Abstract
Nineteen linezolid-resistant Staphylococcus epidermidis and two Staphylococcus aureus isolates recovered from two medical institutions in northeast Ohio and an S. aureus cfr index strain previously collected in the same facilities during the 2007 SENTRY Antimicrobial Surveillance Program were investigated for the genetic basis of oxazolidinone resistance and the location of cfr. S. aureus isolates were typed by pulsed-field gel electrophoresis (PFGE), spa typing, and multilocus sequence typing (MLST). The location of cfr was determined by Southern blotting and hybridization. Plasmid sequencing was performed using the 454 Life Sciences (Roche) GS-FLX DNA platform. The two S. aureus isolates showed unique PFGE patterns but were multilocus sequence type 5 (ST5) and spa type t002, whereas the S. aureus index strain was ST239 and t037. Southern blot and hybridization experiments showed that cfr was plasmid located and that the S. epidermidis isolates, one of the S. aureus isolates, and the S. aureus index strain shared an identical cfr-carrying plasmid (39.3 kb). Sequencing results confirmed these findings. A 10-kb fragment containing cfr showed the highest identity (99.9%) to a 9.5-kb fragment of plasmid pSCFS3 from a bovine Staphylococcus lentus isolate from Germany. In addition, these 39.3-kb plasmids from human S. epidermidis and S. aureus exhibited BglII restriction profiles very similar to that observed for plasmid pSCFS3. The cfr-carrying plasmid detected in the remaining S. aureus isolate (7.9 kb) was distinct and showed the highest identity to the chromosomal cfr integrate found in the chromosomal DNA of a Proteus vulgaris isolate from a pig in China.
INTRODUCTION
Linezolid has been widely prescribed to treat serious infections caused by multidrug-resistant (MDR) Gram-positive pathogens since its clinical introduction as the first oxazolidinone in the United States in 2000 and in the United Kingdom in 2001 (1). Linezolid is currently approved by the Food and Drug Administration (FDA) for the treatment of complicated and uncomplicated skin and skin structure infections and for nosocomial and community-acquired pneumonia caused by susceptible organisms. Linezolid also has an FDA indication for the treatment of vancomycin-resistant Enterococcus faecium (VRE) infections, including cases with concurrent bacteremia (2).
This oxazolidinone alters protein synthesis via binding to the 50S ribosomal subunit, with more recent data suggesting that this drug binds to the A site of the peptidyl transferase center (PTC) of the bacterial ribosome and interferes with the positioning of aminoacyl-tRNA, resulting in protein synthesis inhibition (3). Although the prevalence of linezolid resistance remains relatively low among Gram-positive surveillance clinical isolates (4, 5), the resistance mechanisms have been extensively characterized. These mechanisms are mostly comprised of mutations in domain V of 23S rRNA, and alterations in the ribosomal proteins L3 and L4 have also been associated with decreased susceptibility (6). A more recent resistance mechanism, namely, cfr, has been recognized. The gene cfr encodes a methyltransferase that catalyzes the posttranscriptional methylation of nucleotide A2503 (Escherichia coli numbering of rRNA) in the 23S rRNA, causing decreased susceptibility to phenicol, lincosamide, oxazolidinone, pleuromutilin, and streptogramin A (PhLOPSA) compounds (6–8).
Initially detected among staphylococcal species recovered from animal and human sources (9–11), the cfr gene has now been reported among several Gram-positive isolates, such as Enterococcus faecalis (12, 13), Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis (14), and Bacillus spp. (15–17), as well as in isolates of the Gram-negative organisms E. coli (18) and Proteus vulgaris (19) recovered from several different specimen sources collected from animals. However, recent data have demonstrated the Bacillales order as containing the natural reservoirs for the cfr genes (20). With a few exceptions, cfr has exclusively been associated with mobile elements, such as insertion sequences and mobilization genes carried by a variety of plasmids (6). In this study, 19 linezolid-resistant Staphylococcus epidermidis isolates (21) and two S. aureus clinical isolates recovered from two medical institutions located in northeast Ohio were included (i) to phenotypically and genotypically characterize these isolates, (ii) to assess the cfr location, and (iii) to determine the genetic elements responsible for gene mobilization and/or dissemination.
MATERIALS AND METHODS
Bacterial strains.
