Abstract
Human rhinovirus (HRV) is the predominant cause of the common cold, but more importantly, infection may have serious repercussions in asthmatics and chronic obstructive pulmonary disorder (COPD) patients. A cell-based antiviral screen against HRV was performed with a subset of our proprietary compound collection, and an aminothiazole series with pan-HRV species and enteroviral activity was identified. The series was found to act at the level of replication in the HRV infectious cycle. In vitro selection and sequencing of aminothiazole series-resistant HRV variants revealed a single-nucleotide mutation leading to the amino acid change I42V in the essential HRV 3A protein. This same mutation has been previously implicated in resistance to enviroxime, a former clinical-stage antipicornavirus agent. Enviroxime-like compounds have recently been shown to target the lipid kinase phosphatidylinositol 4-kinase III beta (PI4KIIIβ). A good correlation between PI4KIIIβ activity and HRV antiviral potency was found when analyzing the data over 80 compounds of the aminothiazole series, covering a 750-fold potency range. The mechanism of action through PI4KIIIβ inhibition was further demonstrated by small interfering RNA (siRNA) knockdown of PI4KB, which reduced HRV replication and also increased the potency of the PI4KIIIβ inhibitors. Inhibitors from two different structural classes with promising pharmacokinetic profiles and with very good selectivity for PI4KIIIβ were used to dissociate compound-related toxicity from target-related toxicity. Mortality was seen in all dosing groups of mice treated with either compound, therefore suggesting that short-term inhibition of PI4KIIIβ is deleterious.
INTRODUCTION
Human rhinovirus (HRV) is a positive-stranded RNA virus that is a member of the Picornaviridae family with over 133 genotypes classified into three species: HRV-A, HRV-B, and HRV-C (1). HRV is known as the cause of the common cold, but it has been increasingly associated with worsening of symptoms of asthma and chronic obstructive pulmonary disorder (COPD). Eighty to 85% of asthma exacerbations in children (2, 3) and 75% in adults (4) have been associated with viral upper respiratory tract infections (URTI), of which two-thirds are due to HRV. Fifty to 75% of COPD exacerbations are associated with prior viral URTI (5), of which half are due to HRV. Furthermore, in a human experimental HRV challenge model, asthmatics had increased upper and lower respiratory tract symptoms following infection and increased viral loads compared to nonasthmatic subjects infected with the same virus (6). COPD patients who were experimentally infected with HRV had higher viral loads and developed more severe and prolonged lower respiratory symptoms, airflow obstruction and inflammation than did nondiseased controls (7). Asthma and COPD patients appear to be less able to clear the viral infection compared to healthy controls. Overall, this indicates that there is a clear and high medical need for the prevention of HRV-triggered exacerbations in asthma and COPD patients.
Over the last few decades, several direct-acting antiviral inhibitors targeting the HRV capsid and protease and inhibitors of viral replication have been identified and examined for clinical development (reviewed in reference 8). The clinical development of rupintrivir, a 3C protease inhibitor, was halted due to lack of efficacy against naturally acquired infections even though it has broad rhinoviral and enteroviral activity in vitro (9, 10). An orally bioavailable compound that is similar to rupintrivir was not pursued (9), presumably due to economic factors. However, the 3C protease remains an attractive target currently at the exploratory level of drug discovery with the identification of broad-spectrum Michael acceptor inhibitors for example (11).
Enviroxime is an enteroviral inhibitor that acts at the level of RNA replication. Enviroxime had been in clinical development but failed due to poor exposure and lack of efficacy when administered both orally and intranasally (12, 13). Gastrointestinal side effects were seen in clinical trials with oral administration of enviroxime in an induced HRV infection experimental human model (13). Sixty percent of patients receiving enviroxime reported side effects of nausea, vomiting, and stomach pain. The intranasal formulation of enviroxime was tolerated well apart from nasal irritation; however, only limited to no efficacy was seen in experimental infection trials (12, 13). No therapeutic effect of intranasal enviroxime was demonstrated against natural HRV infections (14). The lack of efficacy of enviroxime when administered intranasally could be due to poor solubility. More recently, it was shown that enviroxime acts by preventing viral replication through inhibition of the phosphatidylinositol 4-kinase III beta (PI4KIIIβ) (15).
Inhibitors targeting viral attachment and uncoating have been well studied in the enteroviral field. Of these, the capsid-binding inhibitor pirodavir was not effective in the treatment of natural infections when it was administered intranasally (13, 16). Furthermore, the development of the capsid-binding inhibitor pleconaril was halted because it had clinical efficacy only against drug-susceptible viruses and induced the cytochrome P450 3A4 enzymes (17, 18). Patients whose baseline virus isolate susceptibility was >1 μM did not benefit from pleconaril treatments, and 21% of subjects in this trial were in this category (18). Currently, the only HRV inhibitor in clinical development is vapendavir (BTA798), which is an analog of pirodavir. Preliminary results from a phase II trial showed that oral vapendavir reduced asthma symptoms (19); however, it has been reported that vapendavir is not effective against all HRV genotypes (20).
The overall lack of efficacy of capsid-binding inhibitors against natural infections highlights the genetic diversity seen in the capsid genes and that not all HRV genotypes are susceptible to capsid-binding inhibitors. Wider use of molecular diagnostics has led to a recent reappraisal of HRV genetic diversity, including the discovery of HRV clade or species C that is refractory to traditional virus laboratory propagation procedures (21, 22). Currently, there are more than 33 genotypes of HRV-C based on the nucleotide sequence of the VP1 capsid, with another 28 provisional types based on partial VP4/VP2 sequences (1). Recently, cell culture models for HRV-C infection have been established in primary human nasal epithelial cells and organs (23–25). It was demonstrated that replication of one HRV-C genotype (C15) could be inhibited by rupintrivir but not by pleconaril (25), further demonstrating the need for molecules that have mechanisms that are different from that of the capsid binders.
There is mounting evidence that HRV-C is an important trigger of exacerbations in asthmatics. Most HRV-C disease association studies, which are typically focused on children from asthmatic and/or hospital-based populations, have demonstrated a similarly broad range of clinical outcomes as observed in HRV-A and HRV-B infections (26, 27). However, some studies provide evidence for a more frequent role of HRV-C in lower respiratory tract disease, febrile wheeze in infants, and asthma exacerbations in children (21, 28–30). Moreover, recently it was shown in a birth cohort where nasal mucous samples were collected and screened by PCR and sequencing during scheduled and sick visits during the first year of life that HRV-A and HRV-C were more likely to cause moderate to severe illness than HRV-B (31).
We set out to identify pan-species active HRV inhibitors with both known and new mechanisms of action by screening a subset of our proprietary collection against HRV. Virus-induced cytopathic effect (CPE)-based cell protection assays for HRV-B14 and HRV-A16 were established in 384-well format for primary screening. Then other genotypes of rhinoviruses and enteroviruses were used for profiling in the same assay format. An HRV-C reporter replicon assay was established in order to assess the pan-species activity of our compounds. Hits were binned based on preliminary mechanism of action studies and one pan-active hit with an unknown mechanism of action was explored and is the focus of this study.
MATERIALS AND METHODS
Virus and cell lines.
Virus HRV-B14 (VR284), HRV-A16 (VR283), HRV-A1A (VR1559), HRV-A2 (VR482), HRV-A8 (VR488), HRV-A39 (VR340), HRV-A45 (VR1155), HRV-A54 (VR1661), HRV-A89 (VR1199), HRV-B5 (VR485), CVA21 (coxsackievirus group A) (VR850), enterovirus 68 (EV68) (VR1197), and EV70 (VR836) were obtained from ATCC. H1-HeLa cells were obtained from ATCC (CRL-1958) and maintained in a complete medium, minimum essential medium (MEM) (catalog no. 11095; Invitrogen) supplemented with 10% fetal bovine serum (FBS) (catalog no. SH30396-03; HyClone). MRC-5 cells were obtained from ATCC (CCL-171) and maintained in a complete medium, Dulbecco's modified Eagle medium (DMEM) (catalog no. 11995-065; Invitrogen) supplemented with 10% FBS. A549 (CCL-185), BEAS-2B (CRL-9609), Calu-3 (HTB-55), HeLa (CCL-2), rhabdomyosarcoma (RD; CCL-136), and WI-38 (CCL-75) cells were obtained from ATCC and maintained as specified by ATCC. Normal human bronchial epithelial cells (HBEC) were obtained from Lonza (CC-2540) and propagated in BEBM (bronchial epithelial basal medium) (CC-3171; Lonza) supplemented with BEGM Singlequots (supplements and growth factors) (CC-4175; Lonza) (HBEC growth medium) for undifferentiated single-layer culture according to Lonza's cell culture specifications. During differentiation and once differentiated, HBEC cells were grown in 12-well Transwell plates at the air-liquid interface (ALI) in 0.5× DMEM medium, 1× BEBM medium with BEGM Singlequots supplemented with 0.03 μg/ml of all-trans retinoic acid (ATRA) (catalog no. R2625; Sigma) (HBEC differentiation medium).
