Abstract
Malaria's ability to rapidly adapt to new drugs has allowed it to remain one of the most devastating infectious diseases of humans. Understanding and tracking the genetic basis of these adaptations are critical to the success of treatment and intervention strategies. The novel antimalarial resistance locus PF10_0355 (Pfmspdbl2) was previously associated with the parasite response to halofantrine, and functional validation confirmed that overexpression of this gene lowered parasite sensitivity to both halofantrine and the structurally related antimalarials mefloquine and lumefantrine, predominantly through copy number variation. Here we further characterize the role of Pfmspdbl2 in mediating the antimalarial drug response of Plasmodium falciparum. Knockout of Pfmspdbl2 increased parasite sensitivity to halofantrine, mefloquine, and lumefantrine but not to unrelated antimalarials, further suggesting that this gene mediates the parasite response to a specific class of antimalarial drugs. A single nucleotide polymorphism encoding a C591S mutation within Pfmspdbl2 had the strongest association with halofantrine sensitivity and showed a high derived allele frequency among Senegalese parasites. Transgenic parasites expressing the ancestral Pfmspdbl2 allele were more sensitive to halofantrine and structurally related antimalarials than were parasites expressing the derived allele, revealing an allele-specific effect on drug sensitivity in the absence of copy number effects. Finally, growth competition experiments showed that under drug pressure, parasites expressing the derived allele of Pfmspdbl2 outcompeted parasites expressing the ancestral allele within a few generations. Together, these experiments demonstrate that modulation of Pfmspdbl2 affects malaria parasite responses to antimalarial drugs.
INTRODUCTION
Malaria drug resistance poses a serious threat to treatment and control efforts (1, 2). Loci such as pfcrt, dhfr, and pfmdr1 are all known to play a role in mediating Plasmodium falciparum drug resistance and can do so through mutations or copy number variation (CNV) (3), but the precise mechanisms of resistance to many antimalarial drugs are poorly understood. Additionally, these well-known loci do not fully explain the range of responses observed in resistant parasites, suggesting that other loci may be involved in mediating parasite drug sensitivity (4, 5).
Pfmspdbl2, also called PF10_0355, PF3D7_1036300, and MSP3.8, is a novel antimalarial resistance locus recently identified in a genome-wide association study (GWAS) of 50 global parasite isolates with a high-density single nucleotide polymorphism (SNP) array (6). CNV was also observed at this locus among natural parasite isolates, and parasites with more than one copy of Pfmspdbl2 tended to be more resistant to halofantrine. Overexpression of either the sensitive or the resistant allele of Pfmspdbl2 made parasites less sensitive to halofantrine, mefloquine, and lumefantrine but not to structurally unrelated antimalarials. This validated Pfmspdbl2 as a novel antimalarial resistance locus and confirmed that an increased copy number caused increased resistance to halofantrine and structurally related drugs. However, Pfmspdbl2 was originally identified through association between SNPs and drug resistance, and the possible effects of these mutations on the modulation of the parasite drug response remained unclear.
MSPDBL2 is a merozoite surface protein containing a Duffy binding-like (DBL) domain, as well as a secreted polymorphic antigen associated with merozoites (SPAM) domain (7). The MSPDBL2 DBL domain binds to red blood cells as a dimer, and binding is dependent on specific metal ions (8). Additionally, the Pfmspdbl2 gene is highly polymorphic and variably expressed, appears to be under balancing selection, and is a likely target of host immunity (9, 10). Immunofluorescence studies have shown that MSPDBL2 is located on the merozoite surface in both schizonts and free merozoites (7, 8), despite the lack of a membrane anchor.
Here we further characterize the role of Pfmspdbl2 in mediating parasite responses to antimalarial drugs. We wanted to probe the differential effects of CNV and SNPs on drug sensitivity, and we did this through knockout and allelic replacement experiments. We found that in addition to CNV, SNPs within Pfmspdbl2 also affect parasite drug responses. Additionally, we found large difference in long-term fitness when parasites expressing different Pfmspdbl2 alleles competed directly under drug pressure.