A total of 22 staphylococci were initially included in this investigation, and isolates comprised 19 S. epidermidis (13 from hospital A [April 2008 through May 2009] and 6 from hospital B [October 2006 through July 2007]) (21) and 2 S. aureus (isolates 1609 and 1900 from hospital B). All S. aureus and S. epidermidis isolates included were recovered from multiple blood cultures from unique patients and deemed to be clinically significant by local guidelines. In addition, all bloodstream infections occurred after 48 h of hospitalization. S. epidermidis isolates were previously reported by Bonilla et al. (21) to be genetically related, and these isolates displayed indistinguishable chromosomal and plasmid profiles (22). Therefore, one representative S. epidermidis isolate from each hospital (A [isolate 1243] and B [1519]) was selected for further characterization, unless otherwise specified. Moreover, a cfr index strain (strain 737) recovered from the same hospital B complex facilities during the 2007 SENTRY Antimicrobial Surveillance Program was included for comparison purposes (23).
Antimicrobial susceptibility profile.
Susceptibility testing was performed by broth microdilution, according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (24). S. aureus ATCC 29213 and E. faecalis ATCC 29212 were tested concurrently for quality assurance purposes (25). In addition, the inoculum density was monitored by colony counts to ensure an adequate number of cells for each testing event. MIC interpretations were applied as described in CLSI document M100-S22 (25), except in the cases of retapamulin and tigecycline. MIC results for retapamulin and tigecycline compounds were interpreted according to the epidemiological cutoff values for S. aureus proposed by Traczewski and Brown (i.e., ≤0.5 μg/ml for susceptible, 1 μg/ml for intermediate, and ≥2 μg/ml for resistant) and the FDA (≤0.5 μg/ml for susceptible), respectively (26, 27).
Screening for linezolid resistance mechanisms.
Isolates were screened for the presence of cfr, as well as mutations in the 23S rRNA and ribosomal proteins (L3 and L4), by PCR and sequencing as previously described (17). Amplicons were sequenced on both strands. Deduced amino acid sequences of ribosomal proteins were compared to those from wild-type S. epidermidis ATCC 12228 and S. aureus RN4220 using the Lasergene software package (DNAStar, Madison, WI).
Molecular typing.
S. aureus 1609 and 1900 were subjected to pulsed-field gel electrophoresis (PFGE), as previously described (28). SmaI-digested genomic DNA was resolved in CHEF-DR II (Bio-Rad, Richmond, CA), and PFGE profiles obtained were compared to that of cfr index strain 737 and representatives of U.S. clones using GelCompar II software (Applied Math, Kortrijk, Belgium). Percent similarities were identified on a dendrogram derived from the unweighted-pair group method using arithmetic averages and based on Dice coefficients. Band position tolerance and optimization were set at 1.5% and 0.5%, respectively. Isolates showing similarity coefficients of ≥95% were considered genetically indistinguishable (subtype), while those with similarity coefficients between 80.0 and 94.9% were classified as genetically related (type). The cfr-carrying S. aureus strains 1609 and 1900 were further characterized by spa typing and multilocus sequence typing (MLST). The spa types were assigned through the Ridom web server (http://www.ridom.de/spaserver/), and MLST alleles and sequence types (ST) were identified using the MLST database (http://www.mlst.net).
cfr plasmid transfer, analysis, and sequencing.
Whole genomic DNAs from all 19 S. epidermidis isolates and S. aureus 1900, 1609, and 737 were prepared in 1% agarose blocks and partially digested with S1 endonuclease. DNA fragments were resolved by PFGE using CHEF-DR II (Bio-Rad), and plasmid band sizes were determined by comparison with bacteriophage λ concatemers (New England BioLabs, Ipswich, MA). Plasmid extractions from S. epidermidis 1243 and 1519 and S. aureus isolates 1900, 1609, and 737 were obtained by using the plasmid DNA MIDI kit (Qiagen GmbH, Hilden, Germany). Plasmid transfer into wild-type S. aureus RN4220 was performed by electroporation using Micropulser (Bio-Rad, Richmond, CA). Probable transformants were selected using retapamulin (0.25 μg/ml). The presence of cfr among transformant colonies was confirmed by PCR. Plasmids were digested with restriction enzymes (EarI, NsiI, and AsiI; New England BioLabs) and separated in 1% agarose gels. DNA fragments from agarose gels were transferred to a nylon membrane by Southern blotting. Membranes were hybridized using a digoxigenin-labeled cfr-specific probe (Roche Diagnostics GmbH, Mannheim, Germany).
