Abstract
Selective decontamination of the digestive tract (SDD) selectively eradicates aerobic Gram-negative bacteria (AGNB) by the enteral administration of oral nonabsorbable antimicrobial agents, i.e., colistin and tobramycin. We retrospectively investigated the impact of SDD, applied for 5 years as part of an infection control program for the control of an outbreak with extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in an intensive care unit (ICU), on resistance among AGNB. Colistin MICs were determined on stored ESBL-producing K. pneumoniae isolates using the Etest. The occurrence of both tobramycin resistance among pathogens intrinsically resistant to colistin (CIR) and bacteremia caused by ESBL-producing K. pneumoniae and CIR were investigated. Of the 134 retested ESBL-producing K. pneumoniae isolates, 28 were isolated before SDD was started, and all had MICs of <1.5 mg/liter. For the remaining 106 isolated after starting SDD, MICs ranged between 0.5 and 24 mg/liter. Tobramycin-resistant CIR isolates were found sporadically before the introduction of SDD, but their prevalence increased immediately afterward. Segmented regression analysis showed a highly significant relationship between SDD and resistance to tobramycin. Five patients were identified with bacteremia caused by ESBL-producing K. pneumoniae before SDD and 9 patients thereafter. No bacteremia caused by CIR was found before SDD, but its occurrence increased to 26 after the introduction of SDD. In conclusion, colistin resistance among ESBL-producing K. pneumoniae isolates emerged rapidly after SDD. In addition, both the occurrence and the proportion of tobramycin resistance among CIR increased under the use of SDD. SDD should not be applied in outbreak settings when resistant bacteria are prevalent.
INTRODUCTION
Colistin, a polypeptide antibiotic belonging to the polymyxin group, was initially used for the treatment of a variety of Gram-negative bacterial infections. However, because of its significant side effects and the introduction into clinical practice of less toxic antibiotics, the intravenous formulations of colistin were gradually abandoned in the early 1980s in most parts of the world (1). Oral nonabsorbable compounds, nebulized formulations, and topical preparations of colistin continue to be used for selective decontamination of the digestive tract (SDD); for treatment of cystic fibrosis patients colonized with Pseudomonas aeruginosa; and for therapy of otitis, conjunctivitis, and skin infections (1). With the increasing prevalence of infections caused by multidrug-resistant (MDR) Gram-negative bacteria while the discovery and development of newer, effective antibiotics has declined, colistin has reemerged as the last-resort therapy for many infections. Fortunately, recent clinical data show that the use of colistin is acceptably safe if certain precautions are taken (2). Of great concern, however, is the emergence, all over the world, of colistin resistance in multidrug-resistant clinical strains of Acinetobacter baumannii (3) and P. aeruginosa (4). This study reports on the impact of long-term use of topical colistin as part of a regimen for SDD started during an outbreak involving extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae in an intensive care unit (ICU) on resistance among aerobic Gram-negative bacteria (AGNB).
MATERIALS AND METHODS
Setting.
SDD was applied for 5 years to all patients admitted to the ICU as part of an infection control program for the control of an outbreak involving ESBL-producing K. pneumoniae (Fig. 1). SDD was given as a topical mixture of nonabsorbable antibiotics, including tobramycin, colistin, and amphotericin B (doses of 80, 100, and 500 mg, respectively), applied on the buccal mucosa and as a suspension administered via a nasogastric tube in the gastrointestinal tract four times a day (5). The aim of the SDD treatment was to reduce colonization of the digestive tract with resistant bacteria (5).
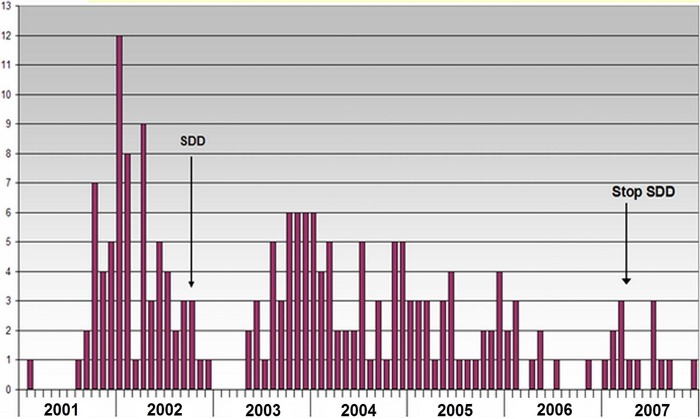
Fig 1.