MATERIALS AND METHODS
Parasite culture, drug testing, and knockout parasite lines.
Parasites were maintained under standard culture conditions (11) in RPMI medium supplemented with 5% human O+ serum and 5% Albumax II. Where indicated, parasites were synchronized with 5% d-sorbitol. Drug testing of knockout and allelic replacement lines was performed by measuring parasite incorporation of tritium-labeled hypoxanthine (12). Drug assays were run with at least two technical replicates of each data point, and 50% inhibitory concentration (IC50) data were averaged over multiple assays (either duplicate or triplicate), as referenced in the figure legends. Drug data from Senegalese parasites (see Fig. 2 for a summary) were generated with a high-throughput SYBR green I-based assay and have been previously reported (13). In all cases, IC50s were calculated with GraphPad Prism (GraphPad, San Diego, CA) by using a four-parameter, log-logistic nonlinear regression of fluorescence intensity versus log10-transformed drug concentrations.
Fig 2.
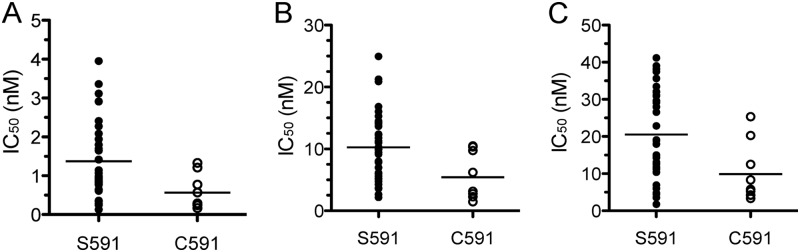
A mutation at position 591 within the SPAM domain of Pfmspdbl2 is associated with parasite responses to halofantrine (A), mefloquine (B), and lumefantrine (C) in 45 Senegalese parasites. SNP calls and drug data are from reference 13; horizontal lines show the mean IC50s of the groups. P = 0.021 for halofantrine, P = 0.016 for mefloquine, and P = 0.014 for lumefantrine (two-tailed unpaired t tests of S591 versus C591).
Pfmspdbl2 knockout parasites were generated by double-crossover recombination in the 3D7 parasite background with the pCC1 vector (14) (A. D. Uboldi et al., unpublished data). See Fig. S1 in the supplemental material for the experimental design and validation of locus disruption.
Whole-genome sequencing and the association between Pfmspdbl2 SNPs and halofantrine sensitivity.
Whole-genome sequencing of 45 culture-adapted parasite lines from Senegal was conducted, and genome-wide association studies were performed previously (13). The SNP Pf_10_001435509 (by PlasmoDB v5.0 coordinates; also known as Pf_10_001434265 in PlasmoDB v5.5-v7.2 and Pf_10_001434268 in PlasmoDB v9.1) encodes a cysteine-to-serine point mutation at position 591 within the SPAM domain of MSPDBL2. SNP calls at this position in 3D7, Dd2, and the Plasmodium reichenowi Oscar strain were verified previously (6).
Allelic replacement parasites.
Allelic replacement vectors were generated by cloning the Pfmspdbl2 gene sequence minus the first 74 nucleotides from Dd2 (derived [DR] allele) and Senegal P26.04 (ancestral [AC] allele) parasites into the pHH1 vector (15). Plasmids were transfected into the Dd2 parasite line, and stable transfectants were selected with 2.5 nM WR99210 (16). Transfectants were twice cycled off WR99210 for 2 weeks each time, and stable integration and loss of episome were confirmed by Southern blotting with ClaI, NotI, and a probed generated with the following primers: F, 5′-GGG GAA AGC ATA TAA TAA TAC TAT AGA TGC-3′; R, 5′-CTT GGA GGA ACA AGA ACC CCC TTA TTA TCA-3′. Protein expression of hemagglutinin (HA)-tagged MSPBDL2 in the Dd2 DR line was measured by Western blotting with anti-HA and anti-lactate dehydrogenase (LDH) monoclonal antibodies with the LI-COR system (LI-COR Biosciences, Lincoln, NE).
