO4 were 0.001 to 2 mM, while those for CuSO4 and ZnCl2 were 0.125 to 128 mM (26). Plates were incubated for 20 h at 37°C under aerobic conditions. The presence of genes responsible for heavy metal resistance (cadmium, copper, zinc) was investigated by PCR and subsequent sequencing of the respective amplicons (Table 2).
Southern blotting for the possible chromosomal localization of erm(T) and associated genes.
Southern blot analysis of genomic DNA after previous digestion with endonuclease I-CeuI (New England BioLabs, Barcelona, Spain) was performed. Agarose plugs with genomic DNA of strains C1902, C2940, and C2941 were digested with I-CeuI (10 U) for 4 h at 37°C. Fragments were separated in a 1% (wt/vol) PFGE agarose gel (18 h, 6 V/cm, and 5–30 s at 14°C) and I-CeuI PFGE digests were transferred to a nylon membrane. Hybridization with probes for erm(T), cadDX, copA, mco, and the 16S rRNA gene and subsequent detection were conducted according to the manufacturer's recommendations (Roche, Madrid, Spain).
Nucleotide sequence accession numbers.
The nucleotide sequences of the sequenced parts of plasmids pUR2941 (20,776 bp), pUR1902 (18,126 bp), and pUR2940 (19,957 bp) have been deposited in the EMBL database under accession numbers HF583290, HF583291, and HF583292, respectively.
RESULTS AND DISCUSSION
PFGE profiles, antimicrobial resistance patterns, and plasmid profiles.
All five strains shared closely related ApaI PFGE profiles, with the porcine strains C1902, C1905, and C1906 exhibiting even indistinguishable fragment patterns (data not shown). MIC values determined for the original strains and their S. aureus RN4220 transformants are shown in Table 1. In addition to the previously described data (5, 8), strain C2940 also revealed resistance to tiamulin (MIC, ≥128 μg/ml), due to the presence of the vga(A) gene, and strain C2941 showed elevated MIC values for spectinomycin (≥512 μg/ml), although it was negative for the presence of the spectinomycin resistance gene spc.
On the basis of plasmid sizes, EcoRI and BglII restriction patterns, and antimicrobial resistance profiles, three different types of erm(T)-carrying plasmids were distinguished. Strains C1902, C1905, and C1906 of porcine origin carried plasmids of ∼22 kb that showed the same restriction patterns. As a representative, plasmid pUR1902 from strain C1902 was included in a further analysis; strain C2940 harbored the ∼25-kb plasmid pUR2940 and strain C2941 harbored the ∼33-kb plasmid pUR2941. MIC testing and PCR analysis of the transformants revealed that, in addition to MLSB resistance due to the erm(T) gene, transformants carrying plasmid pUR1902, pUR2940, or pUR2941 were tetracycline resistant and carried the tet(L) gene. In addition, plasmids pUR1902 and pUR2941 conferred resistance to kanamycin/neomycin and had an aadD gene, while plasmid pUR2940 conferred trimethoprim resistance via the gene dfrK. Moreover, plasmid pUR2940 harbored the additional MLSB resistance gene erm(C) (Table 1).
Analysis of the novel pUR1902, pUR2940, and pUR2941 plasmids.
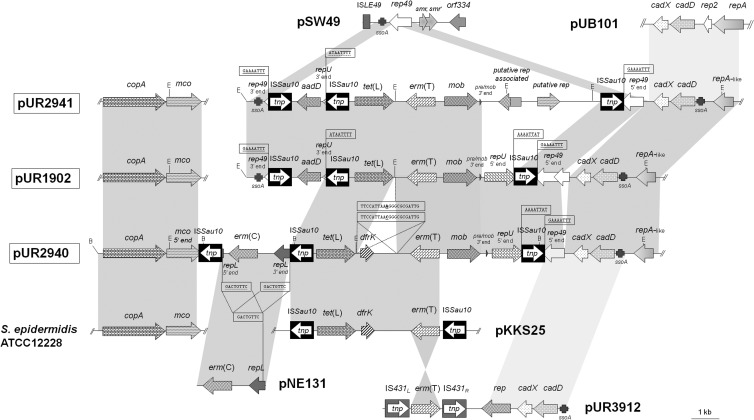
Figure 1 shows maps of the sequenced parts of the three novel multiresistance plasmids in comparison to previously known staphylococcal plasmids that share significant structural similarities. All three plasmids, pUR1902, pUR2940, and pUR2941, had in common the resistance genes erm(T) and tet(L), a plasmid mobilization gene, mob, a 3′-truncated replication gene, rep49, the cadmium resistance cadDX operon, a repA-like gene (70.9% identity to repA of pUB101), and two copies of the insertion sequence ISSau10 (Fig. 1).