Monthly observed numbers of patients harboring extended-spectrum beta-lactamase-producing K. pneumoniae between 2001 and 2008 (n = 197). The ICU was closed from January through April 2003.
Surveillance.
Routine microbiological screening, started in February 2002, was performed for all patients on admission to the ICU and then twice a week. Screening was done by culture of throat and rectal-swab specimens and, in intubated patients, of additional tracheal-fluid samples. When clinically indicated, samples were also obtained from relevant body sites, such as wounds.
Bacteriological methods.
Biochemical tests were routinely performed by using the API 20E system (bioMérieux, Marcy l'Étoile, France). Antimicrobial susceptibility was determined by the agar dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (6). CLSI did not include criteria for colistin susceptibility testing (6). Colistin susceptibility testing was performed by the disc diffusion method (potency of the disc, 10 μg) as recommended in the package insert, where zone diameter breakpoints of ≥11 mm (susceptible), 9 to 10 mm (intermediate), and ≤8 mm (resistant) were used. ESBL confirmation was performed with the double-disc synergy test (7). ESBL-producing K. pneumoniae isolates were stored at −70°C.
Retrospective analysis.
In 2008, a retrospective study was undertaken on existing databases to determine the exact extent of the ESBL-producing K. pneumoniae outbreak, to obtain data on colistin antimicrobial susceptibility among ESBL-producing K. pneumoniae isolates, and to determine the occurrence of pathogens intrinsically resistant to colistin (CIR) (Proteus, Morganella, and Serratia spp.) that were tobramycin resistant (CIR-tR) during the use of SDD. The occurrence of bacteremia caused by ESBL-producing K. pneumoniae and CIR was also investigated. In addition, isolates stored before and after the start of SDD as ESBL-producing K. pneumoniae were retrieved for further analysis. The Vitek 2 Advanced Expert System (bioMérieux, Marcy l'Étoile, France) was used for reidentification of the strains and to determine colistin susceptibility using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (http://www.eucast.org/clinical_breakpoints). ESBL screening and confirmation were performed according to national guidelines (laboratory detection of highly resistant microorganisms [HRMO]; http://www.nvmm.nl). The colisitin MIC was determined by the Etest using EUCAST breakpoints (http://www.eucast.org/clinical_breakpoints). DiversiLab (bioMérieux, Marcy l'Étoile, France), a semiautomated repetitive-sequence-based PCR (REP-PCR) (8), was used to investigate the clonality of the ESBL-producing K. pneumoniae isolates, using >95% similarity as the similarity threshold.
Statistical analysis.
Segmented regression analysis was performed to test the regression slopes pre- and post-SDD and to quantify the difference in tobramycin resistance among CIR by comparing the intercepts at the time of the start of SDD. To further investigate the impact of SDD on tobramycin resistance among CIR isolates from surveillance culture and from clinically indicated cultures, a logistic regression model was fitted to the data. The test outcome (1 or 0) of tobramycin resistance was taken as a response variable. The explanatory variables were sampling date (day), SDD (before, during, and after), and culture (surveillance or clinically indicated). The sudden jump in the proportion of resistant isolates after the start of SDD was incorporated in the regression model, as well as the decrease in the proportion of resistant isolates after stopping SDD. No sudden jump was present here. To avoid numerical problems, it was assumed that there was no difference in the proportion of resistant isolates between the two cultures before the start of SDD. Differences in proportions of resistant isolates are reported as absolute risk differences (RD), while slopes are reported as odds ratios (OR) per year, together with their corresponding 95% confidence intervals (CI) and P values.