Similar hybridization signal results were obtained for S. epidermidis 1243 and 1519 and S. aureus 1609 and 737, suggesting that these isolates shared the same cfr-carrying plasmid or had closely related plasmids. Therefore, a cfr plasmid from S. epidermidis 1243 and S. aureus 737 and 1900 were selected for further sequencing. Plasmid preparations were subjected to DNA sequencing using the 454 Life Sciences (Roche) GS-FLX DNA platform. DNA assembling and analysis were performed by Lasergene. In addition, cfr plasmids from isolates 737, 1609, and 1243 were digested using BglII, and restriction patterns were compared with that of pSCFS3 (9, 29).
Nucleotide sequence accession numbers.
The nucleotide sequences of the cfr-carrying plasmids described herein have been submitted to the EMBL/GenBank/DDBJ sequence databases and assigned accession numbers KC206006, KC222021, and KC561137.
RESULTS AND DISCUSSION
Antimicrobial susceptibility and resistance mechanisms.
S. aureus exhibited linezolid MIC results between 8 and 16 μg/ml, while S. epidermidis showed MICs of ≥128 μg/ml (Table 1). Elevated MICs were noted for chloramphenicol (≥64 μg/ml), clindamycin (>64 μg/ml), virginiamycin (8 to 32 μg/ml), quinupristin-dalfopristin (2 to 8 μg/ml), retapamulin (>8 μg/ml), and tiamulin (>64 μg/ml) among all cfr-positive staphylococcal isolates (Table 1). All S. aureus strains demonstrated wild-type sequences for 23S rRNA and the ribosomal proteins investigated. S. epidermidis isolates also had wild-type sequences for 23S rRNA, but L3 amino acid alterations (H146Q, V154L, A157R) were observed in the representative S. epidermidis strains, as well as a 71G72 insertion in the L4 protein (21).
Table 1.
Antimicrobial susceptibility profiles for one representative S. epidermidis isolate each from hospital A (1243) and B (1519) and S. aureus from hospital B (1609 and 1900)
| Antimicrobial agent | MIC (μg/ml) (susceptibility category)a |
|||
|---|---|---|---|---|
|
S. epidermidis |
S. aureus |
|||
| 1243 | 1519 | 1609 | 1900 | |
| Linezolid | 128 (R) | >128 (R) | 16 (R) | 8 (R) |
| Chloramphenicol | >128 (R) | >128 (R) | >128 (R) | 64 (R) |
| Clindamycin | >64 (R) | >64 (R) | >64 (R) | >64 (R) |
| Virginiamycin | 8 | 16 | 32 | 16 |
| Quinupristin-dalfopristin | 2 (I) | 2 (I) | 8 (R) | 4 (R) |
| Retapamulin | >8 (R) | >8 (R) | >8 (R) | >8 (R) |
| Tiamulin | >64 | >64 | >64 | >64 |
| Tigecycline | 0.25 (S) | 0.12 (S) | 0.25 (S) | 0.12 (S) |
| Tetracycline | 0.25 (S) | 0.5 (S) | 0.5 (S) | 0.25 (S) |
| Vancomycin | 2 (S) | 2 (S) | 1 (S) | 1 (S) |
| Daptomycin | 0.25 (S) | 0.25 (S) | 0.25 (S) | 0.25 (S) |
| Oxacillin | >2 (R) | >2 (R) | >2 (R) | >2 (R) |
| Ciprofloxacin | >4 (R) | >4 (R) | >4 (R) | >4 (R) |
| Erythromycin | 4 (I) | 4 (I) | >4 (R) | >4 (R) |
| Gentamicin | 8 (I) | 8 (I) | >8 (R) | ≤1 (S) |
| Trimethoprim-sulfamethoxazole | 4 (R) | 4 (R) | ≤0.5 (S) | ≤0.5 (S) |
MIC interpretive criteria as published in CLSI document M100-S22, when available (25). Retapamulin MIC results were interpreted according to the microbiological breakpoints proposed by Traczewski and Brown (i.e., ≤0.5 μg/ml for susceptibility, 1 μg/ml for intermediate, and ≥2 μg/ml for resistant) (26). The tigecycline breakpoint for susceptibility was that for S. aureus (≤0.5 μg/ml) approved by the FDA (27). S, susceptible; I, intermediate; R, resistant.