Fig 1.
Comparative schematic presentation of the sequenced segments of the three novel erm(T)-carrying plasmids (pUR2941 [accession number HF583290], pUR1902 [HF583291], and pUR2940 [HF583292]) described in this study and the previously reported plasmids pSW49 (AM040730), pUB101 (AY373761), pKKS25 (FN390947), pNE131 (M12730), and pUR3912 (HE805623), as well as a chromosomal fragment of S. epidermidis ATCC 12228 (AE015929). The arrows indicate the extents and directions of transcription of antimicrobial [erm(T), tet(L), aadD, dfrK] and metal (cadD, cadX, copA, mco) resistance genes as well as genes involved in replication (rep49, repU, repL repA-like, putative rep-associated, putative rep, repA, and rep2), mobilization (mob), and others (ISLE49, smr, smr′, orf334). The 5′ and 3′ ends of the truncated rep49, repU, and repL genes and the 5′ end of the truncated mco gene in pUR2940 are likewise displayed. The different SSO regions detected are indicated as ssoA. The ISSau10 and IS431 copies are shown as black or gray boxes with a white arrow indicating the transposase gene tnp. The 8-bp direct target site duplications at the extremes of ISSau10 are shown in boxes and underlined. The 21-bp sequences of the integration site of the dfrK-carrying region are also boxed. The regions of >90% homology are shown in dark gray, while those between 80 and 90% similarity are displayed in light gray. The EcoRI (E) and BglII (B) cleavage sites are indicated. A size scale in kilobases is displayed in the lower right corner.
Plasmid pUR2940 exhibited a 5,436-bp resistance region that was 99.9% identical to that of plasmid pKKS25 (12) (Fig. 1). This region comprised the resistance genes erm(T), tet(L), and the dfrK, in addition to one ISSau10 copy. A pair of 21-bp imperfect direct duplications (5′-TTCCATTAAC/AGGGCGCGATTG-3′ [the two underlined bases are the ones exchanged in the different sequences]) was found that flanked the 1,590-bp region that comprised the dfrK gene (Fig. 1). As previously suggested (29), this sequence might have served for the integration of the dfrK region into pUR2940. Immediately downstream of the aforementioned ISSau10, a region delimited by another ISSau10 copy was observed. The region between both IS elements was 98.9% identical to the small erm(C)-carrying plasmid pNE131 (Fig. 1). The repL gene of this small plasmid was interrupted by two ISSau10 elements that were located in the same orientation. An 8-bp target site duplication, 5′-GACTGTTC-3′, was detected upstream of the left-hand ISSau10 and downstream of the right-hand ISSau10 (Fig. 1). The erm(C) gene was located between the two repL segments. This region exhibited the typical structural characteristics for the integration of a small plasmid into a larger one via ISs (24). Immediately downstream of this segment, the copper resistance genes copA and Δmco, whose 3′ end was truncated by ISSau10, were detected (Fig. 1).
Two discontinuous regions were sequenced in plasmids pUR1902 and pUR2941: (i) erm(T)-carrying segments of 14,626 bp and 17,276 bp, respectively, obtained from several contiguous EcoRI fragments, and (ii) a copA- and mco-carrying segment of 3,500 bp obtained by PCR mapping (Fig. 1). Despite extensive attempts, these two regions could not be joined by PCR mapping. Except for a single mismatch, the copA-mco segments were identical to that of pUR2940 but harbored the complete mco gene. At the left terminus of the erm(T)-flanking region of both plasmids, a region of 2,702 bp was detected that was absent in pUR2940. This small fragment carried another ISSau10 copy, whose upstream region showed the highest identity to a segment of the Staphylococcus warneri small plasmid pSW49, which includes a single-strand origin of replication, ssoA, and the 3′ end of rep49 (Fig. 1). The region downstream of the ISSau10 contained the aadD gene and the 3′ end of repU, which were identical to the corresponding regions of plasmid pKKS825 from MRSA ST398 (30) (Fig. 1). In addition, plasmid pUR2941 exhibited a unique segment of 4,140 bp in which two putative rep genes of 744 bp and 726 bp were detected.