RESULTS
Between January 2001 and January 2008, 197 ESBL-producing K. pneumoniae-positive patients were identified (Fig. 1). Isolates from 134 out of the 197 identified cases (one isolate per patient) were included in the present analysis. Upon retesting, all isolates were confirmed to be ESBL-producing K. pneumoniae. Most ESBL-producing K. pneumoniae isolates were resistant to tobramycin prior to the introduction of SDD (data not shown). Data on colistin susceptibility, as determined by disc diffusion, could be found for 89 ESBL-producing K. pneumoniae isolates and are shown in Table 1. All isolates tested before the introduction of SDD were found susceptible to colistin by this method. Four isolates (4.5%), all isolated after starting SDD, were resistant (Table 1). The results of the antimicrobial susceptibility testing of stored isolates as performed by the Vitek method revealed that of the 134 tested isolates, all 28 isolates obtained before the start of SDD were colistin susceptible. However, of the remaining 106, isolated after starting SDD, 75 (70.75%) were colistin resistant (Table 1). The results of the Vitek susceptibility testing were confirmed by the Etest method (Table 1). All isolates obtained before SDD had colisitin MICs of <1.5 mg/liter, but the MIC values rose to 24 mg/liter in those tested isolates that were obtained after the introduction of SDD (Fig. 2).
Table 1.
Susceptibilities of ESBL-producing K. pneumoniae isolates to colistin as determined by disc diffusion, Vitek, and Etest
| Isolate groupa | No. of isolatesb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Disc diffusion (n = 89) |
Vitek (n = 134) |
Etest (n = 134) |
|||||||
| S | I | R | S | I | R | S | I | R | |
| Before SDD | 12 | 0 | 0 | 28 | 0 | 0 | 28 | 0 | 0 |
| After SDD | 45 | 28 | 4 | 31 | 0 | 75 | 32 | 0 | 74 |
Isolates are grouped according to whether they were identified before or after the introduction of SDD on the ICU in October 2002.
S, susceptible; I, intermediate; R, resistant.
Fig 2.
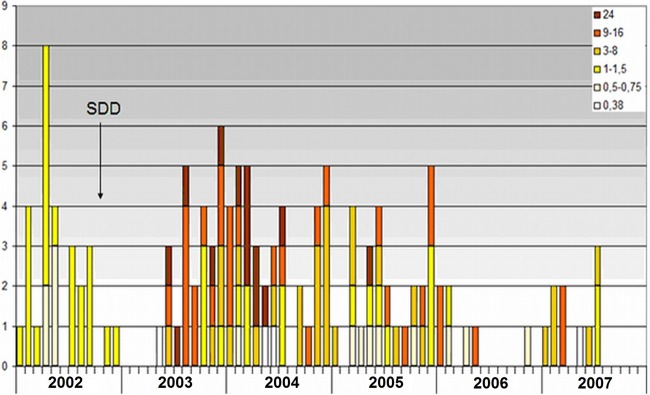
Susceptibility to colistin among 134 extended-spectrum beta-lactamase-producing K. pneumoniae isolates, as retrospectively determined by the Etest method. MICs (mg/liter) are shown.
Typing of the 134 isolates by REP-PCR revealed that the majority were clonally related, as 114 were identical by this method, 28 of which were isolated before the start of SDD and 86 after. Of the remaining 20, 1 cluster contained 5 isolates, 4 clusters each contained 2 isolates, and the remaining 7 isolates had different patterns. Colistin MICs among the ESBL-producing K. pneumoniae isolates that were obtained after the start of SDD and that belonged to the major clone varied between 0.5 and 24 mg/liter. Of the 74 colistin-resistant ESBL-producing K. pneumoniae isolates, 71 belonged to the major clone and 3 had different patterns.
After the ward was reopened in April 2008, ESBL-producing K. pneumoniae isolates were sporadically detected in the ICU; all were colistin susceptible and genotypically unrelated to the outbreak, as investigated by DiversiLab.
Between 2001 and 2008, data on 2,572 CIR could be retrieved (Fig. 3). CIR isolates were found sporadically before the introduction of SDD, but their prevalence increased immediately afterward (Fig. 3). Segmented regression analysis showed a highly significant relationship between the introduction of SDD and resistance to tobramycin, with an RD value for the increase in the proportion of tobramycin-resistant isolates at the start of SDD of 0.41 (95% CI, 0.33 to 0.47; P value < 0.001) (Fig. 4). Figure 5 shows the proportions of resistant isolates for the surveillance cultures (black dots) and the clinically indicated cultures (gray triangles) between 2001 and 2009. After the start of SDD, the proportion of resistant isolates suddenly increases. The increase for the surveillance cultures (RD = 0.51 [95% CI, 0.42 to 0.57; P value < 0.001]) is greater than the increase for the clinically indicated cultures (RD = 0.27 [95% CI, 0.18 to 0.34; P value < 0.001]). The difference between the two is 0.24 (95% CI, 0.15 to 0.32; P value < 0.001). During the SDD period, the slope, in terms of an odds ratio, for the surveillance cultures is as follows: OR = 1.34 (95% CI, 1.21 to 1.48). For the clinically indicated cultures, the slope is steeper (OR = 1.58 [95% CI, 1.43 to 1.76]). The difference is as follows: OR = 0.85 (95% CI, 0.73 to 0.98; P value = 0.023). After stopping SDD, the proportion of resistant isolates in the clinically indicated cultures sharply decreases, while it does not for the surveillance cultures.