Immunofluorescence assays were performed by harvesting mixed-stage parasites growing in culture, pelleting the red blood cells, and washing them with 1× phosphate-buffered saline (PBS) before fixing them in a rotating suspension with 4% paraformaldehyde–0.0075% glutaraldehyde for 30 min at room temperature. After washing, parasites were permeabilized in a rotating suspension with 0.1% Triton X-100 and 3% bovine serum albumin in 1× PBS for 45 min at room temperature. Parasites were then blocked in 3% bovine serum albumin in 1× PBS for at least 1 h, followed by rat anti-HA 3F10 antibody (150 ng/ml) staining overnight at 4°C. After washing, parasites were stained with Alexa 488-conjugated anti-rat antibodies (1:750 dilution) for 1 h at room temperature. Cells were mounted with Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA) and imaged with the 100× objective on a Nikon Eclipse TE300 microscope. Images were obtained with MetaMorph software v7.5 (Sunnyvale, CA) with a Hamamatsu C4742-95 camera and processed with Adobe Photoshop CS 5.0 (Adobe, San Jose, CA).
Growth competition experiments.
Competition experiments were performed by mixing synchronized ring stage cultures of DR and AC parasite lines in equal ratios either in the absence of drug or in the presence of 1 nM halofantrine. Parasites were harvested after 1, 4, 8, or 14 asexual cycles, and genomic DNA was extracted. Ratios of each line were determined by quantitative real-time PCR with a custom TaqMan assay designed to detect the two different alleles of SNP Pf_10_001435509. The sequences of the assay primers used were as follows: F, 5′-GGG TCA TCA TCT CTT GAA CAA CAC T-3′; R, 5′-TCG CTT TCA TTA GCT ATC TGT TCA ATA TCC-3′. The sequences of the probes used were as follows: Allele 1 (AC; VIC), 5′-CAA TTC TAA AGC ACT TCC CTT-3′; Allele 2 (DR; 6-carboxyfluorescein, FAM), 5′-ATT CTA AAG CAC ATC CCT T-3′. Cycle threshold values were normalized to a standard curve of known genomic DNA mixtures of the two parasite lines.
RESULTS
Loss of Pfmspdbl2 makes parasites more sensitive to halofantrine, mefloquine, and lumefantrine.
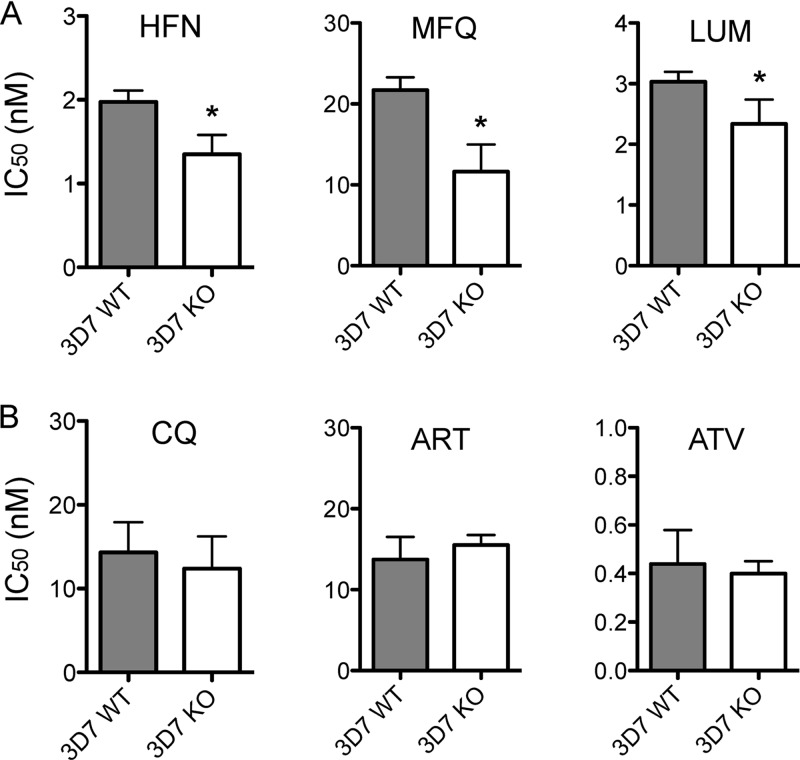
The Pfmspdbl2 locus was successfully disrupted in the 3D7 parasite line (Uboldi et al., unpublished data). See Fig. S1 in the supplemental material for design and validation of locus disruption. Drug testing by tritium-labeled hypoxanthine incorporation showed that parasites in which Pfmspdbl2 had been disrupted were more sensitive than 3D7 wild-type parasites to halofantrine, mefloquine, and lumefantrine (Fig. 1A). Parasites lacking a functional Pfmspdbl2 gene were unchanged in their responses to chloroquine, artemisinin, and atovaquone (Fig. 1B).