Neither the erm(T) gene nor the cadmium nor copper resistance determinants (cadD, cadX, copA, or mco) detected were present on any of the chromosomal DNA fragments obtained after I-CeuI digestion of C1902, C2940, and C2941 genomic DNA. In contrast, the 16S rRNA gene probe yielded the expected positive hybridization results for all chromosomal DNA bands visualized in all three strains (data not shown).
Role of IS elements in the development of novel multiresistance plasmids.
ISSau10-related IS257 and IS431 have been shown to be responsible for the cointegration of small plasmids within larger plasmids or within the chromosomal DNA (24, 31, 32). The different copies of ISSau10 disrupted diverse rep genes of small plasmids as well as the mco gene of pUR2940. It has been suggested that the truncation of the rep genes in the cointegration process of different plasmids is important for the maintenance of the replication system of the original replicon (24). In addition, the presence of two ISSau10 copies in the same orientation with the typical 8-bp direct target site duplications at both ends and bracketing a sequence that closely resembles a small erm(C)-carrying plasmid in pUR2940 strongly suggests that its integration was ISSau10 mediated. IS elements play an important role in the evolution of multidrug resistance plasmids because of their capacity to integrate, split, and undergo homologous recombination with related IS elements. The presence of several copies of the recently described ISSau10 element in the sequenced regions of all three plasmids suggests that these ISSau10 copies play a role in the mosaic structure of plasmids pUR1902, pUR2940, and pUR2941.
Comparative analysis of the erm(T) gene and its immediate upstream and downstream regions.
Comparative analysis of the complete 735-bp erm(T) genes with those deposited in the GenBank/EMBL databases revealed that the erm(T) genes of plasmids pUR2940 and pUR2941 were identical to those of streptococcal plasmids and to that of plasmid pKKS25 of MRSA ST398. In contrast, the erm(T) gene present in plasmid pUR1902 showed a unique sequence that differed from the aforementioned erm(T) genes by a single nucleotide (A416G), resulting in the amino acid change Asn139Ser. A truncated translational attenuator region of erm(T) in the three novel plasmids was observed and showed the same 57-bp deletion as found on plasmid pKKS25 (12). Consequently, the three novel plasmids conferred constitutive clindamycin resistance.
Comparison of the up- and downstream sequences of the erm(T) genes identified a common region of ∼1.4 kb, including the erm(T) gene, that was embedded in largely nonhomologous segments in the erm(T)-carrying streptococcal plasmids and in the plasmids described in this study. Exactly at the junctions between these homologous and nonhomologous segments, imperfect direct repeats of 56 to 58 bp were detected. These imperfect direct repeats were also present up- (396 bp) and downstream (199 bp) of the erm(T) gene in the three novel plasmids (Fig. 2). It is likely that these imperfect direct repeats have played a role in the acquisition of erm(T) via recombination by erm(T)-free precursors of these plasmids. Interestingly, these regions were not present in the corresponding parts of pUR3912 or strain ST398NM01, what may suggest an alternative erm(T) acquisition pathway or subsequent divergence of such regions. Analysis of the erm(T)-lacking MRSA ST398 plasmid pKKS825, which also carries the aadD, tet(L), and dfrK genes, identified one such short region of 59 bp (30). This region in pKKS825 differed by only 1 bp from the 58-bp sequence found downstream of erm(T) in pUR1902, pUR2940, and pUR2941 (Fig. 2) and may represent a suitable acceptor site for an erm(T)-carrying segment.
Fig 2.
