Abstract
An NDM-1 carbapenemase-producing Pseudomonas aeruginosa isolate was recovered from a patient hospitalized in France after a previous hospitalization in Serbia. Genetic studies revealed that the blaNDM-1 gene was surrounded by insertion sequence ISAba125 and a truncated bleomycin resistance gene. This blaNDM-1 region was a part of the variable region of a new complex class 1 integron bearing IS common region 1 (ISCR1). The presence of ISPa7 upstream of this integron suggests insertion in a chromosomally located Tn402-like structure.
TEXT
Since the first description in Klebsiella pneumoniae and Escherichia coli, the NDM-1 carbapenemase has been identified in many different species, including Enterobacteriaceae and Acinetobacter baumannii (1, 2). To date, the Indian subcontinent and, more recently, Balkan countries constitute a reservoir of NDM producers (2–5). The corresponding gene, which is usually plasmid borne, is spreading worldwide in Enterobacteriaceae (2) and also in species isolated from water supplies in India, such as Pseudomonas putida and Pseudomonas pseudoalcaligenes (6). Recent reports have described the occurrence of chromosomally located blaNDM-1 in Acinetobacter baumannii related to transposon Tn125 (7, 8). Before 2012, only one report of two NDM-1-producing Pseudomonas aeruginosa isolates was described. Both were isolated in Serbia (9). To date, the transfer mechanisms, location, and blaNDM-1 genetic environment in P. aeruginosa remain unknown.
Here, we report the genetic environment of the blaNDM-1 gene in the first NDM-1-producing P. aeruginosa isolate from France, HIABP11. The isolate was recovered from a urine culture of a 63-year-old female patient, who was admitted to the infectious unit of the Bégin military hospital (Saint-Mandé, France) for an acute pyelonephritis complicated with renal microabscesses (10). Three months before her admission, the patient was hospitalized in Serbia for a posttraumatic fronto-temporo-parietal subdural hematoma surgical drainage. P. aeruginosa HIABP11 exhibited serotype O11 (monoclonal and polyclonal antisera; Bio-Rad Laboratories, Marne-la-Coquette, France). Susceptibility testing (disk diffusion and Etest) performed and interpreted according to the EUCAST guidelines showed that P. aeruginosa HIABP11 was resistant to all carbapenems (imipenem, meropenem, and doripenem) and antipseudomonal cephalosporins, as well as to aminoglycosides and fluoroquinolones. The isolate remained susceptible to piperacillin-tazobactam (MIC of 12 mg/liter) and colistin (MIC of 2 mg/liter) and intermediate to aztreonam (MIC of 3 mg/liter). Metallo-β-lactamase (MBL) production in the isolate was suggested by positive EDTA (bioMérieux, Marcy l'Etoile, France) and dipicolinic acid (KPC+MBL Confirm ID Pack; Rosco Diagnostica, Taastrup, Denmark) tests with imipenem and meropenem, respectively. PCR experiments were carried out on purified DNA with primers specific for known MBL genes, as previously described (11). Sequencing of amplification products confirmed the presence of the blaNDM-1 gene. Screening for additional β-lactamase genes, 16S RNA methylase genes, and qnr genes was negative. Interestingly, PCR experiments failed to detect the bleomycin resistance gene in P. aeruginosa HIABP11.
Repeated attempts to visualize a plasmid by the Kieser method (12) or to transfer by conjugation a plasmid using the recipient strain E. coli J53, as well as the rifampin-resistant P. aeruginosa strain PU21 (13), were unsuccessful, suggesting a chromosomal location for the blaNDM-1 gene. To gain further insight into the genetic environment, a BamHI library of P. aeruginosa HIABP11 was cloned into the BamHI-restricted plasmid pK18 (Kmr). Transformants of E. coli DH5α were selected on Mueller-Hinton (MH) agar plates containing 30 mg/liter kanamycin and 100 mg/liter ampicillin. In comparison to recipient strain DH5α, all of the recombinant clones tested displayed high resistance to β-lactams, except aztreonam. Sequencing of the whole insert (12,732 bp) from one of these clones allowed characterization of the genetic environment surrounding the blaNDM-1 gene (GenBank accession no. KC170992).