Fig 3.
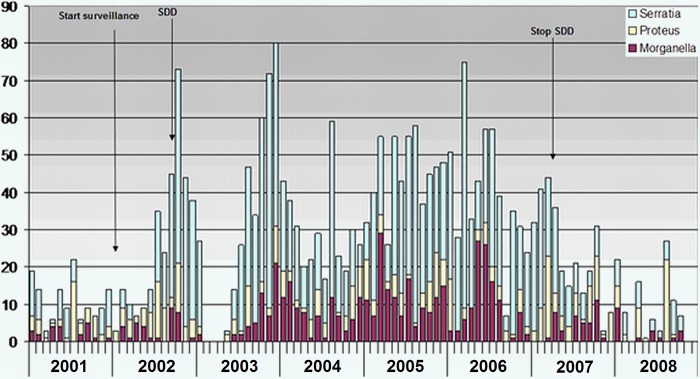
Monthly occurrence of CIR isolates (n = 2,572) between 2001 and 2008.
Fig 4.
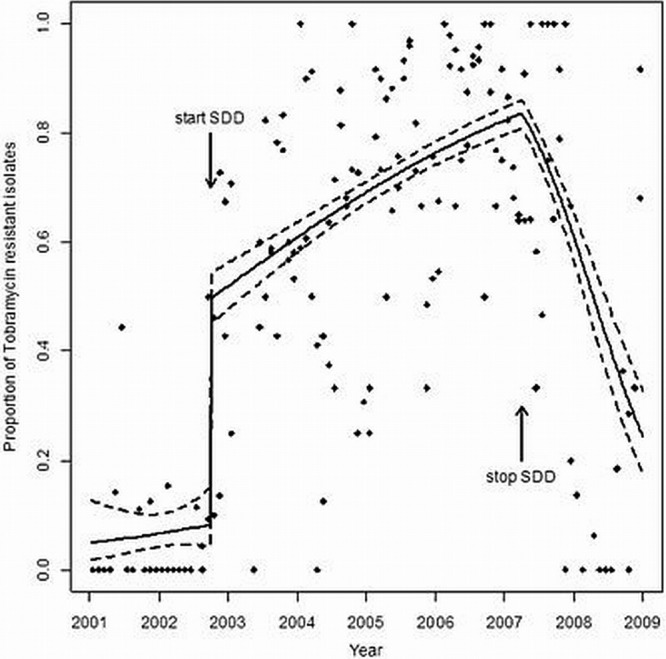
Segmented regression analysis showing the relationship between the introduction of SDD and tobramycin resistance among CIR isolates (black dots). The logistic regression model fit (solid lines) is shown, together with its 95% confidence interval (dashed lines).
Fig 5.