Fig 1.
(A) Drug responses of 3D7 wild-type (WT) and Pfmspdbl2 knockout (KO) lines to halofantrine (HFN), mefloquine (MFQ), and lumefantrine (LUM), measured by tritium-labeled hypoxanthine incorporation. (B) Drug responses of 3D7 WT and KO lines to chloroquine (CQ), artemisinin (ART), and atovaquone (ATV). The values shown are means ± standard deviations of at least two separate assays run on different days. *, P < 0.05 (two-tailed, unpaired t test).
A mutation within the SPAM domain of Pfmspdbl2 is associated with parasite responses to halofantrine, mefloquine, and lumefantrine.
SNP 1434268 (in PlasmoDB v9.1; 1434265 in PlasmoDB v5.5-v7.2 and 1435509 in PlasmoDB v5.0) on chromosome 10 encodes a nonsynonymous cysteine-to-serine mutation at position 591 within the SPAM domain of Pfmspdbl2. Both the P. reichenowi Oscar strain and the P. falciparum 3D7 line have cysteine at position 591 (6), suggesting that cysteine is the ancestral (AC) allele and serine is the derived (DR) allele at this position. We previously tested 45 culture-adapted parasites that were recently isolated from infected patients in Senegal with a SYBR green I-based drug assay (13). Only 9 of the 45 Senegalese parasites tested have cysteine at this position and 36 have serine, resulting in a DR allele frequency of 80% in this population. Parasites with C591 were more sensitive to halofantrine, mefloquine, and lumefantrine than those with S591 were (Fig. 2, P < 0.05 for each drug). C591S was not significantly associated with parasite sensitivity to chloroquine, artemisinin, or atovaquone (data not shown).
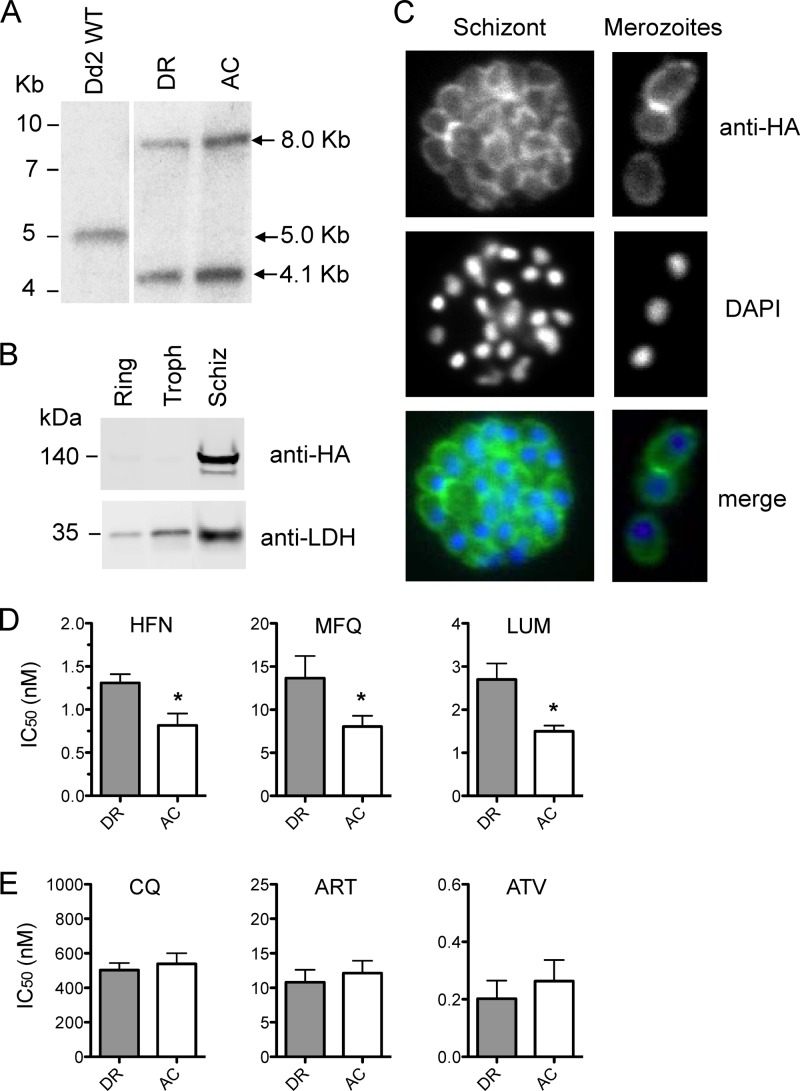
To further investigate the association between the C591S mutation and parasite drug responses, we generated Dd2 parasites where a 3′ portion of the endogenous Pfmspdbl2 locus was replaced with a triple-HA-tagged version of either the DR or the AC allele of this gene. Stable integrants containing the DR or the AC construct were confirmed by Southern blotting (Fig. 3A). Western blotting of lysates from the DR line with anti-HA antibodies showed expression of the