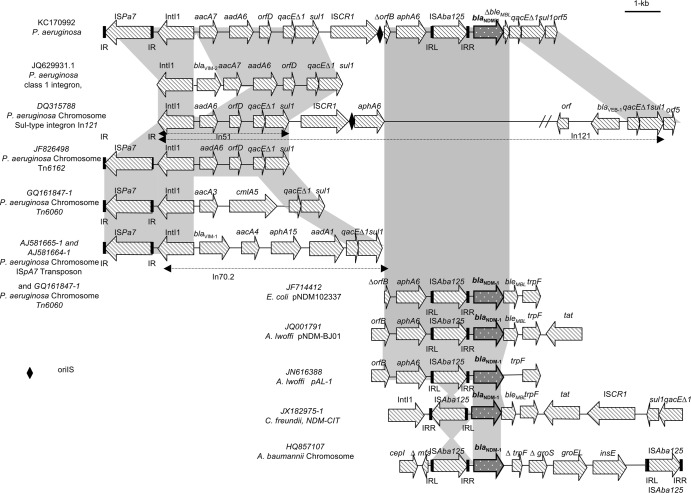
Upstream of the blaNDM-1 gene, four open reading frames (ORFs) were identified (Fig. 1). The first corresponded to orf513, a gene encoding a transposase ISCR, with its origin of replication, oriS, region. ISCR1 insertion sequences are often found beyond but close to the 3′-CS of class 1 integrons, belonging to the IS91 family, and are implicated in gene resistance mobilization (14). Here, the ISCR1 element was immediately followed by the truncated transposase gene insB, ΔorfB (IsABA14; accession no. JQ080305.1), the aminoglycoside resistance gene aphA6, the transposase ISAba125 gene, the blaNDM-1 gene, the truncated bleomycin resistance gene, and the truncated qac/sul 3′ end of a class 1 integron, followed by the orf5 gene. Detailed analysis of the 3′ flanking region of ISAba125 showed an identical promoter region in HIABP11, in plasmids pNDM102337 in E. coli, pNDM-BJ01 and pAL-1 in A. lwoffi, and pNDM-CIT in C. freundii, and in chromosomal sequences in A. baumannii (GenBank accession no. JF714412, JQ001791, JN616388, JX182975, HQ857107, JN872328, and JN872329) (7, 8, 15, 16). The expression of blaNDM-1 was under the control of a promoter whose −35 sequence was located in the right inverted repeat of ISAba125. A ribosomal binding site was found 14 bp upstream of blaNDM-1. Furthermore, in silico analysis indicated that the 64 bp at the start of aphA6 and blaNDM-1 displays 100% identity, as previously described in a chimera blaNDM-1 construction hypothesis (17). ISAba125 has often been described upstream of blaNDM-1, first in A. baumannii and then in A. lwoffi and Enterobacteriaceae. An ISAba125-related mobilization mechanism could be responsible for the en bloc acquisition of both blaNDM-1 and bleMBL, mobilized from the same progenitor (8). In other blaNDM-1-surrounding sequences described in Enterobacteriaceae and Acinetobacter, the blaNDM-1 gene was adjacent to a complete bleomycin resistance gene and a complete or truncated trpF gene (18). A truncated bleomycin resistance gene has not been described so far.
Fig 1.
Schematic representation of the structure of the blaNDM-1-surrounding sequences (12,735 bp) in P. aeruginosa HIABP11 (GenBank accession no. KC170992). Shown are alignments with (i) different chromosomally located transposon structures reported in P. aeruginosa isolates (some of them producing carbapenemase) and (ii) various blaNDM-1-associated genetic structures identified among Enterobacteriacea and Acinetobacter spp. The truncated bleomycin resistance gene is indicated by “ΔbleMBL.” IR, inverted repeat; IRL, left inverted repeat, IRR, right inverted repeat.
The region upstream of ISCR1 corresponds to a classical class 1 integron, with the IntI1 gene (encoding the integrase) followed by a cassette array containing 3 gene cassettes: the genes aacA7 and aadA6 encoding the aminoglycoside-modifying enzymes AAC-6′(I) and AADA6, respectively, and an orfD cassette encoding a putative polypeptide regulated by SdiA, a member of the LuxR family of transcriptional regulators. This last cassette is followed by a truncated qac/sul 3′ conserved sequence (3′-CS). The cassette array is quite related to that of JQ629931.1, which differs by the addition of a blaVIM-2 cassette upstream of the aac7 cassette. The orfD cassette, already described in In1 and In51, was generally associated with blaoxa-2 or aadA6 genes (19, 20). The structure is also close to the chromosomal class 1 integron sequence in Tn6162 with an aacA7 cassette insertion (21) and from In51 (19), already described in P. aeruginosa.