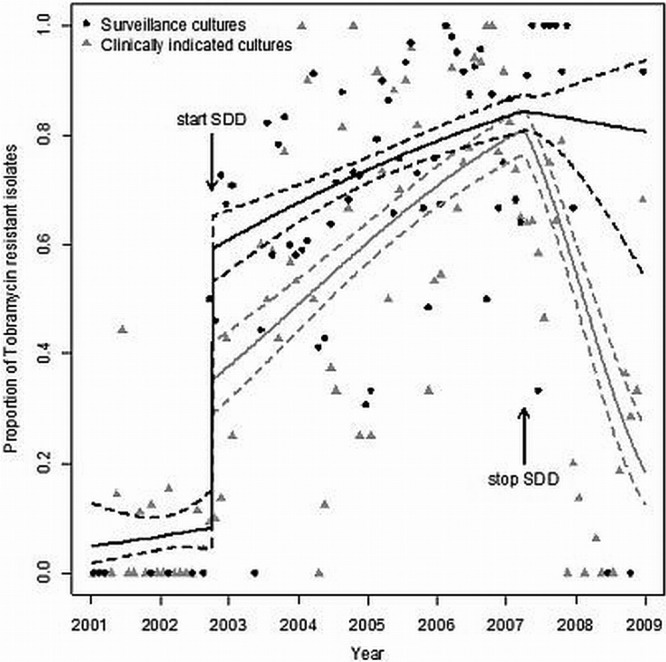
Proportions of tobramycin-resistant isolates from surveillance cultures (black dots) and clinically indicated cultures (gray triangles) between 2001 and 2009. The logistic regression model fit (solid lines) is shown, together with its 95% confidence interval (dashed lines).
Five patients were identified with bacteremia caused by ESBL-producing K. pneumoniae before the introduction of SDD and 9 patients thereafter (Fig. 6). No bacteremia caused by CIR was found before SDD, but its occurrence increased to 26 after the introduction of SDD, 24 of which were caused by Serratia spp. and 2 by Morganella spp. (Fig. 6).
Fig 6.
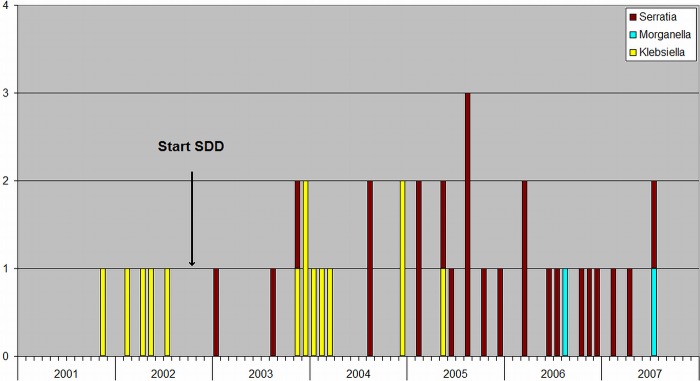
Monthly occurrence of bacteremia caused by CIR (Proteus, Morganella, and Serratia spp.) and extended-spectrum beta-lactamase-producing K. pneumoniae between 2001 and 2008.
DISCUSSION
The main findings of this study are the emergence under SDD of colistin resistance among ESBL-producing K. pneumoniae isolates, the increase in both the prevalence and the proportion of tobramycin resistance among CIR, and the increase in occurrence of bacteremia caused by organisms resistant to SDD, which had been introduced in an outbreak setting.
The potential for the emergence of resistance to colistin among sensitive organisms has been found to be low (9). Nonetheless, increase in colistin-resistant aerobic Gram-negative strains during the use of SDD has been observed (10), and with the increasing systemic use of colistin against multidrug-resistant pathogens, development of resistance to the agent has frequently been reported in recent years in nosocomial strains (11), including MDR K. pneumoniae (12).
In this study, we retrospectively investigated the impact of long-term use of colistin as part of SDD on resistance among ESBL-producing K. pneumoniae and CIR isolates during an outbreak.
Our data show that the routinely used disc diffusion method failed to detect colistin resistance. As shown in Table 1, most strains that were considered intermediate or susceptible to colistin were found to be resistant when retested by the Vitek method. The Vitek method has been found to be reliable in susceptibility testing for colistin in clinical strains, including Klebsiella spp., compared to broth microdilution as the reference method (10). The observed inaccuracy of disc diffusion testing of colistin has also been shown in numerous studies, and therefore, alternative methods should be used (11).