tagged protein in schizonts and the presence of two bands of around 140 kDa (Fig. 3B). The expected size of HA-tagged MSPDBL2 is 90 kDa; however, mass spectrometry analysis confirmed that the two 140-kDa bands identified both corresponded to MSPDBL2 (data not shown). Immunofluorescence assay showed the proper localization of the HA-tagged protein on the merozoite surface (Fig. 3C). Drug testing by incorporation of tritium-labeled hypoxanthine revealed that AC parasites were more sensitive to halofantrine, mefloquine, and lumefantrine than DR parasites were (Fig. 3D) but showed similar responses to chloroquine, artemisinin, and atovaquone (Fig. 3E).
Fig 3.
Pfmspdbl2 mutations in the absence of CNV affect parasite responses to halofantrine and structurally related antimalarials. (A) Southern blot assays of Dd2 wild-type (WT) and Pfmspdbl2 allelic replacement parasites where the endogenous genome locus has been replaced with HA-tagged versions of the DR allele (S591 cloned from Dd2) or the AC allele (C591 cloned from Senegal P26.04). Digestion of the wild-type, untagged Pfmspdbl2 locus with ClaI and NotI generates a single band of 5.0 kb, while digestion of the HA-tagged integrated locus generates two bands of 4.1 and 8.0 kb. (B) Western blotting of ring, trophozoite (Troph), and schizont (Schiz) stage cultures of the DR line with anti-HA and anti-LDH antibodies shows expression of HA-tagged MSPDBL2 in schizonts. (C) Immunofluorescence of a representative schizont and representative merozoites of the DR line with anti-HA antibodies (green) and DAPI staining (blue). (D) Drug responses of DR and AC lines to halofantrine (HFN), mefloquine (MFQ), and lumefantrine (LUM) measured by tritium-labeled hypoxanthine incorporation. (E) Drug responses of DR and AC lines to chloroquine (CQ), artemisinin (ART), and atovaquone (ATV). The values shown are means ± standard deviations of at least three separate assays run on different days. *, P < 0.05 (two-tailed unpaired t test).
Parasites expressing the Pfmspdbl2 DR allele outcompete parasites expressing the AC allele under drug pressure.