Molecular typing.
PFGE analysis revealed that S. aureus 1900, 1609, and 737 clustered within three PFGE types. The PFGE pattern of isolate 1900 (type A1) was similar to that of NRS382 (type A), a USA100 clone representative (similarity coefficient of 94.7% [Fig. 1]). S. aureus 1609 exhibited a unique PFGE pattern, although further typing analysis demonstrated that, similar to USA100 (ST5 and t002), isolates 1900 and 1609 were ST5 (clonal complex 5 [CC5]) and spa t002. The index S. aureus 737 was ST239 (CC239) and spa t037 and had a unique PFGE profile, and these typing results are associated with the Brazilian/Hungarian clone.
Fig 1.
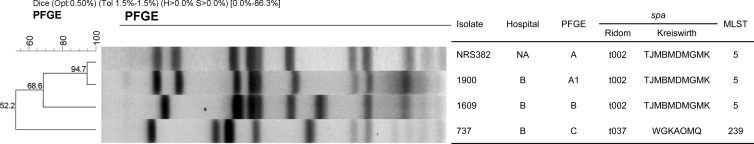
PFGE profiles of cfr-carrying S. aureus 1900 and 1609 (hospital B) compared with 737 (hospital B complex facilities) and a representative strain of the USA100 clone (NRS382). NA, not applicable.
Location of cfr.
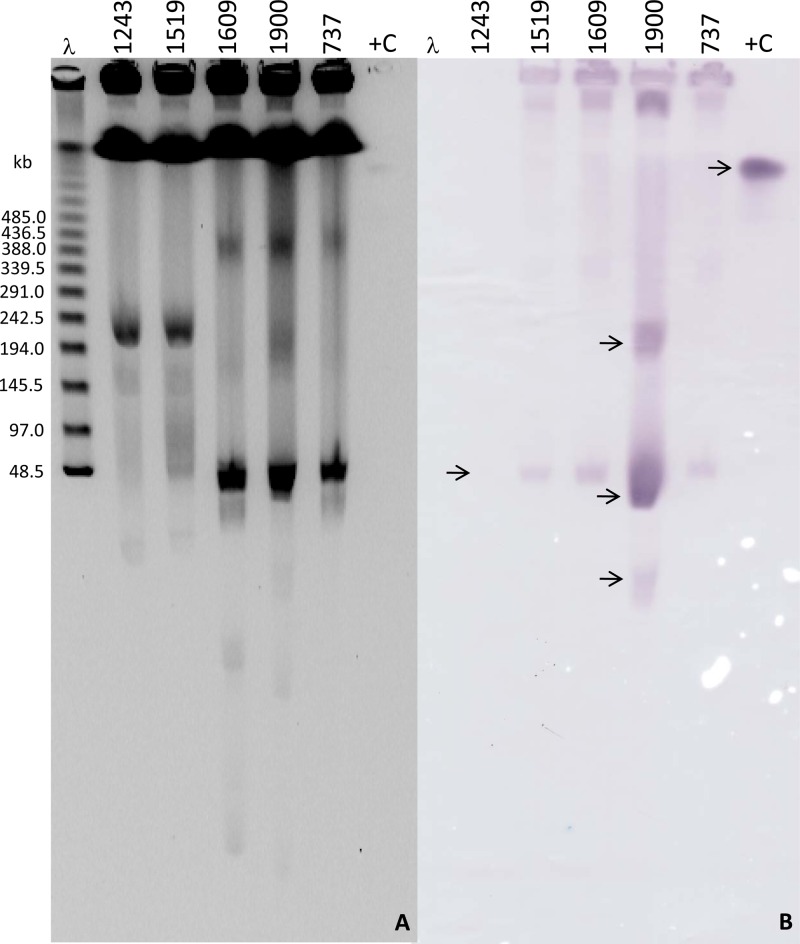
All S. epidermidis isolates showed identical plasmid profiles during S1 endonuclease experiments (data not shown). Southern blotting and hybridization experiments revealed signals from plasmid bands located slightly below the 48.5-kb ladder, suggesting a plasmid size of approximately 40 kb in the S. epidermidis representatives (Fig. 2). Hybridization signals were observed in plasmid bands of similar sizes (∼40-kb) from S. aureus 1609 and 737, while S. aureus 1900 demonstrated signals from three plasmid bands of various sizes (Fig. 2). When plasmid preparations were submitted to restriction digestions prior to Southern blotting and hybridization, the two S. epidermidis representatives of each hospital (A and B) and S. aureus 737 and 1609 displayed equivalent hybridization signal profiles, while S. aureus 1900 showed a distinct pattern with multiple hybridization signals, as for the experiment described above (22). In addition, plasmid DNAs obtained from S. epidermidis isolates (1243 and 1519) and S. aureus 737, 1609, and 1900 were transferred by electroporation to S. aureus RN4220 and results confirmed by PCR. Southern blotting and hybridization experiments performed demonstrated results similar to those observed for the parent isolates, including for the transformant obtained from isolate 1900, which showed multiple hybridization signals (data not shown).