When H1-HeLa cells were used to propagate virus, MEM was supplemented with 5% FBS (assay medium), and infected cells were incubated at 34°C with 5% CO2 (CVA21-infected H1-HeLa cells were incubated at 37°C with 5% CO2) until a complete cytopathic effect (CPE) was observed, typically after 3 days, at which point the virus was collected. The titers of the virus were determined by infecting H1-HeLa cells seeded the day before at 25,000 cells/well with serial dilutions of virus in a 96-well plate. The plates were incubated for 3 days at 34°C (or 37°C for CVA21) with 5% CO2. CPE was monitored by crystal violet staining, and absorbance was measured at 570 nm. The titers of virus were calculated using the 50% tissue culture infective dose (TCID50) method (32). Enterovirus (EV) was amplified in MRC-5 cells in the presence of 5% FBS and 1× nonessential amino acids (NEAA) (assay medium). Infected cells were incubated at 37°C with 5% CO2 until a complete CPE was observed, typically after 4 days. Virus was then collected, and the titers of the virus were determined on MRC-5 cells as described above for HRV.
Animals.
All protocols involving animal experimentation were reviewed and approved by the Institutional Animal Care and Use Committee and in compliance with the Guide for the Care and Use of Laboratory Animals (33) put out by the Canadian Council of Animal Care. Female CD-1 (18 to 20 g) and female SJL Elite mice (16 to 18 g) were purchased from Charles River Laboratories, Inc. Animals were housed under a 12-h light and dark cycle, with free access to food (Agribrands; 18% crude protein) and water.
Viral replication assay.
Activity of inhibitors against HRV, CVA21, and EV was measured using a CPE-based cell protection assay based on the literature (9). Briefly, 18,750 H1-HeLa or MRC-5 cells were seeded in clear 384-well plates (catalog no. GR781098; Greiner) in a volume of 30 μl of assay medium. Inhibitors were serially diluted in dimethyl sulfoxide (DMSO) and diluted further with medium, and 10 μl was added to each well in order to achieve the desired concentration and a final DMSO concentration of 0.6% in the assay. Cells were infected with 30 μl of virus diluted in assay medium in order to achieve the desired multiplicity of infection (MOI) and signal-to-background ratio of five in a final assay volume of 70 μl. For example, HRV-A16 is typically used at an MOI of 0.08, and HRV-B14 is typically used at an MOI of 0.008. HRV-infected cells were incubated at 34°C with 5% CO2 for 72 h. CVA21-infected cells were incubated at 37°C and 5% CO2 for 72 h. EV-infected cells were incubated at 37°C and 5% CO2 for 96 h. CPE was evaluated by measuring cell viability through the addition of 10 μl of a 1:20 mixture of MTS and PMS. MTS stands for 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, and PMS stands for phenazine methosulfate. MTS was from Promega (catalog no. G1111), and PMS was from Sigma (catalog no. P9625). The plates were incubated at 37°C with 5% CO2 for 2.5 h. The plates were read on an Envision plate reader (PerkinElmer) using absorbance at 492 nm for detection. The percent inhibition was calculated for each inhibitor concentration and used to determine the concentration that resulted in 50% inhibition of viral replication (EC50). The cytotoxicity of each compound was assessed in parallel using the same assay without the addition of virus in order to determine the concentration that resulted in 50% inhibition of cell viability (CC50).
The activities of compounds against a panel of enteroviruses were assessed in CPE-based assays in the laboratory of Johan Neyts at the Katholieke Universiteit Leuven (KU Leuven), Leuven, Belgium as described previously (31, 34). Assays used the following virus and cell line combinations: CVB3 Nancy/Vero; CVB4 E2 Edwards/Vero; echovirus 11 Gregory (buffalo green monkey); EV71 BrCr (RD), poliovirus 1 Sabin (buffalo green monkey).
HRV qRT-PCR assay.
HRV quantitative reverse transcription-PCR (qRT-PCR) assay was performed based on the literature (35) using the following probes and primers: 5′ HRV PCR primer, 5′-CTA GCC TGC GTG GC-3′; 3′ HRV PCR primer, 5′-GAA ACA CGG ACA CCC AAA GTA-3′; HRV probe, 5′-/6-FAM/TCC TCC GGC CCC TGA ATG CGG C/6-TAMSp/-3′ where 6-FAM is 6-carboxyfluorescein and 6-TAMSp is 6-carboxytetramethylrhodamine. After infection, total RNA was extracted from infected cells using Qiagen RNeasy 96 kit according to the manufacturer's protocol. qRT-PCR was performed in 96-well plates using 5 μl of extracted RNA, 10 μM each of the 5′ and 3′ HRV PCR primers, 5 μM HRV probe with appropriate amounts of reagents from the TaqMan EZ RT-PCR kit in an Applied Biosystems (ABI) 7500 real-time PCR system. Serial dilutions of in vitro-transcribed HRV-B14 RNA were used to generate a standard curve. HRV replication was expressed as the number of copies per microgram of total RNA by correcting for the total amount of RNA present per well. Total RNA present in each well was measured using the RiboGreen RNA quantification kit (Molecular Probes/Invitrogen).
HBEC viral replication assays.
Undifferentiated HBECs were seeded in growth medium in a 96-well plate 1 day prior to infection at a density of 45,000 cells/well and incubated at 37°C with 5% CO2 overnight. The medium was aspirated and replaced with an HRV-B14 viral suspension of 65 μl in MEM with 5% FBS (in order to infect at the desired MOI) and supplemented with 55 μl of HBEC cell growth medium and adsorbed at 34°C with 5% CO2 for 3 h with gentle rocking. Virus was aspirated, and the cells were washed with phosphate-buffered saline (PBS). Inhibitors were serially diluted in DMSO and added to the infected cells with HBEC growth medium in order to obtain a final DMSO concentration of 0.6% in a 122-μl assay volume. The plates were incubated at 34°C with 5% CO2 for 72 h. In order to measure compound potency, HRV RNA replication in HBEC was determined by qRT-PCR. The percent inhibition was calculated for each inhibitor concentration and used to determine the concentration that results in 50% inhibition of HRV viral replication (EC50).
In order to differentiate HBEC, undifferentiated HBECs were seeded on the apical side of a 12-well Transwell plate (catalog no. 3460; Corning) at a density of 100,000 cells/well in 500 μl of HBEC cell differentiation medium, and 1 ml of HBEC differentiation medium was added to the basolateral side of each well. Transwell plates were incubated at 37°C with 5% CO2, and the medium was changed every 2 days. After 7 days in culture, the medium was removed from the apical side, and the cells were left at the ALI to differentiate. Cellular differentiation was monitored with an EVOM2 epithelial voltohmeter (World Precision Instrument) with an STX2 electrode in order to measure the transepithelial electrical resistance (TEER). After 13 days at the ALI, differentiated HBEC were infected. Before the cells were infected, the cells were washed 5 times with 200 μl of HRV assay medium in order to remove all mucus present on the cells. HRV-B14 was diluted in HRV assay medium, and 200 μl of diluted virus was distributed to the apical side of each well. Inhibitors were serially diluted in HBEC differentiation medium, and 1 ml was distributed to the basolateral side of each well (final DMSO concentration of 0.6%). The plates were incubated for 3 h at 34°C with 5% CO2. Infected cells were then washed twice with HRV assay medium before being put at the ALI. The plates were incubated at 34°C with 5% CO2 for 72 h, and antiviral potency was determined as described above for the HBEC assay in undifferentiated cells.
HRV full-length and subgenomic reporter replicon construction and replication assay (EC50).
HRV-B14 infectious cDNA pWR3.26 (VRMC-7) and HRV-A1 infectious cDNA pR16.11 (VRMC-8) were obtained from ATCC. The aminothiazole series resistance mutation I42V was inserted into HRV-B14 pWR3.26 by site-directed mutagenesis using QuikChange II site-directed mutagenesis kit (Agilent).
Full-length HRV-C15 W10 LUC (LUC stands for luciferase) was synthesized at DNA 2.0. In order to establish a reporter-based replication assay with HRV-C15 W10 (GenBank accession no. GU219984.1), a T7 RNA promoter and a humanized firefly luciferase reporter gene and a 3CD cleavage site (GGGALFQG) were introduced before the 5′ end of VP4. A BstBI restriction site was added at the 3′ end of the 3D polymerase after the 29-nucleotide poly(A) tail in order to permit linearization and in vitro transcription of HRV-C15 W10 LUC. RNA was in vitro transcribed using the T7 Ribomax RNA transcription system (Promega) and transfected into H1-HeLa cells by electroporation following the procedure described previously (36). Transfected cells were immediately seeded at a concentration of 13,000 cells per well in a 384-well white Greiner plate in MEM (lacking phenol red) with 10% FBS and 1× NEAA. Inhibitors were serially diluted and added to cells in order to achieve the desired concentration and a final DMSO concentration of 0.6%. The plates were incubated at 34°C with 5% CO2 for 24 h. Twenty microliters of BrightGlo reagent (Promega) was added to each well, and luminescence was measured using an Envision plate reader. The percent inhibition was calculated for each inhibitor concentration and used to determine the concentration that resulted in 50% inhibition of HRV-C15 W10 replication (EC50).