Sequence alignment of the homologous regions (56 to 59 bp) located upstream (A) and downstream (B) of the erm(T) gene in the three novel MRSA ST398 plasmids (pUR1902, pUR2940, and pUR2941), plasmid pGB2002 (Streptococcus agalactiae), pGB2001 (Streptococcus agalactiae), pGA2000 (Streptococcus pyogenes), p5580 (Streptococcus dysgalactiae), pRW35 (Streptococcus pyogenes), and a possible related precursor, pKKS825 (MRSA ST398) potentially used for the integration of an erm(T)-containing segment into the hypothetical pUR2940 erm(T)-free precursor. Displayed are nucleotides (nt) at the following up- and downstream positions of the erm(T) gene, respectively: nt 339 to 396 and 142 to 199 in pUR2940 (accession number HF583292); nt 524 to 581 and 142 to 197 in pRW35 (EU192194); nt 394 to 451 and 142 to 197 in pGB2002 (JF308629); nt 393 to 450 and 142 to 197 in pGB2001 (JF308630), pGA2000 (JF308631), and p5580 (HE862394); nt 1282 to 1340 downstream of the dfrK gene in pKKS825 (FN377602). Nucleotides in faint gray are those identical in all sequences, nucleotides in black show the identical bases specific for the upstream segments, and those in dark grey show those identical in all downstream regions.
Susceptibility to cadmium, copper, and zinc compounds and the presence of resistance determinants.
Table 1 shows the MIC data for CdSO4, CuSO4, and ZnCl2 of the aforementioned original strains and transformants. An 8-fold increase in the MIC for CdSO4 in S. aureus RN4220 transformants carrying pUR1902, pUR2940, or pUR2941 (0.125 mM) was observed compared with the S. aureus RN4220 strain (0.015 mM). Original strains and transformants carried the cadmium resistance operon cadDX, which consists of the cadD gene, encoding a 209-amino-acid (aa) P-type metal efflux pump involved in cadmium resistance, and cadX, which codes for a 115-aa protein that serves as a transcriptional regulator.
S. aureus RN4220 transformants carrying pUR1902 or pUR2941 exhibited a 2-fold increase in the MIC values for CuSO4, from 8 mM to 16 mM in comparison to the recipient strain. S. aureus RN4220 carrying pUR2940 did not show an increase in the MIC values for CuSO4 in comparison to the recipient strain. The three original strains and their transformants—but not S. aureus RN4220—harbored the copper transport gene copA, which codes for a 660-aa P1-type ATPase protein involved in copper resistance, in addition to the multicopper oxidase gene mco involved in copper homeostasis. However, accordingly, S. aureus RN4220/pUR2940 harbored a truncated Δmco gene, which seemed responsible for the absence of variation in the CuSO4 MIC values, regardless of the presence of the copA gene. Although the copA-mco genes of pUR1902 and pUR2941 increased the MIC values for copper by only one dilution, it should be noted that this MIC difference is in the high concentration range (16 mM corresponds to >2,000 μg/ml) and as such represents a substantial change in the copper MIC.
Strains C2940 and C2941 revealed 4- to 8-fold-higher MIC values for ZnCl2 (8 mM) in comparison with strain C1902, S. aureus RN4220, and the transformants carrying pUR2940, pUR2941, or pUR1902 (1 mM) (Table 1). Accordingly, the presence of the czrC gene, which codes for a 644-aa putative cadmium and zinc transporter, was evidenced by PCR and sequencing in the original S. aureus strains C2940 and C2941 but neither in C1902 nor in the three S. aureus RN4220 transformants.
Simultaneous presence of antimicrobial and metal resistance genes on staphylococcal plasmids.
The presence of cadmium resistance determinants in plasmids of S. aureus of different lineages is relatively common (23, 24, 33). Likewise, genes involved in copper resistance, especially copA and mco or variants (copB and copC), have been detected in different staphylococcal species and other Gram-positive bacteria, such as Listeria monocytogenes and Macrococcus caseolyticus (33–41). In LA-MRSA ST398, cadmium, copper, and other heavy metal resistance genes had been only detected in the chromosomal DNA, either within the SCCmecV(5C2&5)C, which carries the cadmium/zinc resistance gene czrC, or in the novel SCCmecIX and -X, which carry cadmium (cadDX), copper (copB-mco), and arsenic (arsRBC and arsDARBC) resistance elements (26, 42). The gene cluster copA-mco detected in the three plasmids showed the highest percentage of identity to that identified in the chromosomal DNA of Staphylococcus epidermidis ATCC 12228 (92.8%), Staphylococcus haemolyticus JCSC1435 (92.6%) and the novel SCCmecX and SCCmecIX elements of MRSA ST398 strains JCSC6945 (91.9%) and JCSC6943 (91.3%), respectively. The current study presents the first report of LA-MRSA ST398 harboring a plasmid that carries cadmium and copper resistance genes.