Fig 2.
(A) S1 partially digested genomic DNA of representative S. epidermidis isolates from each hospital, A and B (1243 and 1519, respectively), and S. aureus isolates 1609 and 1900 (from hospital B). “737” represents the cfr-carrying S. aureus index strain 737 (from hospital B complex facilities) (23). “λ” and “+C” represent lambda ladder PFGE marker and positive control (diluted PCR product), respectively. (B) Hybridization signals (arrows) obtained with a cfr-specific probe. Hybridization signals for one of the S. epidermidis (1243) are not visible in this particular membrane.
Plasmid sequence analysis.
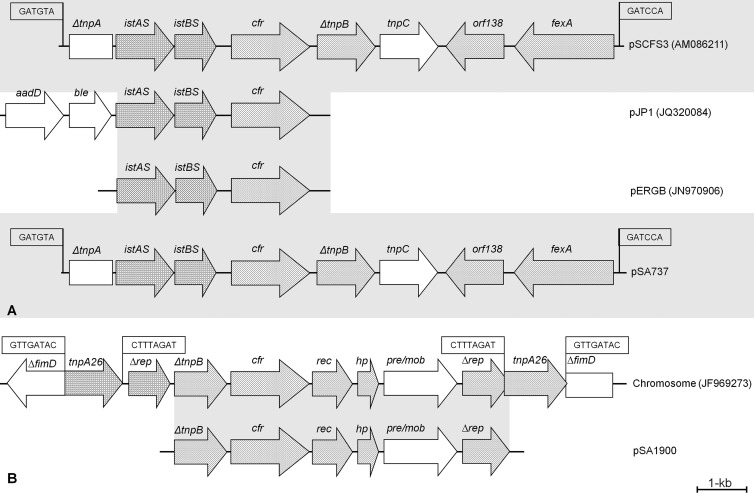
The Southern blotting and hybridization experiments suggested that the S. epidermidis isolates and S. aureus 1609 and 737 shared a same-size cfr-carrying plasmid, which was different from that observed in S. aureus 1900. Therefore, these results led to the selection of cfr plasmids from one S. epidermidis isolate (1243) and two S. aureus isolates (737 and 1900) for further DNA sequencing. Sequencing analysis confirmed that the cfr plasmids from 1243 and 737 were identical, with no variation in nucleotide sequences. These plasmids, designated pSE1243 and pSA737, had a size of 39.3 kb, and approximately 75% of their DNA sequences showed the highest identity (97%) to pSK73 (GenBank accession no. GQ915269), which originated from an S. aureus isolate recovered from a human clinical specimen in 1966 in Sydney, Australia. The pSK73 sequence deposited in GenBank possesses 45 open reading frames (ORF), including resistance determinants [erm(A) (macrolide-lincosamide-streptogramin B resistance) and spc (spectinomycin resistance gene)] and tnpA, tnpB, and tnpC genes associated with a Tn554 element. A second, smaller fragment (∼10 kb) of pSE1243 and pSA737 harbored the cfr gene with a genetic environment identical to that of pSCFS3 (AM086211) and contained ΔtnpA, istAS, and istBS upstream (similar to IS21) of cfr; ΔtnpB, tnpC, orf138, and fexA (similar to Tn558) were located downstream (Fig. 3). Compared with the nucleotide sequence available for pSCFS3 (9,497 bp), pSE1243 and pSA737 displayed only 13 nucleotide differences, which consisted of 10 nucleotides within a noncoding region upstream of istA, a nucleotide change within istA (silent mutation), and one nucleotide mutation each in istB and cfr, causing glycine-to-arginine and alanine-to-asparagine alterations, respectively. The 10-kb fragment of plasmid pSCFS3 was reported to have originated by the insertion of a segment containing an IS21-like element (IS21-558) and the cfr gene into Tn558, thereby truncating both the tnpA and tnpB genes of this transposon (9) (Fig. 3). During these events, the resistance genes erm(A) and spc were deleted and also not observed in pSE1243 or pSA737. These sequence comparisons suggested that pSE1243 and pSA737 (39.3 kb) possess an identical or very similar cfr-containing segment compared to pSCFS3, previously reported to have a similar size (35.7 kb). In fact, analysis of plasmid profiles after restriction digestion with BglII demonstrated that these plasmids had similar patterns, except that pSCFS3 had an extra small fragment (2.1 kb) not observed in pSE1243 and pSA737 (data not shown).
