Abstract
The genetic context of the blaNDM-1 gene in the genome of Pseudomonas aeruginosa MMA83 was investigated. Sequencing of the cosmid selected for the blaNDM-1 gene revealed the presence of two blaNDM-1 copies in the genome of P. aeruginosa MMA83 in a unique genetic environment. Additionally, mating assays, DNA-DNA hybridization, and an S1 nuclease assay strongly suggest that the blaNDM-1 gene in P. aeruginosa MMA83 is chromosome borne.
TEXT
The genetic context of the blaNDM-1 gene has been extensively investigated, mostly among members of the family Enterobacteriaceae (1, 2, 3, 4, 5, 6, 7, 8). From a genetic perspective, the key finding is that the blaNDM-1 gene can be located on plasmids belonging to different incompatibility groups. Although blaNDM-1 genes have the same flanking regions, in a broader sequence context, they reside in diverse genetic environments (6). Although the blaNDM-1 gene is typically found in a plasmid, in certain Escherichia coli and Providencia stuartii isolates, it was chromosome borne (6). The blaNDM-1 gene is considered endemic to the Balkan region, and this gene has been found not only in Enterobacteriaceae (9) but also in Pseudomonas aeruginosa (10). Considering the role of P. aeruginosa in the development of nosocomial infections worldwide and the subsequent complicated clinical management of patients infected with this pathogen, the emergence of NDM-1-positive strains is alarming. This is why the analysis of the genetic context of the blaNDM-1 gene, not only in Enterobacteriaceae but also in all other NDM-1-positive isolates, is crucial and could reveal associations with other genes that confer antimicrobial resistance and potential routes of dissemination. The objective of our study was to identify the genetic location and genetic context of the blaNDM-1 gene in P. aeruginosa MMA83, one of the NDM-1-positive clinical isolates from Serbia.
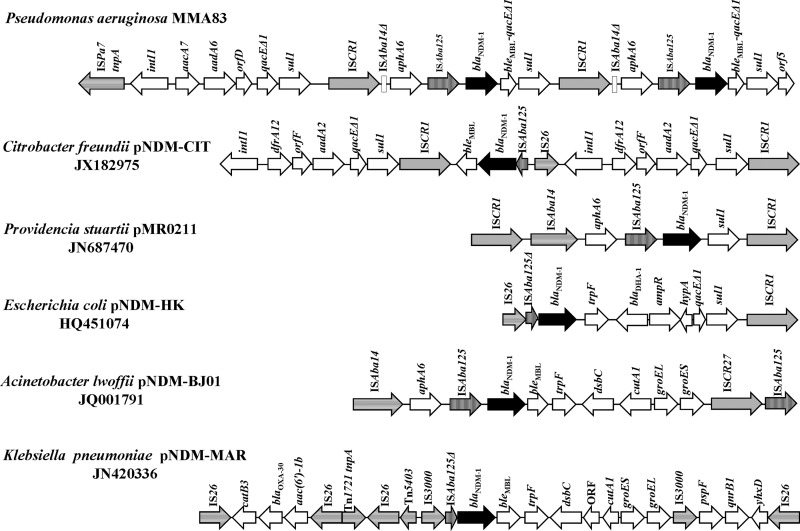
To determine the genetic location of the blaNDM-1 gene in P. aeruginosa MMA83, transformation, mating assays, and DNA-DNA hybridization were performed. Heat shock transformation of E. coli DH5α was performed with total DNA isolated from P. aeruginosa MMA83 (selection based on 100 μg/ml ampicillin). Triparental mating assays with P. aeruginosa MMA83 as a donor, E. coli(pRK2013) as a helper, and two recipients, E. coli DH5α (selection based on 100 μg/ml ampicillin) and an imipenem-sensitive isolate of Pseudomonas mendocina (selection based on 32 μg/ml imipenem), did not result in any transconjugant (11). Counterselection for recipients was based on glycerol for P. mendocina and sucrose for E. coli DH5α as the sole carbon sources. This result suggested that the gene(s) conferring resistance to β-lactams was not present on a conjugative plasmid(s) in MMA83. To find out if MMA83 is a plasmid-free strain, total DNA was isolated in agarose blocks and treated with S1 nuclease, which is used to visualize large, supercoiled plasmids (12). The results obtained confirmed that MMA83 does not carry any plasmid (see Fig. S1 in the supplemental material). Also, pulsed-field gel electrophoresis and subsequent DNA-DNA hybridization of a blaNDM-1 gene probe (1,059-bp EcoRI fragment) (11) with native (not digested) and S1 nuclease-treated total MMA83 DNA revealed the location of the signal in the region where chromosomal DNA migrates (see Fig. S1). These results strongly suggest that the blaNDM-1 gene is chromosome borne rather than plasmid located in MMA83. This is an unusual but not unique hallmark, since chromosomally located blaNDM-1 genes have been described previously (6). To uncover the genetic context of the blaNDM-1 gene, two cosmid libraries of MMA83 were constructed with Gigapack III Gold packaging extract (Stratagene, Amsterdam, Netherlands). Total DNA was partially digested with either BamHI or EcoRI and cloned into the pLAFR3 cosmid (11). Cosmid libraries were screened for the presence of β-lactamases by plating on tetracycline (20 