Elevated concentrations of cadmium in feedstuffs may occur due to the application of sewage sludge or phosphate fertilizers with high levels of cadmium into agricultural soils (43). Even though the accepted maximum levels of cadmium in feedstuffs for livestock are regulated (43), the possibility that such intake might be in part responsible for the selection of cadmium resistance determinants in the present bacterial population of pigs cannot be excluded. In humans, smoking cigarettes is an additional important source of cadmium (44). However, the major route of cadmium intake for the nonsmoking and non-occupationally exposed population is through ingestion of contaminated food (including food of animal origin) and water (43, 45). This exposure might also select for cadmium resistance in transferable elements in the human S. aureus population. In contrast, copper and zinc are essential trace elements for animals and humans (46, 47). They are added as feed supplements for livestock, particularly pigs, in great quantities to increase the daily growth rate of piglets (up to 8 to 10 weeks of age), to prevent gastrointestinal infections, and to limit and control cases of postweaning wanting and wasting (26, 28, 46, 47). The presence and maintenance of genetic elements that carry genes for copper and zinc resistance/tolerance in LA-MRSA ST398 are most probably favored by the extensive use of copper and zinc in pig production.
Staphylococcal plasmids that contain antimicrobial and metal resistance genes have been known for a long time. Plasmids of the pI258 type (GenBank accession no. NC_013319) have been disseminated in S. aureus ST30 (48) and contain, in addition to the β-lactamase gene blaZ and the MLSB resistance gene erm(B), genes conferring resistance to cadmium (cadA and cadC), arsenic (arsRBC), and mercury (mer operon), as well as copies of the IS431 element. In the current study, all three novel plasmids conferred a multidrug resistance phenotype, carried tetracycline and MLSB resistance genes, as well as kanamycin/neomycin and/or trimethoprim resistance genes. Due to the elevated use of antimicrobial agents, in particular tetracyclines, macrolides, lincosamides, and aminoglycosides (1, 49) in veterinary medicine and food animal production, the colocation of antimicrobial resistance and heavy metal resistance genes on the same plasmid may favor their maintenance and dissemination under the selective pressure imposed by the use of either antimicrobial agents or heavy metals.
ACKNOWLEDGMENTS
The work conducted at the University of La Rioja was financially supported by Projects SAF2009-08570 and SAF2012-35474 from the Ministerio de Economía y Competitividad of Spain. The work conducted at the Friedrich-Loeffler-Institut was financially supported by the German Federal Ministry of Education and Research (BMBF) through the German Aerospace Center (DLR), grant number 01KI1014D (MedVet-Staph). Elena Gómez-Sanz is supported by a fellowship from the Gobierno de La Rioja of Spain.
We thank Kerstin Meyer and Vivian Hensel for excellent technical assistance.