The results of this study demonstrate an association between prolonged use of colistin as part of SDD and the emergence of colistin resistance among ESBL-producing K. pneumoniae isolates. The majority of resistant isolates were clonally related, as evidenced by REP-PCR typing, indicating horizontal transmission. All strains isolated before the start of SDD that were colistin susceptible belonged to the same clone as those isolated thereafter that were colistin resistant. In addition, 3 isolates, all obtained after the start of SDD, that were unrelated to the major clone were also colistin resistant, suggesting that resistance emerged through selection pressure. Interestingly, the clonally related strains that were isolated after the start of SDD exhibited a wide range of colistin MIC values, ranging between 0.5 and 24 mg/liter (Fig. 2), a finding that has also been described in a recent study (12) where colistin-resistant K. pneumoniae carbapenemase (KPC)-producing K. pneumoniae strains obtained from patients admitted to the same ward and belonging to the same clone showed different MIC values. This can possibly be explained by the presence of heteroresistant subpopulations within the colistin-susceptible MDR strains after exposure to colistin, as has recently been shown in clinical strains of Acinetobacter (28) and K. pneumoniae (13). Combined with the fact that the majority of strains involved in our study were tobramycin resistant, these data suggest a potential role of monotherapy with colistin in the emergence of resistance.
This study also shows an increase in both the occurrence of CIR and resistance to tobramycin among these isolates during SDD use (Fig. 3 and 4). This may represent selection pressure under SDD. Horizontal spread, however, cannot be excluded, since no genotypic analysis of these isolates has been performed. CIR-tR consisted mainly of Serratia spp., which can be explained by the fact that these species are known to produce aminoglycoside-modifying enzymes that may affect the activity of tobramycin (http://www.eucast.org/expert_rules), rendering them resistant to both SDD components.
As in other studies, detection of antibiotic-resistant nosocomial pathogens was more frequently achieved by surveillance cultures than by clinical cultures alone (14–16). Figure 5 shows that the increase in the proportion of tobramycin-resistant CIR isolates was greater for the surveillance cultures than for the clinically indicated cultures (an increase in RD of 0.24; P value < 0.001). The figure also shows, in accordance with other studies (17, 18), that the increasing rates of tobramycin resistance were not, at least during the study period, returned to baseline by cessation of SDD.
SDD has been used as a method designed to prevent infection in critically ill patients. In settings with low levels of circulating antibiotic-resistant organisms, SDD has been found not to be associated with increased selection or induction of antibiotic resistance (19, 20). However, in settings with high levels of endemic, multidrug-resistant Gram-negative bacteria, SDD was associated with increased selection of such pathogens (19). In the present study, SDD had been started in an outbreak setting, when enhanced infection control measures were found insufficient to control the outbreak. The aim of SDD was to reduce colonization of the digestive tract with resistant bacteria (5), which, in theory, could have a synergistic effect with infection control measures (20). Reductions of bacterial loads at places frequently contacted by nursing staff would reduce the likelihood of cross-transmission (20). Only a few studies have investigated SDD in such settings (5). The results of studies of SDD in outbreak situations when SDD was introduced in our hospital were conflicting. Some had shown that the outbreak with multiresistant Gram-negative bacilli, including Klebsiella spp., could be controlled (21, 22), while in others, SDD alone failed to reduce the incidence of acquisition of outbreak strains (23). In a recent study assessing the effectiveness of SDD for eradicating carbapenem-resistant K. pneumoniae oropharyngeal and gastrointestinal carriage, it was concluded that in outbreaks caused by carbapenem-resistant K. pneumoniae infections that are uncontrolled by routine infection control measures, SDD could provide additional infection containment (24). SDD was also found beneficial in decreasing the intestinal reservoir in ICU patients with A. baumannii colonization, and no development of resistance to colistin was detected in the strains isolated in the course of SDD (25). In another recent study, however, oral colistin did not prevent colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae and appeared to select for colistin-resistant strains or to induce colistin resistance (26). The results from the present study provide arguments against the use of SDD in outbreak settings when resistant bacteria are prevalent. Not only did SDD fail to control the ESBL-producing K. pneumoniae outbreak, it also led to a dramatic increase in resistance, and its introduction was associated with an increase in the occurrence of bacteremia caused by organisms resistant to SDD.
A possible explanation is the selection under SDD for resistant bacteria, which became predominant in the ICU environment, enhancing their likelihood for cross-transmission. This has previously been shown for vancomycin-resistant enterococci (VRE) when their colonization pressure, once high, became the major variable affecting VRE acquisition (27).