μg/ml, selection for cosmid) and ampicillin (100 μg/ml). The cosmid selected from the BamHI cosmid library conferring resistance to β-lactams was designated pLAFRBamNDM, and the cosmid selected from the EcoRI library was designated pLAFREcoNDM. pLAFRBamNDM encompasses only one large BamHI fragment, so the order of genes in pLAFRBamNDM is the same as in the genome of strain MMA83, from which it originated. Afterwards, pLAFRBamNDM was digested with BamHI and XbaI, in which it produced three fragments of 8,292 bp (BamHI-XbaI fragment), 6,451 bp (XbaI-XbaI fragment), and 4,541 bp (XbaI-BamHI fragment). These fragments were cloned into a pBluescript vector (Kanr) digested with the same enzymes. The clones obtained were confirmed by restriction enzyme analysis and subsequently sequenced by primer walking. Cosmid pLAFREcoNDM was digested with EcoRI and PstI, and plasmid libraries were constructed in pBluescript (Kanr). The resulting constructs were sequenced and used as a control in the process of aligning and stacking sequences derived from BamHI-XbaI, XbaI-XbaI, and XbaI-BamHI fragments of pLAFRBamNDM. The order of the open reading frames in fragments from pLAFREcoNDM was the same as that in pLAFRBamNDM, which confirmed that the order of genes in pLAFRBamNDM was the same as in the genome of strain MMA83. Surprisingly, these results revealed that P. aeruginosa MMA83 carries two copies of the blaNDM-1 gene (Fig. 1). Moreover, hybridization of total DNA digested with different restriction enzymes with a blaNDM-1 gene probe revealed the presence of signals at positions predicted based on the sequence of a cloned BamHI fragment of 19.2 kb (see Fig. S1 in the supplemental material). Studies published thus far describe the presence of only one copy of the blaNDM-1 gene in the chromosome or plasmids of bacteria. To our knowledge, ours is the first observation and report of two blaNDM-1 gene copies. Amplification of genes conferring resistance to antibiotics, as a common adaptive mechanism in bacteria, could be related to a gene dosage effect (13). The genetic environment of the blaNDM-1 gene in the genome of MMA83 showed certain similarities to but, in general, was different from the nucleotide sequences of the other bacterial strains analyzed. ISAba125 insertion sequences were identified immediately upstream of the blaNDM-1 genes (Fig. 1). As observed in other blaNDM-1 gene environments, ISAba125 provides the −35 promoter sequence for the blaNDM-1 gene (6). The ISCR1 mobile element lies upstream of both copies of the blaNDM-1 gene and is followed by the gene encoding type VI aminoglycoside phosphotransferase. Since the regions downstream of both ISCR1 elements are identical, it is possible that the two copies of the blaNDM-1 gene were the result of genetic duplication via ISCR1-mediated gene mobilization. ISCR1 undergoes rolling-circle transposition, in which a single IS element can mobilize the sequences to which it is attached and subsequent homologous recombination may result in gene duplication (14). The genetic organization downstream of the blaNDM-1 genes in MMA83 is the same, encompassing the fusion of a bleomycin resistance gene and the qacEΔ1 gene, followed by the sul1 gene. The regions upstream of both blaNDM-1 gene copies in MMA83 are also identical and show similarity to the regions upstream of the blaNDM-1 gene in pNDM-BJ01 and pMR0211 (4, 5). In addition, regions downstream of the blaNDM-1 genes in MMA83 show some similarity to regions of pMR0211 and pNDM-BJ01 downstream from the blaNDM-1 gene but are not identical to them (Fig. 1). The first ISCR1 insertion sequence was preceded by a class 1 integron harboring aacA7, aadA6, orfD, and qacEΔ1 and sul1 resistance genes. This is not atypical, since ISCR1 is commonly associated with class 1 integrons (14). Moreover, although the blaNDM-1 gene is associated with other bla genes, in the case of P. aeruginosa MMA83, no other β-lactamase-encoding gene was found in the vicinity of two blaNDM-1 genes.