Footnotes
Published ahead of print 29 April 2013
REFERENCES
- 1. Kadlec K, Feßler AT, Hauschild T, Schwarz S. 2012. Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 18:745–755 [DOI] [PubMed] [Google Scholar]
- 2. Argudín MA, Tenhagen BA, Fetsch A, Sachsenröder J, Käsbohrer A, Schroeter A, Hammerl JA, Hertwig S, Helmuth R, Bräunig J, Mendoza MC, Appel B, Rodicio MR, Guerra B. 2011. Virulence and resistance determinants of German Staphylococcus aureus ST398 isolates from nonhuman sources. Appl. Environ. Microbiol. 77:3052–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feßler A, Scott C, Kadlec K, Ehricht R, Monecke S, Schwarz S. 2010. Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65:619–625 [DOI] [PubMed] [Google Scholar]
- 4. Feßler AT, Kadlec K, Hassel M, Hauschild T, Eidam C, Ehricht R, Monecke S, Schwarz S. 2011. Characterization of methicillin-resistant Staphylococcus aureus isolates from food and food products of poultry origin in Germany. Appl. Environ. Microbiol. 77:7151–7157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gómez-Sanz E, Torres C, Lozano C, Fernández-Pérez R, Aspiroz C, Ruiz-Larrea F, Zarazaga M. 2010. Detection, molecular characterization, and clonal diversity of methicillin-resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathog. Dis. 7:1269–1277 [DOI] [PubMed] [Google Scholar]
- 6. Kadlec K, Ehricht R, Monecke S, Steinacker U, Kaspar H, Mankertz J, Schwarz S. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156–1164 [DOI] [PubMed] [Google Scholar]
- 7. Lozano C, López M, Gómez-Sanz E, Ruiz-Larrea F, Torres C, Zarazaga M. 2009. Detection of methicillin-resistant Staphylococcus aureus ST398 in food samples of animal origin in Spain. J. Antimicrob. Chemother. 64:1325–1326 [DOI] [PubMed] [Google Scholar]
- 8. Lozano C, Rezusta A, Gómez P, Gómez-Sanz E, Báez N, Martin-Saco G, Zarazaga M, Torres C. 2012. High prevalence of spa types associated with the clonal lineage CC398 among tetracycline-resistant methicillin-resistant Staphylococcus aureus strains in a Spanish hospital. J. Antimicrob. Chemother. 67:330–334 [DOI] [PubMed] [Google Scholar]
- 9. Tannock GW, Luchansky JB, Miller L, Connell H, Thode-Andersen S, Mercer AA, Klaenhammer TR. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid 31:60–71 [DOI] [PubMed] [Google Scholar]
- 10. Teng LJ, Hsueh PR, Ho SW, Luh KT. 2001. High prevalence of inducible erythromycin resistance among Streptococcus bovis isolates in Taiwan. Antimicrob. Agents Chemother. 45:3362–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DiPersio LP, DiPersio JR, Frey KC, Beach JA. 2008. Prevalence of the erm(T) gene in clinical isolates of erythromycin-resistant group D Streptococcus and Enterococcus. Antimicrob. Agents Chemother. 52:1567–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kadlec K, Schwarz S. 2010. Identification of a plasmid-borne resistance gene cluster comprising the resistance genes erm(T), dfrK, and tet(L) in a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 54:915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen J, Yu Z, Michel FC, Jr, Wittum T, Morrison M. 2007. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 73:4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gómez-Sanz E, Torres C, Lozano C, Zarazaga M. 2013. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp. Immunol. Microbiol. Infect. Dis. 36:83–94 [DOI] [PubMed] [Google Scholar]
- 15. Vandendriessche S, Kadlec K, Schwarz S, Denis O. 2011. Methicillin-susceptible Staphylococcus aureus ST398-t571 harbouring the macrolide-lincosamide-streptogramin B resistance gene erm(T) in Belgian hospitals. J. Antimicrob. Chemother. 66:2455–2459 [DOI] [PubMed] [Google Scholar]
- 16. Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, Lipkin WI, Lindsay JA, DeLeo FR, Lowy FD. 2012. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 3(2):e00027–12. 10.1128/mBio.00027-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCarthy AJ, van Wamel W, Vandendriessche S, Larsen J, Denis O, Garcia-Graells C, Uhlemann AC, Lowy FD, Skov R, Lindsay JA. 2012. Staphylococcus aureus CC398 clade associated with human-to-human transmission. Appl. Environ. Microbiol. 78:8845–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3(1):e00305–11. 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mediavilla JR, Chen L, Uhlemann AC, Hanson BM, Rosenthal M, Stanak K, Koll B, Fries BC, Armellino D, Schilling ME, Weiss D, Smith TC, Lowy FD, Kreiswirth BN. 2012. Methicillin-susceptible Staphylococcus aureus ST398, New York and New Jersey, USA. Emerg. Infect. Dis. 18:700–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rasigade JP, Laurent F, Hubert P, Vandenesch F, Etienne J. 2010. Lethal necrotizing pneumonia caused by an ST398 Staphylococcus aureus strain. Emerg. Infect. Dis. 16:1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valentin-Domelier AS, Girard M, Bertrand X, Violette J, François P, Donnio PY, Talon D, Quentin R, Schrenzel J, van der Mee-Marquet N. 2011. Methicillin-susceptible ST398 Staphylococcus aureus responsible for bloodstream infections: an emerging human-adapted subclone? PLoS One 6:e28369. 10.1371/journal.pone.0028369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gómez-Sanz E, Kadlec K, Feßler AT, Billerbeck C, Zarazaga M, Schwarz S, Torres C. 2013. Analysis of a novel erm(T)- and cadDX-carrying plasmid from methicillin-susceptible Staphylococcus aureus ST398-t571 of human origin. J. Antimicrob. Chemother. 68:471–473 [DOI] [PubMed] [Google Scholar]