The whole structure from the IntI1 gene to the second truncated 3′-CS (downstream of the blaNDM-1 and truncated bleomycin resistance genes and upstream of the orf5 gene) corresponds to a new complex class 1 integron, with duplication of truncated 3′-CS. Duplication of 3′-CS regions, already described in many integrons, such as In121 (22) and In34 (23), is classically observed in ISCR1 elements (24). The region from the integrase to the first truncated qac/sul 3′-CS is the classic integron structure, followed by a second copy of the 3′-CS. Between the 2 copies of the 3′-CS is the ISCR, followed by the variable region that contains resistance genes, including blaNDM-1. Thus, the mobilization of genetic sequences surrounding blaNDM-1 into this integron occurred probably by a rolling-circle replication event involving the ISCR1 element and starting at the oriIS site (Fig. 1), which could have mobilized from a plasmid or a transposon the DNA fragment encompassing all the genes from insB to the truncated bleomycin resistance gene.
Upstream of the 5′ end of this complex class 1 integron, a copy of ISPa7 insertion sequence with inverted terminal repeats of 17 bp flanked by direct repeats of 4 bp was identified. This structure is identical to sequences found in Tn6060 and the ISPa7 transposon from VIM-1 metallo-β-lactamase-producing P. aeruginosa isolates from Italy and Australia (24, 25). In these sequences, ISPa7 is inserted between two 19-bp repeats typical of the extremities of Tn402-like element. This transposon is known to have a unique targeting mechanism with a strong preference for insertion into or close to a res site (26, 27).
The average GC content of the sequence is 53.6%, with three GC islands: a first region from ISPa7 to sul1 (positions 1 to 5951) with a GC content of 57.6%; a second region, including only orf513 (positions 6999 to 7703) with a GC content of 56.5%; and a third region from blaNDM-1 to orf5 with a GC content of 62%.
Analyzing the genetic environment of blaNDM-1 and regarding to the presence of ISPa7, we suggest that blaNDM-1 was mobilized into a new complex class 1 integron, which is a part of a chromosomally located Tn402-like structure. This is the first report describing the genetic environment of blaNDM-1 in P. aeruginosa. The presence of the association of ISPa7 upstream of a new CR1-borne sul-type integron and a blaNDM-1 region, classically found in plasmids in E. coli or in A. lwoffi, highlights the high plasticity of the P. aeruginosa genome (21). The ability of P. aeruginosa to accumulate overproduction of intrinsic resistance mechanisms, as well as the combination of various genetic determinants, including nonplasmid lateral exchange of resistance regions (21), results in multidrug-resistant or extensively drug-resistant isolates. Multilocus sequence typing, conducted according to published protocols (28; http://pubmlst.org/paeruginosa/), assigned HIABP11 to sequence type 235 (ST235), which has been encountered in clinical and environmental isolates of different countries (29, 30), including Serbia (31). ST235 is the primary founder of the successful epidemic clonal complex 235 (CC235), associated with various β-lactamase genes, including PER-1 extended-spectrum β-lactamase and VIM-type metallo-β-lactamases (32, 33). Detection of NDM-1 carbapenemase in a CC235 Pseudomonas aeruginosa isolate underscores its potential to spread in the hospital environment.
Nucleotide sequence accession number.
The nucleotide sequence of the blaNDM-1 region in P. aeruginosa HIABP11 has been registered in GenBank under accession no. KC170992.
ACKNOWLEDGMENT
The National Reference Center for Antibiotic Resistance was funded by the French Ministry of Health (InVS).
Footnotes
Published ahead of print 22 April 2013
REFERENCES
- 1. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol. 19:588–595 [DOI] [PubMed] [Google Scholar]
- 3. Halaby T, Reuland AE, al Naiemi N, Potron A, Savelkoul PHM, Vandenbroucke-Grauls CMJE, Nordmann P. 2012. A case of New Delhi metallo-β-lactamase 1 (NDM-1)-producing Klebsiella pneumoniae with putative secondary transmission from the Balkan region in the Netherlands. Antimicrob. Agents Chemother. 56:2790–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Livermore DM, Walsh TR, Toleman MA, Woodford N. 2011. Balkan NDM-1: escape or transplant? Lancet Infect. Dis. 11:164. 10.1016/S1473-3099(11)70048-2 [DOI] [PubMed] [Google Scholar]
- 5. Struelens MJ, Monnet DL, Magiorakos AP, O'Connor S, Giesecke J, European NDM-1 Survey Participants 2010. New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill. 15:19716 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19716 [DOI] [PubMed] [Google Scholar]
- 6. Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 7. Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Göttig S, Hunfeld KP, Seifert H, Wolfgang W, Higgins P. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J. Antimicrob. Chemother. 66:1998–2001 [DOI] [PubMed] [Google Scholar]