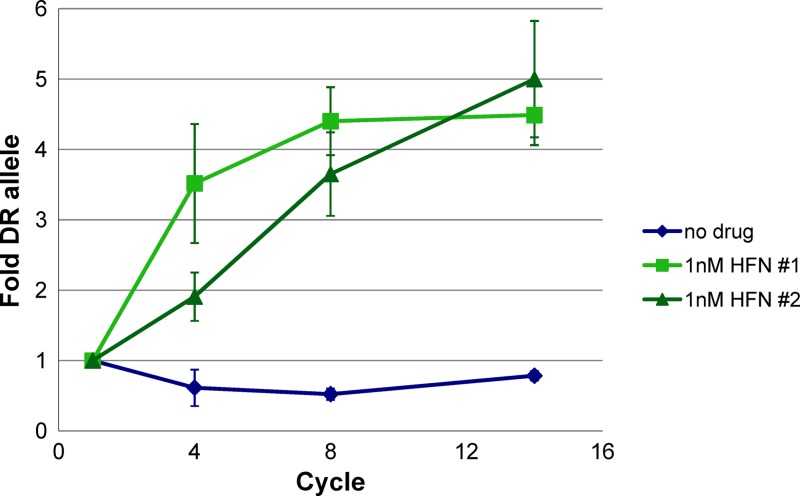
We were interested in determining if the observed differences between DR and AC drug responses affect the fitness of parasites when they are cultured over many cycles. DR and AC parasite lines were synchronized, and ring stage parasites were mixed in a 1:1 ratio. Parasite mixtures were allowed to grow in normal medium containing no drug or in medium containing 1 nM halofantrine, and the ratio of DR to AC parasites was determined with a TaqMan SNP assay after 1, 4, 8, and 14 asexual replication cycles (Fig. 4). The ratio of the two parasite lines was largely unchanged in the absence of drug pressure, remaining close to the 1:1 starting ratio. However, when parasites were incubated with 1 nM halofantrine, DR parasites quickly outcompeted AC parasites, reaching an almost 5-fold excess after eight replication cycles. These results show that the Pfmspdbl2 DR allele has no associated fitness cost in the absence of drug pressure and furthermore suggest that modest IC50 differences may have large effects on parasite fitness when parasites compete under drug pressure.
Fig 4.
Pfmspdbl2 DR allele parasites outcompete AC allele parasites in the presence of halofantrine. Parasites were mixed at equal ratios and grown with no drug or 1 nM halofantrine (HFN). Ratios of DR and AC parasites were determined after 1, 4, 8, and 14 asexual cycles by quantitative real-time PCR with TaqMan probes and are shown as fold abundance of the DR allele. Two biological replicates (#1 and #2) are shown, and error bars show standard deviations of technical replicates.
DISCUSSION
Drug resistance poses a serious threat to efforts aimed at eliminating malaria from regions where it is endemic, and considerable attention is currently focused on understanding the molecular basis of resistance. Nonetheless, precise mechanisms of antimalarial drug action and drug resistance remain poorly defined for many antimalarials (17). Transgenic parasite lines, created and tested under controlled and standardized laboratory conditions, allow the precise study of how subtle genetic changes can contribute to drug resistance. Even if the effects are modest, elucidation of the impacts of individual loci on drug responses can deepen our understanding of how parasites may eventually become clinically resistant.
We wanted to know how CNVs and SNPs within Pfmspdbl2 alter parasite susceptibility to antimalarials, in order to better understand the role of this locus in mediating P. falciparum drug responses. Previous work showed that an increased Pfmspdbl2 copy number made parasites more resistant to halofantrine and structurally related antimalarials (6). Consistent with this result, we found that parasites lacking expression of Pfmspdbl2 were more sensitive to halofantrine, mefloquine, and lumefantrine but not to structurally unrelated drugs. In the absence of Pfmspdbl2 expression, parasites showed no growth defects compared to wild-type parasites, suggesting that this gene is not essential for asexual parasite growth but its absence makes parasites more sensitive to a specific class of antimalarials. These findings further support a role for Pfmspdbl2 in modulating parasite drug responses, as decreased drug sensitivity seems to scale with gene copy number in a manner similar to that of CNVs at other drug resistance loci such as pfmdr1 and pfgchI (18, 19).