Fig 3.
(A) Schematic representation of DNA sequences surrounding cfr detected in S. aureus 737 and 1609 and S. epidermidis 1243, as well as those from pSCFS3 (9), pJP1 (14), and pERGB (35). (B) Representation of DNA sequence surrounding cfr detected in S. aureus 1900, as well as the P. vulgaris chromosomal DNA fragment containing cfr (19). The 6- and 8-bp repeats are boxed.
The cfr-carrying plasmid detected in S. aureus 1900 (pSA1900) was 7.9 kb and had five complete (including cfr) reading frames, of which four were possibly involved in plasmid replication (Δrep) and mobilization (rec, hp, and pre/mob). This context showed complete identity to the flanking regions of the cfr gene detected in the chromosome of a P. vulgaris isolate from a pig in China (JF969273) (13). Interestingly, hybridization experiments using a cfr-specific probe exhibited signals from three distinct plasmids on S. aureus 1900 (Fig. 2), which indicates that a smaller and circular plasmid form was sequenced and that likely this fragment was also inserted in other larger plasmids detected in S. aureus 1900 (Fig. 2).
More detailed plasmid analysis demonstrated that cfr was plasmid located in all isolates, and further sequencing results disclosed that the genetically unrelated S. aureus 737 (ST239) and 1609 (ST5) and S. epidermidis 1243 shared a closely related cfr-carrying plasmid, which was then identified as similar to the previously reported pSCFS3 from bovine S. lentus (9). Similar findings have been reported by Kehrenberg et al. (29), who described the detection of pSCFS3 in other lineages (ST9 and ST398) of livestock-associated S. aureus recovered from swine during a survey among 367 farms in Germany. pSCFS3-like plasmids seem to be nonconjugative (29); however, they have been shown to be transferable in vitro into another staphylococci in this study and elsewhere (9, 29). Although pSCFS3-like plasmids are considered nonconjugative elements, these findings suggest that they can be transferred within staphylococci by other mechanisms. Moreover, it has been reported that nonconjugative staphylococcal plasmids can be mobilized in vivo by the copresence of a plasmid or chromosomally located conjugative machinery (30–32). In addition, mechanisms other than conjugation, such as mobilization, transduction, or rolling-circle replication, can also promote plasmid transfer (33).
In contrast, S. aureus 1900 carrying pSA1900 had a distinct cfr plasmid compared with pSCFS3 or with pSA737 and pSE1243. However, the genetic context of cfr observed in pSA1900 was similar to that described for the chromosome of a P. vulgaris isolate from China, where cfr seemed to have been mobilized by transposases (IS26 elements) (19). Three hybridization signals were observed during the experiments performed in this study to determine the cfr location in S. aureus 1900 and the respective transformant. Therefore, these results suggest that the small plasmid containing cfr (pSA1900) may have been replicated within the cell by the plasmid replication (Δrep) and mobilization (rec, hp, and pre/mob) genes observed or by a transduction mechanism (33). However, it is unclear whether this small cfr plasmid can be transferred within staphylococcal species, and further investigations are needed to clarify the mobilization of pSA1900.