In conclusion, our study adds evidence to existing data showing the emergence of colistin resistance in ESBL-producing K. pneumoniae strains exposed to prolonged treatment with colistin. The strains were resistant to tobramycin, which was combined with colistin in the SDD regimen, suggesting a potential role for monotherapy in resistance and emphasizing the need for further studies to determine whether combination therapy of colistin with other agents prevents the emergence of resistance (11). This study further provides evidence against the use of SDD in outbreak settings and supports the rational use of colistin, combined with a monitoring surveillance program for colistin resistance and the strict and timely implementation of infection control rules, as a main factor in avoiding the emergence and spread of resistance.
ACKNOWLEDGMENTS
We thank Jan van de Kassteele (Department of Statistics, Mathematical Modeling and Data Logistics, National Institute for Public Health and the Environment, RIVM, The Netherlands) for his statistical assistance.
Footnotes
Published ahead of print 29 April 2013
REFERENCES
- 1. Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 40:1333–1341 [DOI] [PubMed] [Google Scholar]
- 2. Falagas ME, Rafailidis PI. 2009. Nephrotoxicity of colistin: new insight into an old antibiotic. Clin. Infect. Dis. 48:1729–1731 [DOI] [PubMed] [Google Scholar]
- 3. Ko KS, Suh JY, Kwon KT, Jung S-I, Park K-H, Kang CI, Chung DR, Peck KR, Song J-H. 2007. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J. Antimicrob. Chemother. 60:1163–1167 [DOI] [PubMed] [Google Scholar]
- 4. Tan TY, Ng SY. 2006. The in-vitro activity of colistin in gram-negative bacteria. Singapore Med. J. 47:621–624 [PubMed] [Google Scholar]
- 5. van der Voort PHJ, van Saene HKF. (ed). 2008. Selective digestive tract decontamination in intensive care medicine: a practical guide to controlling infection. Springer-Verlag Italia, Milan, Italy [Google Scholar]
- 6. National Committee for Clinical Laboratory Standards 2000. Performance standards for antimicrobial susceptibility testing: 10th informational supplement M100-S10. NCCLS, Wayne, PA [Google Scholar]
- 7. Livermore DM, Brown DF. 2001. Detection of beta-lactamase-mediated resistance. J. Antimicrob. Chemother. 48(Suppl 1):59–64 [DOI] [PubMed] [Google Scholar]
- 8. Carretto E, Barbarini D, Farina C, Grosini A, Nicoletti P, Manso E, APSI Acinetobacter Study Group Italy 2008. Use of the DiversiLab semiautomated repetitive-sequence-based polymerase chain reaction for epidemiologic analysis on Acinetobacter baumannii isolates in different Italian hospitals. Diagn. Microbiol. Infect. Dis. 60:1–7 [DOI] [PubMed] [Google Scholar]
- 9. Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. 2005. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int. J. Antimicrob. Agents 25:11–25 [DOI] [PubMed] [Google Scholar]
- 10. Lo-Ten-Foe JR, de Smet AM, Diederen BM, Kluytmans JA, van Keulen PH. 2007. Comparative Evaluation of the VITEK 2, disk diffusion, Etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob. Agents Chemother. 51:3726–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin. Microbiol. Rev. 21:449–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin. Infect. Dis. 53:373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meletis G, Tzampaz E, Sianou E, Tzavaras I, Sofianou D. 2011. Colistin heteroresistance in carbapenemase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 66:946–947 [DOI] [PubMed] [Google Scholar]
- 14. Ben-David D, Maor Y, Keller Regev-Yochay NG, Tal I, Shachar D, Zlotkin A, Smollan G, Rahav G. 2010. Potential role of active surveillance in the control of a hospital-wide outbreak of carbapenem-resistant Klebsiella pneumoniae infection. Infect. Control Hosp. Epidemiol. 31:620–626 [DOI] [PubMed] [Google Scholar]
- 15. Calfee D, Jenkins SG. 2008. Use of active surveillance cultures to detect asymptomatic colonization with carbapenem-resistant Klebsiella pneumoniae in intensive care unit patients. Infect. Control Hosp. Epidemiol. 29:966–968 [DOI] [PubMed] [Google Scholar]