Subgenomic replicons of HRV-B14 and HRV-A16 were constructed based on the literature (36). Briefly, using full-length cDNA of each virus, capsid VP4-VP2 and part of VP3 genes were replaced by a humanized firefly luciferase reporter gene. The 71 C-terminal amino acids from VP3 were kept, and a 3CD cleavage site (GGGALFQG) was introduced between the reporter gene and the remainder of VP3, generating the subgenomic HRV-B14Luc and HRV-A16Luc DNA constructs. The HRV-B14Luc and HRV-A16Luc subgenomic replicons were linearized with SacI and MluI restriction enzymes, respectively. RNA was transcribed using the T7 Ribomax RNA production system (Promega) and transfected by electroporation into H1-HeLa cells by the same protocol used for HRV-C15 W10 LUC. The assay conditions used for HRV-C15 W10 LUC were used to generate EC50 with HRV-B14Luc and HRV-A16Luc.
Mechanism of action studies.
Time of addition and time of removal assays based on the literature (10) were performed using the CPE-based antiviral HRV-A16 assay described above with the following modifications where the compounds were added or removed at time −1, −0.5, 0, 0.5, 1, 2, 3, 4, 5, 6, 7, 24, and 48 h postinfection. H1-HeLa cells were seeded at 25,000 cells/well in a 96-well plate 18 h before the experiment and infected with HRV-A16 at an MOI of 5 by adsorbing the virus for 2 h at 34°C with 5% CO2. The infected cells were then washed twice with PBS, and then medium or medium with compound at 10× EC50 and a final DMSO concentration of 0.6% was added to the cells. The assay was terminated 48 h postinfection by the addition of 20 μl of MTS-PMS (1/20), the plates were incubated at 37°C with 5% CO2 for 3.5 h, and the absorbance at 492 nm was read on an Envision plate reader (PerkinElmer).
An assay to determine whether the compounds stabilize the virus against heat inactivation was developed based on the literature (37). Briefly, the HRV-A16 virus (2.5 × 105 TCID50/ml) was incubated with compounds at a concentration of 10× EC50 or 100× EC50 for 1 h at room temperature in a volume of 300 μl. One hundred microliters was removed from each sample for unheated control, and the remaining samples were heated at 56°C for 6 min. Samples were quickly cooled by diluting 1/10 in room temperature MEM with 5% FBS. The titers of the virus were determined in all samples by serial dilution and by infecting H1-HeLa cells seeded the day before at 25,000 cells/well in a 96-well plate. Virus was adsorbed for 2 h at 34°C with 5% CO2, the cells were then washed twice with PBS, and then medium was added to the cells. The plates were incubated for 3 days at 34°C with 5% CO2, and the titers of the virus were calculated as indicated above.
HRV variants were isolated in vitro in the presence of increasing compound concentrations as described previously (38). Briefly, 6.5 × 105 H1-HeLa cells were seeded in 12-well plates and initially infected with HRV-B14 at an MOI of 0.1 in the presence of 3.5× the EC50 of compound 1 or an analog from the aminothiazole series. Supernatants were collected when the CPE reached at least 50%. The supernatant (0.2 to 0.5 ml) was used to reinfect fresh H1-HeLa cells. Virus was adsorbed for 2 h at 34°C with 5% CO2, the cells were washed twice with PBS, higher concentrations of compound (1× to 3×) were added to the cells, and the cells were monitored for the development of CPE. The resistance study was performed over 30 days, reaching a concentration of 20× EC50. All supernatants were clarified by filtration (0.2 μm) and were used immediately or stored at −80°C for subsequent rounds of infection, population sequence analysis, or antiviral susceptibility testing. Viral RNA was extracted from the supernatant using the Qiamp viral RNA extraction kit (Qiagen) according to the manufacturer's protocol. The entire genome of HRV-B14 was amplified using the one-step RT-PCR system with Platinum Taq HiFi kit (Invitrogen) and sequenced.
Kinase assays.
PI4KIIIβ and PI4KIIIα activities were assessed in kinase assays as described previously (39). Assays for an extensive panel of lipid and protein kinases were performed by Invitrogen Corporation (SelectScreen Kinase Screening Service).
qRT-PCR for lipid kinase.
RNA was isolated from cells using the RNeasy 96 kit or RNeasy minikit (Qiagen). The total RNA concentration was determined with the RiboGreen RNA quantification kit (Molecular Probes/Invitrogen) using a standard curve of rRNA. The TaqMan EZ RT-PCR kit (ABI) was used to perform qRT-PCRs on an ABI 7500 real-time PCR system. Lipid kinase RNA transcripts were quantified using the TaqMan gene expression assay kits (ABI) containing gene-specific probes and primer sets. Serial dilutions of untreated cellular RNA from all the cell lines were used to generate a standard curve for each gene-specific expression analysis to determine relative changes in transcript levels.
PI4KIIIβ immunoblotting.
Cells were lysed in K buffer (20 mM HEPES [pH 7.4], 150 mM NaCl, 5 mM MgCl2) supplemented with 0.1% Triton X-100 and protease inhibitor cocktail (catalog no. 118336170001; Roche). Protein concentration was determined using Bradford reagent, and equal amounts of protein (30 μg) were resolved by electrophoresis using a 12% SDS-polyacrylamide gel (Nupage Novex Tris-acetate minigel; Life Technologies) and then electroblotted onto a nitrocellulose membrane (Hybond-ECl; GE Healthcare). The membrane was blocked with 3% bovine serum albumin (BSA) in PBS–0.1% Tween 20 and probed with a polyclonal antibody against PI4KIIIβ obtained from Abgent (catalog no. AP8030A). After extensive washing in PBS–0.1% Tween 20, blots were probed with the appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and washed, and protein was detected by enhanced chemiluminescence (ECL plus Western blotting detection system; GE Healthcare).
siRNA knockdown.
On-Targetplus SMARTpool small interfering RNAs (siRNAs) were obtained from Dharmacon/Thermo Scientific (PI4KA, catalog no. L-006776-00; PI4KB, catalog no. L-006777-00; PIK3CD, catalog no. L-006775-00; PIK3C2A, catalog no. L-006771-00) and irrelevant siRNA (IRR) was obtained from Qiagen (GL2 luciferase, catalog no. 1022070). The transfection agent was DharmaFECT 4 from Dharmacon/Thermo Scientific, and Opti-MEM I reduced serum medium (Invitrogen) was used to dilute siRNA and transfection agent. siRNAs (10 nM) were transfected according to the manufacturer's protocol 24 h after seeding H1-HeLa cells at 10,000 cells/well in assay medium in 96-well plates. After 72 h of incubation at 37°C with 5% CO2, the cells were transfected with siRNA a second time. The cells were then infected with HRV-B14 (MOI of 0.05 or 0.005) with or without compounds serially diluted in DMSO (final concentration of 1%). A plate of mock-infected cells was prepared in parallel in duplicate for determination of the level of knockdown of lipid kinases by qRT-PCR and cytotoxicity. Plates of infected or mock-infected cells were incubated at 34°C with 5% CO2. Seventy-two hours postinfection, viral replication and cytotoxicity were assessed by the addition of 20 μl of MTS-PMS (1/20) and incubation at 37°C with 5% CO2 for 3.5 h, and the absorbance at 492 nm was read on an Envision plate reader (PerkinElmer). While very little siRNA-dependent cytotoxicity was observed, the viral replication data were corrected for cytotoxicity before data were expressed as percent HRV infection compared to treatment with IRR siRNA.
PK in CD-1 and SJL mice.
For studies of CD-1 mice, dosing suspensions of T-00127-HEV-1 and compound 2 were prepared in 1% N-methyl-2-pyrrolidone, 0.3% Tween 80, and 0.5% methylcellulose in water. Single doses of 5, 50, and 250 mg/kg of body weight were administered orally at a dosing volume of 10 ml/kg. Blood samples (100 μl) were obtained via mandibular cheek puncture and collected into lithium heparinized microcuvettes (Sarstedt) at various time points (0.25, 0.5, 1, 2, 4, 6, 8, and 24 h). The samples were centrifuged at 8,000 rpm and 4°C for 8 min, and plasma samples were transferred and frozen at −20°C pending analysis. At each time point, plasma compound concentrations from 4 different animals were averaged, and a single composite pharmacokinetic (PK) profile was generated. PK parameters were determined following noncompartmental analysis with ToxKin 3.3.0 software (Boehringer Ingelheim, Unilog AG).