In summary, previous studies describing outbreaks caused by Cfr-producing organisms have demonstrated that the vast majority of isolates were clonally related and that reduction of linezolid use and infection control measures have been effective in controlling the dissemination of clinical isolates (21, 34). However, this genetic investigation indicates that pSCFS3-like plasmids are widely disseminated and can be transferred within staphylococci, warranting continued surveillance for resistance dissemination. In addition, S. epidermidis may act as a possible reservoir for resistance genes, since this species is still often considered a contaminant in clinical settings and determinants may spread unnoticed.
ACKNOWLEDGMENTS
We thank the following staff members at JMI Laboratories, Inc. (North Liberty, IA): S. Benning, M. Castanheira, S. Farrell, and A. Costello for technical support and manuscript assistance.
This study was sponsored by Pfizer Inc., Collegeville, PA.
JMI Laboratories, Inc. (R.E.M., L.M.D., and R.N.J.), received research and educational grants from 2009 to 2012 from the following: American Proficiency Institute (API), Anacor, Astellas, AstraZeneca, Bayer, Cempra, Cerexa-Forest, Contrafect, Cubist, Daiichi, Dipexium, Enanta, Furiex, GlaxoSmithKline, Johnson & Johnson (Ortho McNeil), LegoChem Biosciences Inc., Meiji Seika Kaisha, Merck, Nabriva, Novartis, Pfizer (Wyeth), Rempex, Rib-X Pharmaceuticals, Seachaid, Shionogi, The Medicines Co., Theravance, ThermoFisher, and some other corporations. Some JMI employees are advisors or consultants for Astellas, Cubist, Pfizer, Cempra, Cerexa-Forest, Johnson & Johnson, and Theravance. The work conducted at the Friedrich-Loeffler-Institut was financially supported by German Federal Ministry of Education and Research (BMBF) grant number 01KI1014D (MedVet-Staph). S.S. received research grants from Pfizer Animal Health and Bayer Animal Health. M.D.H. and J.P.Q. were employees and shareholders of Pfizer Inc. at the time the study was conducted.
Footnotes
Published ahead of print 9 April 2013
REFERENCES
- 1. Ager S, Gould K. 2012. Clinical update on linezolid in the treatment of Gram-positive bacterial infections. Infect. Drug Resist. 5:87–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pfizer 2010. Zyvox package insert. Pfizer, New York, NY: http://www.pfizer.com/products/rx/rx_product_zyvox.jsp [Google Scholar]
- 3. Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U. S. A. 105:13339–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flamm RK, Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2012. LEADER surveillance program results for 2010: an activity and spectrum analysis of linezolid using 6801 clinical isolates from the United States (61 medical centers). Diagn. Microbiol. Infect. Dis. 74:54–61 [DOI] [PubMed] [Google Scholar]
- 5. Ross JE, Farrell DJ, Mendes RE, Sader HS, Jones RN. 2011. Eight-year (2002–2009) summary of the linezolid (Zyvox® Annu. Appraisal of Potency and Spectrum; ZAAPS) program in European countries. J. Chemother. 23:71–76 [DOI] [PubMed] [Google Scholar]
- 6. Shaw KJ, Barbachyn MR. 2011. The oxazolidinones: past, present, and future. Ann. N. Y. Acad. Sci. 1241:48–70 [DOI] [PubMed] [Google Scholar]
- 7. Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mendes RE, Deshpande L, Rodriguez-Noriega E, Ross JE, Jones RN, Morfin-Otero R. 2010. First report of staphylococcal clinical isolates in Mexico with linezolid resistance caused by cfr: evidence of in vivo cfr mobilization. J. Clin. Microbiol. 48:3041–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mendes RE, Deshpande LM, Costello A, Farrell DJ. 2012. Molecular epidemiology of Staphylococcus epidermidis clinical isolates from U. S. hospitals. Antimicrob. Agents Chemother. 56:4656–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diaz L, Kiratisin P, Mendes R, Panesso D, Singh KV, Arias CA. 2012. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob. Agents Chemother. 56:3917–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, Du XD, Dai L, Zhang W, Zhang Q, Shen J. 2012. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob. Agents Chemother. 56:1650–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu C, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J. Antimicrob. Chemother. 67:1824–1827 [DOI] [PubMed] [Google Scholar]