We focused on a specific mutation (C591S) within MSPDBL2 for two reasons. First, in the original global GWAS that identified Pfmspdbl2 as a novel antimalarial resistance locus, the genome-wide significant association signal mapped to the 3′ portion of the gene and spanned the C591S mutation (6). Second, C591S was the mutation within Pfmspdbl2 that was most strongly associated with parasite sensitivity to halofantrine in a recent whole-genome sequence-based GWAS (13), although this association did not reach genome-wide significance. Importantly, only 20% of the 45 sequenced Senegalese parasites in that study retained the AC allele at position 591. This suggests that selection of some sort, such as the use of halofantrine or perhaps more likely the structurally related antimalarials mefloquine and lumefantrine, has driven the DR allele to high prevalence in this population.
Other studies have documented cases where drug resistance-associated mutations confer a fitness disadvantage in the absence of drug pressure (20, 21), but this does not seem to be the case with Pfmspdbl2. This could be due to the modest change in drug response conferred by the C591S mutation or perhaps because compensatory mutations that restore parasite fitness have occurred. Because we used genomic DNA from natural parasite isolates to generate the allelic replacement constructs used here, the AC parasite line contains additional mutations within Pfmspdbl2 besides C591S. While it remains possible that mutations other than or in addition to C591S could underlie the changes in drug response that we observed, these results confirm that mutations within Pfmspdbl2, in the absence of CNV, also mediate parasite drug responses.
The precise mechanism by which MSPDBL2 affects antimalarial drug responses remains unknown. MSPDBL2 localizes to the merozoite surface, despite lacking a membrane anchor. Other merozoite surface proteins are known to form complexes (22), and it seems possible that MSPDBL2 could act in concert with other surface proteins. Additionally, we wondered whether MSPDBL2 binds to halofantrine directly; however, attempts to investigate this were confounded by high rates of nonspecific drug binding to a reference protein (data not shown). The C591S mutation resides within the SPAM domain of MSPDBL2; the SPAM motif has yet to be characterized and could have a function different from that of the DBL domain, which appears to mediate binding to erythrocytes (8). Alternately, perhaps parasites are better able to bind and invade red blood cells in the presence of certain antimalarials when they have more copies of Pfmspdbl2 or when the protein is in a specific conformation.
Overall, these experiments demonstrate that genetic manipulation of Pfmspdbl2 affects drug sensitivity of P. falciparum. Similar to other malaria parasite drug resistance genes, we find effects of both CNVs and SNPs in modulating parasite drug responses. Importantly, our findings demonstrate that large differences in parasite fitness can be revealed when parasites are grown over multiple generations under drug pressure, as they likely would in the natural setting of infected patients.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bradley Coleman, Manoj Duraisingh, Jeffrey Dvorin, and Ulf Ribacke for providing reagents, helping design experiments and interpret results, and helpful discussion. We also thank Daniel Park and Pardis Sabeti for help analyzing whole-genome sequencing data.
This work was supported by the Bill and Melinda Gates Foundation.
Footnotes
Published ahead of print 15 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02574-12.