Dosing suspensions were prepared daily as described above for SJL Elite mice. The mice were dosed orally (10 ml/kg) once a day (q.d.) for 7 days at 50 and 250 mg/kg. Blood samples (100 μl) from groups of 4 animals were obtained via mandibular cheek puncture on days 1, 3, and 7 at 0.5, 1, 4, 8 and 24 h and processed as described above.
In order to extract plasma, an aliquot of plasma (5 or 10 μl) was mixed with 350 μl of acetonitrile-water (1:9, vol/vol)–2% formic acid. The mixture was loaded onto Phenomenex Strata X-C 30- mg/1-ml cartridges (Phenomenex) previously conditioned with 1 ml of methanol followed by 1 ml of water–2% formic acid. The cartridges were washed with 1 ml of water–2% formic acid, 3 times with 1 ml of acetonitrile-water (1:9, vol/vol)–2% formic acid, and 500 μl of methanol. Compound 2 and T-00127-HEV1 were eluted with 1 ml of methanol–5% ammonium hydroxide. The eluate was evaporated under a nitrogen stream using Turbo Vap evaporator (Zymark) at 45°C. The residue was reconstituted in 100 to 150 μl of acetonitrile-water (1:3, vol/vol), and 2 μl was injected for liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) analysis.
Both drugs were analyzed using LC-MS/MS on a TSQ Vantage system (Thermo Scientific) with an electrospray ionization source (ESI) in positive ionization mode. T-00127-HEV1 was separated on an Xbridge C8 column (2.1 by 30 mm; 5 μm; Waters, Milford, MA, USA), while compound 2 was separated using an Atlantis dC18 column (2.1 by 30 mm; 5 μm; Waters). A generic fast gradient method was applied to the two compounds but using different mobile phases. For T-00127-HEV1, the mobile phases consisted of mobile phase A (water–0.1% formic acid) and mobile phase B (acetonitrile-methanol [1:1, vol/vol]–0.1% formic acid). For compound 2, mobile phase B was replaced by acetonitrile–0.1% formic acid. The gradient was initially held for 0.1 min at 10% mobile phase B, followed by a 1.2-min linear ramp to 98% mobile phase B with a 1.0-min hold. The total LC run time was 3 min, including 0.7-min equilibration time between injections. The flow rate was 0.4 ml/min. The TSQ Vantage system was operated in selected reaction monitoring (SRM) mode. The SRM transitions monitored were m/z 412 to m/z 114 for T-00127-HEV1 and m/z 359 to m/z 206 for compound 2.
RESULTS
Screening of a diverse subset of the proprietary collection identifies pan-HRV species active aminothiazole series with a new mechanism of action.
A cell-based antiviral screen against HRV was completed with a 98,000-compound clustered subset of the Boehringer Ingelheim compound collection that was cherry-picked from the full proprietary collection using diversity and drug-like properties as criteria. Specifically, 98,097 compounds were evaluated for antiviral activity against HRV-B14 with an average plate Z′ of 0.9 and average plate Z of 0.76, where Z (full plate) and Z′ (controls only) are screening window coefficients that are calculated as follows: Z = 1 − [(3SD of sample + 3SD of control)/|mean of sample − mean of control|]. The overall hit rate was 1.4% at greater than 20% inhibition. Single-point confirmation of primary hits was done on 1,361 compounds against HRV-B14 and HRV-A16 as well as cytotoxicity on H1-HeLa cells. A total of 167 compounds matched the hit selection criteria against both HRV-B14 and HRV-A16 and were noncytotoxic (less than 25% inhibition). Twenty hits exhibited activity against an initial panel of six HRVs. These 20 HRV antiviral screen hits underwent further profiling and prospecting to eliminate hits based on cytotoxicity against a panel of cell lines, lack of productive structure-activity relationship (SAR), and lack of potential for activity against the initial panel of six HRV genotypes. This resulted in the removal of six hits. None of the 20 hits displayed activity in the in vitro assays of HRV 3Dpol RNA polymerization or HRV 3C protease (data not shown). Furthermore, preliminary mechanism of action studies were performed aiming at identifying and removing 13 traditional capsid binding-like inhibitors that interfere with attachment and uncoating of the virus (time of addition, time of removal, heat stabilization, and activity against HRV-A45 and HRV-B5). These capsid-binding inhibitors were removed due to the lack of activity against the initial panel of 6 HRVs.
One series remained after this exercise, exemplified in Fig. 1A by compounds 1 and 2. This aminothiazole series appeared to have a unique mechanism of action acting later in the HRV infectious cycle at 6 h postinfection (in a single-cycle viral replication assay). This aminothiazole series was optimized for activity against HRV and had no cytotoxicity as demonstrated by compound 1 with an antiviral potency ranging from 110 to 480 nM against 10 HRV genotypes of species A and B, and the CC50 was >64 μM (Table 1). Design, synthesis, and chemical optimization of this aminothiazole series is described elsewhere (65). Compounds within this aminothiazole series proved active against subgenomic luciferase reporter HRV-A16Luc and HRV-B14Luc replicons as well as a full-length HRV-C15 W10 LUC reporter replicon (exemplified by compound 2 in Table 2), further demonstrating that this series is acting at the intracellular replication stage of the HRV infectious cycle.
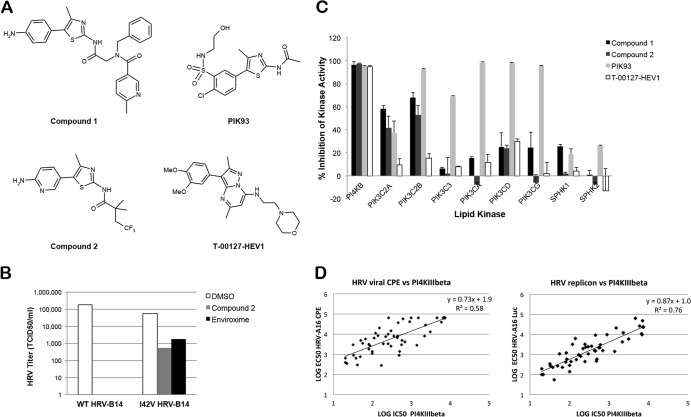
Fig 1.
Antiviral effects of aminothiazole series of compounds that appear to target PI4KIIIβ. (A) Structures of example compounds compared to PIK93 and T-00127-HEV1. (B) Titer of wild-type HRV-B14 and I42V HRV-B14 mutant after being in the presence of compounds at 10× EC50 for 24 h (n = 1). (C) Inhibitory effects of compounds (10 μM) on in vitro activities of lipid kinases (averages of two experiments with standard deviations [error bars]). (D) Correlation of HRV-A16 antiviral and replicon activity and in vitro PI4KIIIβ activity.
Table 1.
Antiviral activity of compound 1 and T-00127-HEV1 against panel of HRV-A and HRV-B
| Parametera | Compound 1 | T-00127-HEV1 |
|---|---|---|
| Antiviral potency and virus | ||
| HRV species A EC50 (nM) | ||
| HRV-A1A | 390 ± 10 | 2,900 ± 800 |
| HRV-A2 | 360 ± 10 | 2,900 ± 800 |
| HRV-A8 | 110 ± 60 | 600 ± 200 |
| HRV-A16 | 480 ± 80 | 1,800 ± 900 |
| HRV-A39 | 150 ± 10 | 1,700 ± 400 |
| HRV-A45 | 150 ± 10 | 1,500 ± 900 |
| HRV-A54 | 190 ± 10 | 2,000 ± 800 |
| HRV-A89 | 210 ± 20 | 2,100 ± 900 |
| HRV species B EC50 (nM) | ||
| HRV-B5 | 120 ± 50 | 800 ± 100 |
| HRV-B14 | 400 ± 100 | 2,100 ± 600 |
| H1-HeLa CC50 (nM) | >64,000 | >60,000 |
The EC50 and CC50 data are averages of three to eight experiments (± standard deviations).
Table 2.
Antiviral activity of compounds against HRV in H1-HeLa and nondifferentiated and differentiated HBEC
| Antiviral potency and virus or cella | Compound 1 | Compound 2 | PIK93 | T-00127-HEV1 |
|---|---|---|---|---|
| Viral CPE assay EC50 (nM) | ||||
| HRV-B14 | 400 ± 100 | 500 ± 200 | 590 | 2,100 ± 600 |
| HRV-A16 | 480 ± 80 | 190 ± 50 | 590 | 1,800 ± 900 |
| Replication assay EC50 (nM) | ||||
| HRV-B14Luc | 240 ± 40 | 120 ± 30 | NDb | 600 ± 400 |
| HRV-A16Luc | 130 ± 10 | 40 ± 20 | ND | 200 ± 100 |
| HRV-C15Luc | ND | 60 ± 20 | ND | 200 ± 100 |
| Toxicity CC50 (nM) | ||||
| H1-HeLa | >64,000 | >66,000 | 23,000 | >64,000 |
| HBEC HRV-B14 EC50 (nM) | ||||
| Undifferentiated | 60 ± 10 | ND | ND | ND |
| Differentiated | 300 ± 20 | ND | ND | ND |
The EC50 and CC50 data are averages of three to eight experiments (± standard deviations) except for PIK93, which was tested once in this assay format.