- 23. Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67:3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwarz S, Feßler AT, Hauschild T, Kehrenberg C, Kadlec K. 2011. Plasmid-mediated resistance to protein biosynthesis inhibitors in staphylococci. Ann. N. Y. Acad. Sci. 1241:82–103 [DOI] [PubMed] [Google Scholar]
- 25. Hasman H, Franke S, Rensing C. 2006. Resistance to metals used in agricultural production, p 99–114 In Aarestrup FM. (ed), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC [Google Scholar]
- 26. Cavaco LM, Hasman H, Stegger M, Andersen PS, Skov R, Fluit AC, Ito T, Aarestrup FM. 2010. Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrob. Agents Chemother. 54:3605–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI document M100-S22. CLSI, Wayne, PA [Google Scholar]
- 29. Kadlec K, Schwarz S. 2009. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L) Antimicrob. Agents Chemother. 53:776–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kadlec K, Schwarz S. 2009. Novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob. Agents Chemother. 53:3589–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leelaporn A, Firth N, Paulsen IT, Skurray RA. 1996. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J. Bacteriol. 178:6070–6073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Firth N, Skurray RA. 1998. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist. Updat. 1:49–58 [DOI] [PubMed] [Google Scholar]
- 33. Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 35. Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Highlander SK, Hultén KG, Qin X, Jiang H, Yerrapragada S, Mason EO, Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7:99. 10.1186/1471-2180-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sitthisak S, Howieson K, Amezola C, Jayaswal RK. 2005. Characterization of a multicopper oxidase gene from Staphylococcus aureus. Appl. Environ. Microbiol. 71:5650–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577–1593 [DOI] [PubMed] [Google Scholar]
- 39. Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. 10.1186/1471-2164-11-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. 2009. Complete genome sequence of Micrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci. J. Bacteriol. 191:1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li S, Skov RL, Han X, Larsen AR, Larsen J, Sørum M, Wulf M, Voss A, Hiramatsu K, Ito T. 2011. Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 55:3046–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. European Food Safety Authority 2004. Opinion of the Scientific Panel on Contaminants in the Food Chain on a request from the Commission related to cadmium as undesirable substance in animal feed. EFSA J. 72:1–24 [Google Scholar]
- 44. Ashraf MW. 2012. Levels of heavy metals in popular cigarette brands and exposure to these metals via smoking. Sci. World J. 2012:729430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ciobanu C, Slencu BG, Cuciureanu R. 2012. Estimation of dietary intake of cadmium and lead through food consumption. Rev. Med. Chir. Soc. Med. Nat. Iasi 116:617–623 [PubMed] [Google Scholar]
- 46. European Commission 2003. Opinion of the Scientific Committee for Animal Nutrition on the Use of Copper in Feedingstuffs, p 1–47 Health & Consumer Protection Directorate General, Directorate C—Scientific Opinions, C2 Management of Scientific Committees; Scientific Co-operation and Networks, Brussels, Belgium [Google Scholar]
- 47. European Commission 2003. Opinion of the Scientific Committee for Animal Nutrition on the Use of Zinc in Feedingstuffs, p 1–33 Health & Consumer Protection Directorate General, Directorate C—Scientific Opinions, C2 Management of Scientific Committees; Scientific Co-operation and Networks, Brussels, Belgium [Google Scholar]
- 48. Stephens AJ, Huygens F, Inman-Bamber J, Price EP, Nimmo GR, Schooneveldt J, Munckhof W, Giffard PM. 2006. Methicillin-resistant Staphylococcus aureus genotyping using a small set of polymorphisms. J. Med. Microbiol. 55:43–51 [DOI] [PubMed] [Google Scholar]
- 49. Prescott JF. 2006. History of antimicrobial usage in agriculture: an overview, p 19–28 In Aarestrup FM. (ed), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC [Google Scholar]