Fig 1.
Comparative schematic representations of the blaNDM-1 gene's genetic context in the genomes of Enterobacteriaceae and Acinetobacter and the genetic environment of the blaNDM-1 gene in P. aeruginosa MMA83. Insertion sequence transposase genes are drawn as arrows filled with different patterns. The box in the scheme of the MMA83 blaNDM-1 gene's genetic context represents the left end of ISAba14. The selection of the genetic environments represented was based on the presence of ISCR1 and relationships to different bacterial genera.
This study provides new data on the complexity and diversity of genetic features associated with the blaNDM-1 gene, which are of great importance for further analyses of dissemination routes and mechanisms, including clonal dispersions of the blaNDM-1 gene within clinical isolates.
Nucleotide sequence accession number.
The nucleotide sequence of the BamHI fragment (19,272 bp) obtained from the pLAFRBamNDM cosmid is available in GenBank/EMBL under accession number HF546976.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant 173019 from the Ministry of Education, Science and Technological Development of the Republic of Serbia.
We are grateful to Myra (Macpherson) Poznanovic, official native English editor of the scientific journal Archives of Biological Sciences (Belgrade), for proofreading the manuscript.
Footnotes
Published ahead of print 22 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02312-12.
REFERENCES
- 1. Bonnin RA, Poirel L, Carattoli A, Nordmann P. 2012. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One 7:e34752. 10.1371/journal.pone.0034752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dolejska M, Villa L, Poirel L, Nordmann P, Carattoli A. 2013. Complete sequencing of an IncHI plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 68:34–39 [DOI] [PubMed] [Google Scholar]
- 3. Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989. 10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56:1698–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGann P, Hang J, Clifford RJ, Yang Y, Kwak YI, Kuschner RA, Lesho EP, Waterman PE. 2012. Complete sequence of a novel 178-kilobase plasmid carrying blaNDM-1 in a Providencia stuartii strain isolated in Afghanistan. Antimicrob. Agents Chemother. 56:1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, Hishinuma A, Arakawa Y, Kuroda M. 2011. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6:e25334. 10.1371/journal.pone.0025334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. 2012. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15, and qnrB1 genes. J. Antimicrob. Chemother. 67:1645–1650 [DOI] [PubMed] [Google Scholar]
- 9. Mirovic V, Tomanovic B, Lepsanovic Z, Jovcic B, Kojic M. 2012. Isolation of Klebsiella pneumoniae producing NDM-1 metallo-β-lactamase from the urine of an outpatient baby boy receiving antibiotic prophylaxis. Antimicrob. Agents Chemother. 56:6062–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jovcic B, Lepsanovic Z, Suljagic V, Rackov G, Begovic J, Topisirovic L, Kojic M. 2011. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 55:3929–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Staskawicz B, Dahlbeck D, Keen N, Napoli C. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large megaplasmids. Anal. Biochem. 226:235–240 [DOI] [PubMed] [Google Scholar]
- 13. Normark S, Edlund T, Grundström T, Bergström S, Wolf-Watz H. 1977. Escherichia coli K-12 mutants hyperproducing chromosomal beta-lactamase by gene repetitions. J. Bacteriol. 132:912–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.