ND, not determined.
In addition to the HRV antiviral assays that were established in immortalized cell lines (H1-HeLa, HeLa, WI-38, rhabdomyosarcoma, and BEAS-2B), HRV-B14 antiviral assays with a qRT-PCR readout were also developed in both undifferentiated and differentiated primary human bronchial epithelial cells (HBEC). The aminothiazole series shows antiviral potency in the same range in the primary HBEC (exemplified by compound 1 in Table 2).
Compounds are acting through the lipid kinase phosphatidylinositol 4-kinase III beta.
In order to aid the elucidation of the mechanism of action of this series, selection of compound-resistant variants of HRV-B14 was achieved by passaging HRV-B14 in H1-HeLa cells in the presence of two compounds from this series for 30 days reaching 20× EC50. Population sequencing revealed that the resistant variants all contained a single-nucleotide mutation encoding the substitution I42V in the HRV 3A protein. This same mutation has been previously reported to confer resistance to enviroxime, an antipicornaviral agent that acts at the stage of viral RNA replication (40). The I42V mutation was reverse engineered into the full-length HRV-B14 replicon, and virus was produced. Surprisingly, no shift in antiviral potency was seen in the comparison of wild-type viruses to mutant viruses in the regular HRV CPE-based assay. Published data suggested that the mutations that seemed to confer resistance to enviroxime did not confer a very high resistance level (41). In order to assess the resistance potency of the mutation, a different approach was taken. The mutant virus, as well as the wild-type HRV-B14 virus, was grown in the presence or absence of compounds for 24 h at 10× EC50. After a 2-h adsorption and wash, the titers of the produced progeny virus were then determined on H1-HeLa cells. The I42V mutant virus grew to the same titer as the wild-type virus in the absence of compounds. In the presence of compound, the mutant virus produced 1,000× more virus than the wild-type virus, confirming that the mutations did in fact confer resistance to the aminothiazole series (Fig. 1B). However, the lack of a shift in antiviral potency suggested that the resistance level was low, pointing toward a cellular target.
In order to determine the pharmacological profile of these compounds, an Invitrogen protein and lipid kinase screen was run. The aminothiazole series showed very high inhibitory activity against the lipid kinase PI4KIIIβ (Fig. 1C). The 50% inhibitory concentrations (IC50s) of 80 HRV compounds, which cover a broad potency range and represent all key elements of the structure-activity relationship, were determined in PI4KIIIα and PI4KIIIβ assays, and a good correlation between PI4KIIIβ activity, HRV antiviral potency, and HRV replicon activity was observed (HRV-A16 [Fig. 1D]) (HRV-B14 data not shown). No correlation was seen with PI4KIIIα activity, and indeed the majority of compounds were not active (data not shown; see Table 3 for examples of compounds). While this work was being conducted, a study was published showing that the target of enviroxime-like compounds is PI4KIIIβ (15). The compound identified in this study (T-00127-HEV) was synthesized and had comparable activity to compounds 1 and 2 of the aminothiazole series in all antiviral and kinase assays (Tables 1, 2, and 3).
Table 3.
Specificity of compound 2 and T-00127-HEV1 for PI4KIIIβ compared to a panel of lipid and protein kinases
| Specificitya | Compound 2 | T00127HEV1 |
|---|---|---|
| PI4KIIIB IC50 (nM) | 48 ± 5 | 140 ± 10 |
| PI4KIIIA IC50 (nM) | >10,000 | >10,000 |
| Lipid kinase panel | 2/8b | 0/8 |
| PIK3C2A IC50 (nM) | 12,000 | >10,000 |
| PIK3C2B IC50 (nM) | 5,600 | >10,000 |
| Protein kinase panel | 2/277 | 0/277 |
| STK17A IC50 (nM) | 2,100 | >10,000 |
| MSK2 IC50 (nM) | 4,600 | >10,000 |
The PI4KIIIβ and PI4KIIIα EC50 and CC50 data are averages of two experiments (± standard deviations). Invitrogen kinase screening was performed in duplicate at a single point of 10 μM and in dose response when active at 10 μM.
x/number of kinases tested indicates that compound showed activity greater than 50% against x kinases.
Notably, prior to our identification of this series, a variety of lipid kinase inhibitors had been tested in our HRV antiviral assays, and only PIK93 was shown to be active (Table 2 and data not shown). PIK93 is a relatively nonspecific lipid kinase inhibitor that has been shown to inhibit enteroviral infection (Table 2 and Fig. 1C) (42, 43). The lack of activity of the other specific protein and lipid kinase inhibitors demonstrates the importance of specificity for PI4KIIIβ for inhibiting HRV.
Confirmation of mechanism of action of aminothiazole series through siRNA knockdown of PI4KIIIβ.
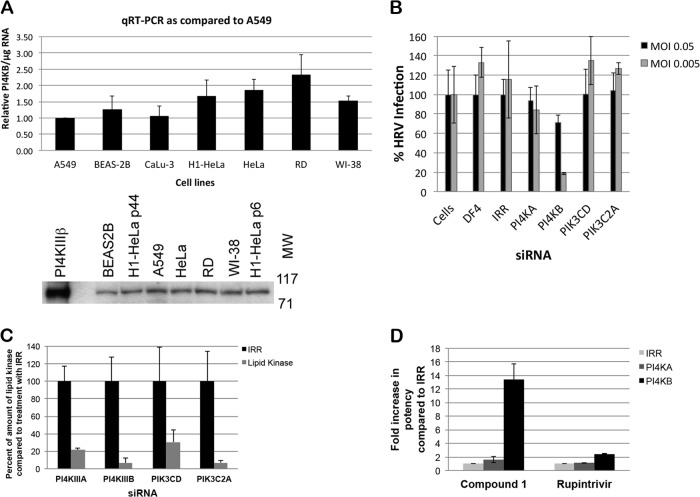
The expression of PI4KIIIβ was confirmed by monitoring the presence of RNA and protein in all cell lines tested (Fig. 2A). The mechanism of action of the aminothiazole series through PI4KIIIβ inhibition was further demonstrated by siRNA knockdown of PI4KIIIβ, which reduced the ability of HRV to cause a cytopathic effect on H1-HeLa cells (Fig. 2B and C). This effect was more evident at lower MOIs than higher MOIs (30% protection at an MOI of 0.05 and 80% protection at an MOI of 0.005) and was not observed with siRNA knockdown of other lipid kinases, even though all lipid kinases were knocked down by at least 80% as determined by gene-specific qRT-PCR (data not shown). Moreover, PI4KIIIβ knockdown increased the potency of compounds from this aminothiazole series by 13-fold (Fig. 2C).
Fig 2.
Confirmation of mechanism of action of aminothiazole series through siRNA knockdown of PI4KIIIβ. (A) Expression of PI4KIIIβ determined by qRT-PCR and Western blot analysis. The qRT-PCR results are averages of four samples with standard deviations (error bars). Two passages of H1-HeLa cells were examined by Western blotting (passage 6 and 44). The positions of molecular weight in kDa are shown to the right of the Western blot. (B) Effect of knockdown of various lipid kinases or irrelevant gene (IRR) via siRNA transfection on HRV infection of H1-HeLa cells at two multiplicities of infection (MOIs). Data are averages of four samples with standard deviations. The data are representative of three experiments. (C) Expression of lipid kinases as determined by qRT-PCR after siRNA treatment for 6 days (averages of four samples with standard deviations). (D) Effect of compound 1 targeting PI4KIIIβ and the protease inhibitor rupintrivir on HRV infection in siRNA-transfected H1-HeLa cells (MOI of 0.05). Data are averages of four samples with standard deviations. The data are representative of three experiments.
Compounds identified with high PI4KIIIβ specificity and enteroviral activity and good pharmacokinetics.
Compound 2 was completely profiled in a panel of 277 protein kinases and 8 lipid kinases (not including PI4KIIIα and -β) (Table 3). As found with other members of this series, an excellent specificity for PI4KIIIβ versus other lipid and protein kinases was observed. For the few instances where minor inhibitory activity (∼50%) was observed at 10 μM in the Invitrogen panel screen, the full concentration response testing was performed and IC50s on the order of 2 to 5 μM were obtained for the protein kinases (STK17A and MSK2 [Table 3]) and IC50s on the order of 6 to 12 μM were obtained for the lipid kinases (PIK3C2A and PIK3C2B [Table 3]).