REFERENCES
- 1. malERA Consultative Group on Drugs 2011. A research agenda for malaria eradication: drugs. PLoS Med. 8:e1000402. 10.1371/journal.pmed.1000402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sibley CH, Barnes KI, Watkins WM, Plowe CV. 2008. A network to monitor antimalarial drug resistance: a plan for moving forward. Trends Parasitol. 24:43–48 [DOI] [PubMed] [Google Scholar]
- 3. Ekland EH, Fidock DA. 2007. Advances in understanding the genetic basis of antimalarial drug resistance. Curr. Opin. Microbiol. 10:363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. 2004. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 52:985–997 [DOI] [PubMed] [Google Scholar]
- 5. Kinga Modrzynska K, Creasey A, Loewe L, Cezard T, Trindade Borges S, Martinelli A, Rodrigues L, Cravo P, Blaxter M, Carter R, Hunt P. 2012. Quantitative genome re-sequencing defines multiple mutations conferring chloroquine resistance in rodent malaria. BMC Genomics 13:106. 10.1186/1471-2164-13-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Tyne D, Park DJ, Schaffner SF, Neafsey DE, Angelino E, Cortese JF, Barnes KG, Rosen DM, Lukens AK, Daniels RF, Milner DA, Johnson CA, Shlyakhter I, Grossman SR, Becker JS, Yamins D, Karlsson EK, Ndiaye D, Sarr O, Mboup S, Happi C, Furlotte NA, Eskin E, Kang HM, Hartl DL, Birren BW, Wiegand RC, Lander ES, Wirth DF, Volkman SK, Sabeti PC. 2011. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet. 7:e1001383. 10.1371/journal.pgen.1001383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh S, Soe S, Weisman S, Barnwell JW, Pérignon JL, Druilhe P. 2009. A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS One 4:e5410. 10.1371/journal.pone.0005410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hodder AN, Czabotar PE, Uboldi AD, Clarke OB, Lin CS, Healer J, Smith BJ, Cowman AF. 2012. Insights into Duffy binding-like domains through the crystal structure and function of the merozoite surface protein MSPDBL2 from Plasmodium falciparum. J. Biol. Chem. 287:32922–32939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ochola LI, Tetteh KKA, Stewart LB, Riitho V, Marsh K, Conway DJ. 2010. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol. Biol. Evol. 27:2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amambua-Ngwa A, Tetteh KK, Manske M, Gomez-Escobar N, Stewart LB, Deerhake ME, Cheeseman IH, Newbold CI, Holder AA, Knuepfer E, Janha O, Jallow M, Campino S, Macinnis B, Kwiatkowski DP, Conway DJ. 2012. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 8:e1002992. 10.1371/journal.pgen.1002992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trager W, Jensen JB. 1976. Human malaria parasites in continuous culture. Science 193:673–675 [DOI] [PubMed] [Google Scholar]
- 12. Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16:710–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park DJ, Lukens AK, Neafsey DE, Schaffner SF, Chang HH, Valim C, Ribacke U, Van Tyne D, Galinsky K, Galligan M, Becker JS, Ndiaye D, Mboup S, Wiegand RC, Hartl DL, Sabeti PC, Wirth DF, Volkman SK. 2012. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc. Natl. Acad. Sci. U. S. A. 109:13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maier AG, Braks JA, Waters AP, Cowman AF. 2006. Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol. Biochem. Parasitol. 150:118–121 [DOI] [PubMed] [Google Scholar]
- 15. Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, Bannister LH, Mitchell GH, Blackman MJ. 2005. Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog. 1:241–251. 10.1371/journal.ppat.0010029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fidock DA, Wellems TE. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U. S. A. 94:10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Neill PM, Barton VE, Ward SA. 2010. The molecular mechanism of action of artemisinin—the debate continues. Molecules 15:1705–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kidgell C, Volkman SK, Daily J, Borevitz JO, Plouffe D, Zhou Y, Johnson JR, Le Roch KG, Sarr O, Ndir O, Mboup S, Batalov S, Wirth DF, Winzeler EA. 2006. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2:e57. 10.1371/journal.ppat.0020057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. 2006. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194:528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayward R, Saliba KJ, Kirk K. 2005. pfmdr1 mutations associated with chloroquine resistance incur a fitness cost in Plasmodium falciparum. Mol. Microbiol. 55:1285–1295 [DOI] [PubMed] [Google Scholar]
- 21. Babiker HA. 2009. Seasonal fluctuation of drug-resistant malaria parasites: a sign of fitness cost. Trends Parasitol. 25:351–352 [DOI] [PubMed] [Google Scholar]
- 22. Kadekoppala M, Holder AA. 2010. Merozoite surface proteins of the malaria parasite: the MSP1 complex and the MSP7 family. Int. J. Parasitol. 40:1155–1161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.