- 16. Lucet J-C, Decre D, Fichelle A, Joly-Guillou M-L, Pernet M, Deblangy C, Kosmann M-J, Regnier B. 1999. Control of a prolonged outbreak of extended-spectrum β-lactamase-producing Enterobacteriaceae in a university hospital. Clin. Infect. Dis. 29:1411–1418 [DOI] [PubMed] [Google Scholar]
- 17. Enne VI, Livermore DM, Stephens P, Hall LM. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325–1328 [DOI] [PubMed] [Google Scholar]
- 18. Willemsen I, Cooper B, van Buitenen C, Winters M, Andriesse G, Kluytmans J. 2010. Improving quinolone use in hospitals by using a bundle of interventions in an interrupted time series analysis. Antimicrob. Agents Chemother. 54:3763–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Smet AM, Kluytmans JA, Cooper BS, Mascini EM, Benus RF, van der Werf TS, van der Hoeven JG, Pickkers P, Bogaers-Hofman D, van der Meer NJ, Bernards AT, Kuijper EJ, Joore JC, Leverstein-van Hall MA, Bindels AJ, Jansz AR, Wesselink RM, de Jongh BM, Dennesen PJ, van Asselt GJ, te Velde LF, Frenay IH, Kaasjager K, Bosch FH, van Iterson M, Thijsen SF, Kluge GH, Pauw W, de Vries JW, Kaan JA, Arends JP, Aarts LP, Sturm PD, Harinck HI, Voss A, Uijtendaal EV, Blok HE, Thieme Groen ES, Pouw ME, Kalkman CJ, Bonten MJ. 2009. Decontamination of the digestive tract and oropharynx in ICU patients. N. Engl. J. Med. 360:20–31 [DOI] [PubMed] [Google Scholar]
- 20. Oostdijk EA, Wittekamp BH, Brun-Buisson C, Bonten MJ. 2012. Selective decontamination in European intensive care patients. Intensive Care Med. 38:533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brun-Buisson C, Legrand P, Rauss A, Richard C, Montravers F, Besbes M, Meakins JL, Soussy CJ, Lemaire F. 1989. Intestinal decontamination for control of nosocomial multiresistant gram-negative bacilli. Study of an outbreak in an intensive care unit. Ann. Intern. Med. 110:873–881 [DOI] [PubMed] [Google Scholar]
- 22. Taylor ME, Oppenheim BA. 1991. Selective decontamination of the gastrointestinal tract as an infection control measure. J. Hosp. Infect. 17:271–278 [DOI] [PubMed] [Google Scholar]
- 23. Decré D, Gachot B, Lucet JC, Arlet G, Bergogne-Bérézin E, Régnier B. 1998. Clinical and bacteriologic epidemiology of extended-spectrum beta-lactamase-producing strains of Klebsiella pneumoniae in a medical intensive care unit. Clin. Infect. Dis. 27:834–844 [DOI] [PubMed] [Google Scholar]
- 24. Saidel-Odes L, Polachek H, Peled N, Riesenberg K, Schlaeffer F, Trabelsi Y, Eskira S, Yousef B, Smolykov R, Codish S, Borer A. 2012. A randomized, double-blind, placebo-controlled trial of selective digestive decontamination using oral gentamicin and oral polymyxin E for eradication of carbapenem-resistant Klebsiella pneumoniae carriage. Infect. Control Hosp. Epidemiol. 33:14–19 [DOI] [PubMed] [Google Scholar]
- 25. Agustí C, Pujol M, Argerich MJ, Ayats J, Badía M, Domínguez MA, Corbella X, Ariza J. 2002. Short-term effect of the application of selective decontamination of the digestive tract on different body site reservoir ICU patients colonized by multi-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 49:205–208 [DOI] [PubMed] [Google Scholar]
- 26. Strenger V, Gschliesser T, Grisold A, Zarfel G, Feierl G, Masoud L, Hoenigl M, Resch B, Müller W, Urlesberger B. 2011. Orally administered colistin leads to colistin-resistant intestinal flora and fails to prevent faecal colonisation with extended-spectrum β-lactamase-producing enterobacteria in hospitalised newborns. Int. J. Antimicrob. Agents. 37:67–69 [DOI] [PubMed] [Google Scholar]
- 27. Bonten MJ, Slaughter S, Ambergen AW, Hayden MK, van Voorhis J, Nathan C, Weinstein RA. 1998. The role of “colonization pressure” in the spread of vancomycin-resistant enterococci: an important infection control variable. Arch. Intern. Med. 158:1127–1132 [DOI] [PubMed] [Google Scholar]
- 28. Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob. Agents Chemother. 52:351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]