In view of the limitations of the current HRV pharmacological models, we assessed the potential in vivo efficacy of the aminothiazole series in a coxsackievirus (CVB4) mouse pancreatitis model in SJL mice. In order to distinguish between compound-related toxicity and target-related toxicity, the structurally distinct compound 2 and T-00127-HEV1 were chosen for further profiling. Based on the reported profiles of other PI4KIIIβ inhibitors (15) and the profiles of compounds from our series, which have antiviral activity against coxsackievirus A21 (CVA21) and enterovirus 70 (EV70) (Table 4 and data not shown), it was expected that such compounds would have antiviral activity against all enteroviruses. Compound 2 and T-00127-HEV1 were evaluated in a larger panel of CPE-based assays and were found active against each virus of the panel of enteroviruses (Table 4). Specifically, compound 2 has an EC50 of 120 nM against CVB4. Furthermore, both compound 2 and T-00127-HEV1 showed a maximum concentration of compound in serum (Cmax) of >10 μM following a single oral dose of 50 mg/kg in CD-1 mice (Table 5). Overall, compound 2 and T-00127-HEV1 are potent and selective compounds that would allow for a toxicological assessment of PI4KIIIβ inhibition.
Table 4.
Antiviral activity of compound 2 and T-00127-HEV1 across a panel of enteroviruses
| Virus (cell line) and parametera | Compound 2 | T00127HEV1 |
|---|---|---|
| CVA21 (H1-HeLa) | ||
| EC50 (nM) | 400 ± 200 | 4,800 ± 100 |
| CC50 (nM) | >66,000 | >64,000 |
| CVB3 (Vero) | ||
| EC50 (nM) | 280 | 4,900 |
| CC50 (nM) | >50,000 | >50,000 |
| CVB4 (Vero) | ||
| EC50 (nM) | 120 | 710 |
| CC50 (nM) | >50,000 | >50,000 |
| Echovirus 11 (BGM) | ||
| EC50 (nM) | <23 | 860 |
| CC50 (nM) | >50,000 | >50,000 |
| EV71 (RD) | ||
| EC50 (nM) | 170 | 4,400 |
| CC50 (nM) | >50,000 | >50,000 |
| Poliovirus 1 (BGM) | ||
| EC50 (nM) | 130 | 1,200 |
| CC50 (nM) | >50,000 | >50,000 |
The CVA21 EC50 and CC50 data are averages of three experiments (± standard deviation). All other data are from one experiment.
Table 5.
Cmax at day 1 and observations of daily oral dosing of compound 2 and T-00127-HEV1 in CD-1 and SJL micea
| Compound and dose (mg/kg) |
Cmax (μM) |
AUC0–∞ (μM) |
Tmax (h) |
Observation(s) in SJL mice | |||
|---|---|---|---|---|---|---|---|
| CD-1 mice | SJL mice | CD-1 mice | SJL mice | CD-1 mice | SJL mice | ||
| Compound 2 | |||||||
| 5 | 0.5 ± 0.1 | ND | 3 ± 1 | ND | 0.5 | ND | |
| 50 | 12 ± 6 | 15 ± 8 | 70 ± 40 | 160 ± 50 | 0.5 | 1.4 | Ruffled fur, weakness, difficulty breathing |
| 250 | 60 ± 10 | 50 ± 30 | 510 ± 70 | ND | 0.9 | ND | Euthanized |
| T-00127-HEV1 | |||||||
| 5 | 1.8 ± 0.2 | ND | 4 ± 1 | ND | 0.3 | ND | |
| 50 | 13 ± 6 | 19 ± 1 | 40 ± 20 | 200 ± 100 | 0.5 | 0.6 | Ruffled fur, weakness |
| 250 | 50 ± 20 | 40 ± 10 | 300 ± 40 | 3,000 ± 3,000 | 0.5 | 3.5 | Ruffled fur, weakness, difficulty breathing |
Cmax, maximum concentration of compound in serum; AUC0–∞, area under the concentration-time curve from 0 h to infinity; Tmax, time to maximum concentration of compound in serum; ND, not determined.
Repeat dosing in SJL mice suggests that short-term inhibition of PI4KIIIβ is deleterious.
Compound 2 and T-00127-HEV1 underwent repeat dosing studies in SJL mice, and PK and tolerability were assessed over 7 days. The SJL mice were dosed orally once per day at 50 and 250 mg/kg. As evidenced in the survival graph (Fig. 3), effects were seen on survival with both compounds from different structural classes. Moreover, all animals dosed with 250 mg of compound 2 per kg were euthanized 4 h after dosing for ethical reasons. At all tested doses with both compounds, animals showed signs of ruffled fur, weakness, and, at higher concentrations, difficulty breathing (Table 5). The deleterious effects were dose related as evidenced in the pharmacokinetic oral daily dosing profile (Table 5). In addition, toxicity was related to potency in that T-00127-HEV1 is 3× less potent than compound 2 and less mortality was observed. In the absence of an adequate safety margin, we did not pursue further in vivo experiments.
Fig 3.
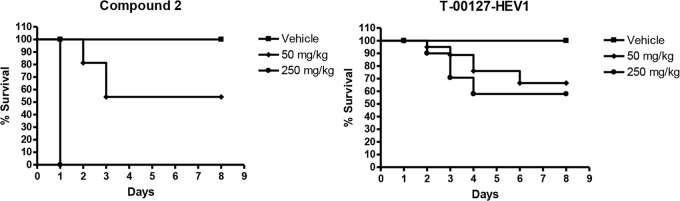
Repeat dosing in SJL mice suggests that short-term inhibition of PI4KIIIβ is deleterious. Survival graphs after compound 2 and T-00127-HEV1 were dosed at 50 and 250 mg/kg over 7 days (n = 20 for each dose).
DISCUSSION
In order to identify highly potent compounds with broad HRV antiviral activity, we screened a subset of our proprietary collection with the goal of identifying novel targets. We identified an aminothiazole series of compounds that prevented HRV replication by inhibiting PI4KIIIβ. Hijacking of lipid kinases has become a common theme in viral pathogenesis (reviewed in references 44 and 45). While we were conducting this research, several groups published on the link between enteroviral replication and PI4KIIIβ including the identification of nonspecific (e.g., PIK93) and specific PI4KIIIβ inhibitors (e.g., T-00127-HEV1) (15, 42, 46, 47). Moreover, PI4KIIIα or PI4KIIIβ has been shown to be essential for the replication of other viruses such as hepatitis C virus, West Nile virus, and coronavirus (39, 48–53).
The identification of this novel target for drug discovery is balanced by the fact that these lipid kinases have multiple and not necessarily interchangeable roles in the cell (reviewed in reference 54). Type III PI4Ks are involved in membrane trafficking, and specifically PI4KIIIβ is involved in regulating the transport and exit of cargo from the Golgi complex (55, 56). Moreover, the yeast homolog of PI4KIIIβ, PIK1, is an essential gene (57). A recent study suggests that PI4KIIIβ preserves lysosomal identity, and the absence of PI4KIIIβ or kinase activity leads to abnormal formation of tubular structures at the lysosomal surface through which lysosomal constituents are lost (58). The rationale and the exact role of phosphatidylinositol 4-phosphate (PI4P) lipids in enterovirus replication remain unknown, but PI4KIIIβ appears to be involved in creating a site for viral RNA replication. Different viruses, even within the Picornaviridae family, appear to use different strategies to recruit PI4KIIIβ (42, 46, 47).
As has been found by others (40, 59), we identified resistance mutations in HRV's 3A protein upon sequential passaging of virus in the presence of increasing concentrations of compounds from our PI4KIIIβ inhibitor series. The I42V resistance mutation in 3A was the first evidence that the mechanism of action of the compound series under investigation was similar to the mechanism of action of enviroxime. While we demonstrated that the reverse-engineered I42V HRV produced 1,000 times more virus than the wild-type virus in the presence of compound, another group recently demonstrated that similar mutations in the 3A gene of CVB3 enable these mutant viruses to replicate efficiently in cells depleted of PI4KIIIβ (60). Moreover, the 3A mutants did not use other PI4K isoforms or did not recruit PI4P to the site of viral replication, although PI4KIIIβ was still recruited by the 3A mutants in the presence and absence of PI4KIIIβ inhibitors.
Notably, no cytotoxicity was seen on H1-HeLa cells either by chemical inhibition of PI4KIIIβ or upon siRNA knockdown for any of the tested lipid kinases over 7 days. This has also been noted by other groups (15, 42, 46). Additionally, we were able to demonstrate antiviral activity without impact on cell viability in primary HBECs treated for 3 days with specific PI4KIIIβ inhibitors. In order to evaluate pharmacologic intervention in vivo, we wanted to explore inhibition in a pharmacodynamic model of CVB4 pancreatitis. Data were obtained from Lexicon Pharmaceuticals that the PI4KB mutant line (TF1995) is embryonic lethal in the homozygous state; however, the effect of inhibiting PI4KIIIβ over a shorter duration was not known (Lexicon Pharmaceuticals, personal communication). Compound 2 and T-00127-HEV1 were identified to be compounds active against enterovirus with good pharmacokinetics. Importantly, they are from two different structural classes. We hypothesized that this would allow in vivo assessment of the target-related tolerability by dissociating compound-related toxicity from target-related toxicity.