- 15. Zhang WJ, Wu CM, Wang Y, Shen ZQ, Dai L, Han J, Foley SL, Shen JZ, Zhang Q. 2011. The new genetic environment of cfr on plasmid pBS-02 in a Bacillus strain. J. Antimicrob. Chemother. 66:1174–1175 [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Schwarz S, Shen Z, Zhang W, Qi J, Liu Y, He T, Shen J, Wu C. 2012. Co-location of the multiresistance gene cfr and the novel streptomycin resistance gene aadY on a small plasmid in a porcine Bacillus strain. J. Antimicrob. Chemother. 67:1547–1549 [DOI] [PubMed] [Google Scholar]
- 17. Dai L, Wu CM, Wang MG, Wang Y, Wang Y, Huang SY, Xia LN, Li BB, Shen JZ. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob. Agents Chemother. 54:3953–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, He T, Schwarz S, Zhou D, Shen Z, Wu C, Wang Y, Ma L, Zhang Q, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J. Antimicrob. Chemother. 67:1094–1098 [DOI] [PubMed] [Google Scholar]
- 19. Wang Y, Wang Y, Wu CM, Schwarz S, Shen Z, Zhang W, Zhang Q, Shen JZ. 2011. Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J. Antimicrob. Chemother. 66:2521–2526 [DOI] [PubMed] [Google Scholar]
- 20. Hansen LH, Planellas MH, Long KS, Vester B. 2012. The order Bacillales hosts functional homologs of the worrisome cfr antibiotic resistance gene. Antimicrob. Agents Chemother. 56:3563–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bonilla H, Huband MD, Seidel J, Schmidt H, Lescoe M, McCurdy SP, Lemmon MM, Brennan LA, Tait-Kamradt A, Puzniak L, Quinn JP. 2010. Multicity outbreak of linezolid-resistant Staphylococcus epidermidis associated with clonal spread of a cfr-containing strain. Clin. Infect. Dis. 51:796–800 [DOI] [PubMed] [Google Scholar]
- 22. Mendes RE, Bonilla H, Deshpande LM, Huband MD, Jones RN, Quinn JP. 2010. IS21-558 insertion sequences associated with cfr dissemination in Staphylococcus epidermidis and Staphylococcus aureus recovered from two medical facilities in Ohio, abstr. C2-1500b. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother., 12 to 15 September 2010, Boston, MA [Google Scholar]
- 23. Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clinical and Laboratory Standards Institute 2012. M07-A9. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard: ninth edition. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 25. Clinical and Laboratory Standards Institute 2012. M100-S22. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 26. Traczewski MM, Brown SD. 2008. Proposed MIC and disk diffusion microbiological cutoffs and spectrum of activity of retapamulin, a novel topical antimicrobial agent. Antimicrob. Agents Chemother. 52:3863–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfizer 2011Tygacil package insert. Pfizer, New York, NY: http://www.tygacil.com [Google Scholar]
- 28. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 53:779–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Udo EE, Grubb WB. 1990. Excision of a conjugative plasmid from the staphylococcal chromosome. J. Med. Microbiol. 33:227–234 [DOI] [PubMed] [Google Scholar]
- 31. Udo EE, Jacob LE. 1998. Conjugative transfer of high-level mupirocin resistance and the mobilization of non-conjugative plasmids in Staphylococcus aureus. Microb. Drug Resist. 4:185–193 [DOI] [PubMed] [Google Scholar]
- 32. Zhu W, Clark N, Patel JB. 2013. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob. Agents Chemother. 57:212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O'Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 1:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sánchez García M, De la Torre MA, Morales G, Pelaez B, Tolon MJ, Domingo S, Candel FJ, Andrade R, Arribi A, Garcia N, Martinez Sagasti F, Fereres J, Picazo J. 2010. Clinical outbreak of linezolid-resistant Staphylococcus aureus in an intensive care unit. JAMA 303:2260–2264 [DOI] [PubMed] [Google Scholar]
- 35. Gopegui ER, Juan C, Zamorano L, Perez JL, Oliver A. 2012. Transferable multidrug resistance plasmid carrying cfr associated with tet(L), ant(4′)-Ia, and dfrK genes from a clinical methicillin-resistant Staphylococcus aureus ST125 strain. Antimicrob. Agents Chemother. 56:2139–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]