- 8. Poirel L, Bonnin R, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1087–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jovcic B, Lepsanovic Z, Suljagic V, Rackov G, Begovic J, Topisirovic L, Kojic M. 2011. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 55:3929–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flateau C, Janvier F, Delacour H, Males S, Ficko C, Andriamanantena D, Jeannot K, Mérens A, Rapp C. 2012. Recurrent pyelonephritis due to NDM-1 metallo-beta-lactamase producing Pseudomonas aeruginosa in a patient returning from Serbia, France, 2012. Euro Surveill. 17:20311 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20311 [PubMed] [Google Scholar]
- 11. Jeannot K, Poirel L, Robert-Micoud M, Cholley P, Nordmann P, Plésiat P. 2012. IMP-29, a novel IMP-type metallo-β-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56:2187–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36 [DOI] [PubMed] [Google Scholar]
- 13. Jacoby GA. 1974. Properties of R plasmids deterining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrobial Agents Chemother. 6:239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang YR, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob. Agents Chemother. 56:1698–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Wu C, Zhang Q, Qi J, Liu H, Wang Y, He T, Ma L, Lai J, Shen Z, Liu Y, Shen J. 2012. Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:E37152. 10.1371/journal.pone.0037152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Toleman MA, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:2773–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dortet L, Nordmann P, Poirel L. 2012. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naas T, Poirel L, Nordmann P. 1999. Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489:445–451 [DOI] [PubMed] [Google Scholar]
- 20. Stokes HW, Hall RM. 1992. The integron In1 in plasmid R46 includes two copies of the oxa2 gene cassette. Plasmid 28:225–234 [DOI] [PubMed] [Google Scholar]
- 21. Martinez E, Marquez C, Ingold A, Merlino J, Djordjevic SP, Stokes HW, Chowdhury PR. 2012. Diverse mobilized class 1 integrons are common in the chromosomes of pathogenic Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 56:2169–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naas T, Aubert D, Lambert T, Nordmann P. 2006. Complex genetic structures with repeated elements, a sul-type class 1 integron, and the blaVEB extended-spectrum β-lactamase gene. Antimicrob. Agents Chemother. 50:1745–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Partridge SR, Hall RM. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chowdhury PR, Merlino J, Labbate M, Cheong EY, Gottlieb T, Stokes HW. 2009. Tn6060, a transposon from a genomic island in a Pseudomonas aeruginosa clinical isolate that includes two class 1 integrons. Antimicrob. Agents Chemother. 53:5294–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riccio ML, Pallecchi L, Docquier JD, Cresti S, Catania MR, Pagani L, Lagatolla C, Cornaglia G, Fontana R, Rossolini GM. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamali-Moghaddam M, Sundström L. 2000. Transposon targeting determined by resolvase. FEMS Microbiol. Lett. 186:55–59 [DOI] [PubMed] [Google Scholar]
- 27. Minakhina S, Kholodii G, Mindlin S, Yurieva O, Nikiforov V. 1999. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33:1059–1068 [DOI] [PubMed] [Google Scholar]
- 28. Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42:5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maatallah M, Cheriaa J, Backhrouf A, Iversen A, Grundmann H, Do T, Lanotte P, Mastouri M, Elghmati MS, Rojo F, Mejdi S, Giske CG. 2011. Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One 6:e25617. 10.1371/journal.pone.0025617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slekovec C, Plantin J, Cholley P, Thouverez M, Talon D, Bertrand X, Hocquet D. 2012. Tracking down antibiotic-resistant Pseudomonas aeruginosa isolates in a wastewater network. PLoS One 7:e49300. 10.1371/journal.pone.0049300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Libisch B, Poirel L, Lepsanovic Z, Mirovic V, Balogh B, Pászti J, Hunyadi Z, Dobák A, Füzi M, Nordmann P. 2008. Identification of PER-1 extended-spectrum beta-lactamase producing Pseudomonas aeruginosa clinical isolates of the international clonal complex CC11 from Hungary and Serbia. FEMS Immunol. Med. Microbiol. 54:330–338 [DOI] [PubMed] [Google Scholar]
- 32. Empel J, Filczak K, Mrówka A, Hryniewicz W, Livermore DM, Gniadkowski M. 2007. Outbreak of Pseudomonas aeruginosa infections with PER-1 extended-spectrum β-lactamase in Warsaw, Poland: further evidence for an international clonal complex. J. Clin. Microbiol. 45:2829–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sardelic S, Bedenic B, Colinon-Dupuich C, Orhanovic S, Bosnjak Z, Plecko V, Cournoyer B, Rossolini GM. 2012. Infrequent finding of metallo-β-lactamase VIM-2 in carbapenem-resistant Pseudomonas aeruginosa strains from Croatia. Antimicrob. Agents Chemother. 56:2746–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]