In preparation for evaluation in the pharmacodynamic model of CVB4 pancreatitis in SJL mice, repeat daily oral dosing of compound 2 and T-00127-HEV1 was performed over 7 days. Mortality was seen in all dosing groups except vehicle controls, therefore suggesting that short-term inhibition of PI4KIIIβ is deleterious in mice. No deleterious effects were seen in the prior single-dose pharmacokinetic experiment with CD-1 mice, and the Cmaxs are similar for both compounds at 50 mg/kg in both CD-1 and SJL mice. However, the CD-1 mice were treated with compound 2 for only 4 h, and it is possible that other effects would have been seen upon longer treatment. We chose to not conduct a repeat dosing study in CD-1 mice for ethical reasons.
The background of SJL mice might also point to why this strain was more sensitive to treatment with the P4KIIIβ inhibitors than CD-1 mice. SJL mice have mutations in the dysferlin gene that increase susceptibility to spontaneous myopathy (61). There are also human patient populations with mutations in the dysferlin gene, and these limb girdle muscular dystrophy patients are often diagnosed in late adolescence/young adulthood (62). Muscle weakness and disease can be further exacerbated in SJL mice by administration of fausudil, a Rho kinase inhibitor, where decreased forelimb and hindlimb grip strength and decreased horizontal activity is reported (63). Similar effects were seen upon administration of compound 2 and T-00127-HEV1. However, it is important to note that neither compound inhibits Rho kinase up to the tested concentration of 10 μM (Invitrogen kinase panel). While the mortality seen in SJL mice could be associated with a genetic factor, deleterious effects should not have been seen with any strain tested. Recently, it was shown that selective PI4KIIIβ inhibitors from Novartis prevented proliferation of bone marrow cells and lymphocytes in vitro (64). No observations were noted in rats dosed orally over 4 days. It is possible that the above deleterious results of PI4KIIIβ inhibition are species specific and not relevant to humans. However, these results underline the potential problems associated with pursuing this cellular target for antiviral therapy. The relevance to humans and the exact mechanism of the observed toxicity should be further examined.
Overall, the same ill effects were seen with both structural classes when administered orally, suggesting that short-term inhibition of PI4KIIIβ is deleterious. It is not known how much PI4KIIIβ activity and PI4P lipids are required for rhinoviral replication and if it would be possible to reduce systemic effects through intranasal administration. However, our data raise doubts on the safety of inhibiting PI4KIIIβ in order to prevent HRV-induced exacerbations.
ACKNOWLEDGMENTS
We acknowledge Armando De Palma and Stijn Delmotte for assistance in screening the compounds against a wider panel of enteroviruses. We acknowledge Lexicon Pharmaceuticals for the information on the PI4KB mutant line. We thank Ralph Gareus from TaconicArtemis GmbH for helpful discussions. We thank Michael Cordingley, Richard Bethell, Patrick Robinson, Elaine Wang, and Peter White for project support and helpful discussions, Louie Lamorte for critical review of the manuscript, and Ywe Looper, Stephanie Warner, and Julie Edwards for patent and scientific administration support.
This work was supported by Boehringer Ingelheim.
Johan Neyts and Pieter Leyssen are employees of the Rega Institute, KU Leuven. All other authors were employees of Boehringer Ingelheim when this work was performed.
Footnotes
Published ahead of print 6 May 2013
REFERENCES
- 1. Simmonds P, McIntyre C, Savolainen-Kopra C, Tapparel C, Mackay IM, Hovi T. 2010. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J. Gen. Virol. 91:2409–2419 [DOI] [PubMed] [Google Scholar]
- 2. Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, Symington P, O'Toole S, Myint SH, Tyrrell DA. 1995. Community study of role of viral infections in exacerbations of asthma in 9–11 year old children. BMJ 310:1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Olenec JP, Kim WK, Lee WM, Vang F, Pappas TE, Salazar LE, Evans MD, Bork J, Roberg K, Lemanske RF, Jr, Gern JE. 2010. Weekly monitoring of children with asthma for infections and illness during common cold seasons. J. Allergy Clin. Immunol. 125:1001–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atmar RL, Guy E, Guntupalli KK, Zimmerman JL, Bandi VD, Baxter BD, Greenberg SB. 1998. Respiratory tract viral infections in inner-city asthmatic adults. Arch. Intern. Med. 158:2453–2459 [DOI] [PubMed] [Google Scholar]
- 5. Seemungal T, Harper-Owen R, Bhowmik A, Moric I, Sanderson G, Message S, Maccallum P, Meade TW, Jeffries DJ, Johnston SL, Wedzicha JA. 2001. Respiratory viruses, symptoms, and inflammatory markers in acute exacerbations and stable chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 164:1618–1623 [DOI] [PubMed] [Google Scholar]
- 6. Message SD, Laza-Stanca V, Mallia P, Parker HL, Zhu J, Kebadze T, Contoli M, Sanderson G, Kon OM, Papi A, Jeffery PK, Stanciu LA, Johnston SL. 2008. Rhinovirus-induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL-10 production. Proc. Natl. Acad. Sci. U. S. A. 105:13562–13567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mallia P, Message SD, Gielen V, Contoli M, Gray K, Kebadze T, Aniscenko J, Laza-Stanca V, Edwards MR, Slater L, Papi A, Stanciu LA, Kon OM, Johnson M, Johnston SL. 2011. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am. J. Respir. Crit. Care Med. 183:734–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rohde G. 2009. Drug targets in rhinoviral infections. Infect. Disord. Drug Targets 9:126–132 [DOI] [PubMed] [Google Scholar]
- 9. Patick AK, Brothers MA, Maldonado F, Binford S, Maldonado O, Fuhrman S, Petersen A, Smith GJ, III, Zalman LS, Burns-Naas LA, Tran JQ. 2005. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 49:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patick AK, Binford SL, Brothers MA, Jackson RL, Ford CE, Diem MD, Maldonado F, Dragovich PS, Zhou R, Prins TJ, Fuhrman SA, Meador JW, Zalman LS, Matthews DA, Worland ST. 1999. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 43:2444–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan J, George S, Kusov Y, Perbandt M, Anemuller S, Mesters JR, Norder H, Coutard B, Lacroix C, Leyssen P, Neyts J, Hilgenfeld R. 2013. 3C protease of enterovirus 68: structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 87:4339–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayden FG, Gwaltney JM., Jr 1982. Prophylactic activity of intranasal enviroxime against experimentally induced rhinovirus type 39 infection. Antimicrob. Agents Chemother. 21:892–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Phillpotts RJ, Jones RW, Delong DC, Reed SE, Wallace J, Tyrrell DA. 1981. The activity of enviroxime against rhinovirus infection in man. Lancet i:1342–1344 [DOI] [PubMed] [Google Scholar]
- 14. Miller FD, Monto AS, Delong DC, Exelby A, Bryan ER, Srivastava S. 1985. Controlled trial of enviroxime against natural rhinovirus infections in a community. Antimicrob. Agents Chemother. 27:102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arita M, Kojima H, Nagano T, Okabe T, Wakita T, Shimizu H. 2011. Phosphatidylinositol 4-kinase III beta is a target of enviroxime-like compounds for antipoliovirus activity. J. Virol. 85:2364–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayden FG, Hipskind GJ, Woerner DH, Eisen GF, Janssens M, Janssen PA, Andries K. 1995. Intranasal pirodavir (R77,975) treatment of rhinovirus colds. Antimicrob. Agents Chemother. 39:290–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayden FG, Herrington DT, Coats TL, Kim K, Cooper EC, Villano SA, Liu S, Hudson S, Pevear DC, Collett M, McKinlay M. 2003. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: results of 2 double-blind, randomized, placebo-controlled trials. Clin. Infect. Dis. 36:1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pevear DC, Hayden FG, Demenczuk TM, Barone LR, McKinlay MA, Collett MS. 2005. Relationship of pleconaril susceptibility and clinical outcomes in treatment of common colds caused by rhinoviruses. Antimicrob. Agents Chemother. 49:4492–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biota Holdings Limited 28 March 2012. HRV phase IIb study achieves primary endpoint. Biota Holdings Limited, Notting Hill, Victoria, Australia: http://www.biota.com.au/uploaded/154/1021819_20hrvphaseiibstudyachieve.pdf [Google Scholar]
- 20. Feil SC, Hamilton S, Krippner GY, Lin B, Luttick A, McConnell DB, Nearn R, Parker MW, Ryan J, Stanislawski PC, Tucker SP, Watson KG, Morton CJ. 2012. An orally available 3-ethoxybenzisoxazole capsid binder with clinical activity against human rhinovirus. ACS Med. Chem. Lett. 3:303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 45:3655–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McErlean P, Shackelton LA, Andrews E, Webster DR, Lambert SB, Nissen MD, Sloots TP, Mackay IM. 2008. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS One 3:e1847. 10.1371/journal.pone.0001847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ashraf S, Brockman-Schneider R, Bochkov YA, Pasic TR, Gern JE. 2013. Biological characteristics and propagation of human rhinovirus-C in differentiated sinus epithelial cells. Virology 436:143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bochkov YA, Palmenberg AC, Lee WM, Rathe JA, Amineva SP, Sun X, Pasic TR, Jarjour NN, Liggett SB, Gern JE. 2011. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat. Med. 17:627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hao W, Bernard K, Patel N, Ulbrandt N, Feng H, Svabek C, Wilson S, Stracener C, Wang K, Suzich J, Blair W, Zhu Q. 2012. Infection and propagation of human rhinovirus C in human airway epithelial cells. J. Virol. 86:13524–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang T, Wang W, Bessaud M, Ren P, Sheng J, Yan H, Zhang J, Lin X, Wang Y, Delpeyroux F, Deubel V. 2009. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One 4:e6355. 10.1371/journal.pone.0006355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwane MK, Prill MM, Lu X, Miller EK, Edwards KM, Hall CB, Griffin MR, Staat MA, Anderson LJ, Williams JV, Weinberg GA, Ali A, Szilagyi PG, Zhu Y, Erdman DD. 2011. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J. Infect. Dis. 204:1702–1710 [DOI] [PubMed] [Google Scholar]
- 28. Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, Martin AC, Khoo SK, Cox DW, Geelhoed GC, McMinn PC, Goldblatt J, Gern JE, Le Souef PN. 2011. Association between human rhinovirus C and severity of acute asthma in children. Eur. Respir. J. 37:1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller EK, Edwards KM, Weinberg GA, Iwane MK, Griffin MR, Hall CB, Zhu Y, Szilagyi PG, Morin LL, Heil LH, Lu X, Williams JV. 2009. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 123:98–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piralla A, Rovida F, Campanini G, Rognoni V, Marchi A, Locatelli F, Gerna G. 2009. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J. Clin. Virol. 45:311–317 [DOI] [PubMed] [Google Scholar]
- 31. Lee WM, Lemanske RF, Jr, Evans MD, Vang F, Pappas T, Gangnon R, Jackson DJ, Gern JE. 2012. Human rhinovirus species and season of infection determine illness severity. Am. J. Respir. Crit. Care Med. 186:886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 33. Canadian Council of Animal Care 1993. Guide to the care and use of experimental animals, vol 1, 2nd ed Canadian Council of Animal Care, Ottawa, Ontario, Canada [Google Scholar]
- 34. De Palma AM, Thibaut HJ, van der Linden L, Lanke K, Heggermont W, Ireland S, Andrews R, Arimilli M, Al-Tel TH, De Clercq E, van Kuppeveld F, Neyts J. 2009. Mutations in the nonstructural protein 3A confer resistance to the novel enterovirus replication inhibitor TTP-8307. Antimicrob. Agents Chemother. 53:1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. 2008. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McKnight KL, Lemon SM. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andries K, Dewindt B, Snoeks J, Willebrords R, van Eemeren K, Stokbroekx R, Janssen PA. 1992. In vitro activity of pirodavir (R 77975), a substituted phenoxy-pyridazinamine with broad-spectrum antipicornaviral activity. Antimicrob. Agents Chemother. 36:100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Binford SL, Weady PT, Maldonado F, Brothers MA, Matthews DA, Patick AK. 2007. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 51:4366–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vaillancourt FH, Brault M, Pilote L, Uyttersprot N, Gaillard ET, Stoltz JH, Knight BL, Pantages L, McFarland M, Breitfelder S, Chiu TT, Mahrouche L, Faucher AM, Cartier M, Cordingley MG, Bethell RC, Jiang H, White PW, Kukolj G. 2012. Evaluation of phosphatidylinositol-4-kinase IIIalpha as a hepatitis C virus drug target. J. Virol. 86:11595–11607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heinz BA, Vance LM. 1995. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J. Virol. 69:4189–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heinz BA, Vance LM. 1996. Sequence determinants of 3A-mediated resistance to enviroxime in rhinoviruses and enteroviruses. J. Virol. 70:4854–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss WA, Williams RL, Shokat KM. 2006. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell 125:733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Altan-Bonnet N, Balla T. 2012. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 37:293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Delang L, Paeshuyse J, Neyts J. 2012. The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication. Biochem. Pharmacol. 84:1400–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greninger AL, Knudsen GM, Betegon M, Burlingame AL, DeRisi JL. 2012. The 3A protein from multiple picornaviruses utilizes the Golgi adaptor protein ACBD3 to recruit PI4KIIIbeta. J. Virol. 86:3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sasaki J, Ishikawa K, Arita M, Taniguchi K. 2012. ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 31:754–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vaillancourt FH, Pilote L, Cartier M, Lippens J, Liuzzi M, Bethell RC, Cordingley MG, Kukolj G. 2009. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology 387:5–10 [DOI] [PubMed] [Google Scholar]
- 49. Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, Cornellataracido I, Baryza J, Tallarico J, Joberty G, Bantscheff M, Schirle M, Bouwmeester T, Mathy JE, Lin K, Compton T, Labow M, Wiedmann B, Gaither LA. 2009. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J. Virol. 83:10058–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J. Virol. 85:8870–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin-Acebes MA, Blazquez AB, Jimenez de Oya N, Escribano-Romero E, Saiz JC. 2011. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS One 6:e24970. 10.1371/journal.pone.0024970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang N, Ma P, Lang J, Zhang Y, Deng J, Ju X, Zhang G, Jiang C. 2012. Phosphatidylinositol 4-kinase IIIbeta is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry. J. Biol. Chem. 287:8457–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Balla A, Balla T. 2006. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 16:351–361 [DOI] [PubMed] [Google Scholar]
- 55. Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. 1999. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1:280–287 [DOI] [PubMed] [Google Scholar]
- 56. Toth B, Balla A, Ma H, Knight ZA, Shokat KM, Balla T. 2006. Phosphatidylinositol 4-kinase IIIbeta regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem. 281:36369–36377 [DOI] [PubMed] [Google Scholar]
- 57. Flanagan CA, Schnieders EA, Emerick AW, Kunisawa R, Admon A, Thorner J. 1993. Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science 262:1444–1448 [DOI] [PubMed] [Google Scholar]
- 58. Sridhar S, Patel B, Aphkhazava D, Macian F, Santambrogio L, Shields D, Cuervo AM. 2013. The lipid kinase PI4KIIIbeta preserves lysosomal identity. EMBO J. 32:324–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arita M, Wakita T, Shimizu H. 2009. Cellular kinase inhibitors that suppress enterovirus replication have a conserved target in viral protein 3A similar to that of enviroxime. J. Gen. Virol. 90:1869–1879 [DOI] [PubMed] [Google Scholar]
- 60. van der Schaar HM, van der Linden L, Lanke KH, Strating JR, Purstinger G, de Vries E, de Haan CA, Neyts J, van Kuppeveld FJ. 2012. Coxsackievirus mutants that can bypass host factor PI4KIIIbeta and the need for high levels of PI4P lipids for replication. Cell Res. 22:1576–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bittner RE, Anderson LV, Burkhardt E, Bashir R, Vafiadaki E, Ivanova S, Raffelsberger T, Maerk I, Hoger H, Jung M, Karbasiyan M, Storch M, Lassmann H, Moss JA, Davison K, Harrison R, Bushby KM, Reis A. 1999. Dysferlin deletion in SJL mice (SJL-Dysf) defines a natural model for limb girdle muscular dystrophy 2B. Nat. Genet. 23:141–142 [DOI] [PubMed] [Google Scholar]
- 62. Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Hamida MB, Bohlega S, Culper EJ, Amato AA, Bossie K, Oeltjen J, Bejaoui K, McKenna-Yasek D, Hosler BA, Schurr E, Arahata K, de Jong PJ, Brown RH., Jr 1998. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20:31–36 [DOI] [PubMed] [Google Scholar]
- 63. Rayavarapu S, Van der Meulen JH, Gordish-Dressman H, Hoffman EP, Nagaraju K, Knoblach SM. 2010. Characterization of dysferlin deficient SJL/J mice to assess preclinical drug efficacy: fasudil exacerbates muscle disease phenotype. PLoS One 5:e12981. 10.1371/journal.pone.0012981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lamarche MJ, Borawski J, Bose A, Capacci-Daniel C, Colvin R, Dennehy M, Ding J, Dobler M, Drumm J, Gaither LA, Gao J, Jiang X, Lin K, McKeever U, Puyang X, Raman P, Thohan S, Tommasi R, Wagner K, Xiong X, Zabawa T, Zhu S, Wiedmann B. 2012. Anti-hepatitis C virus activity and toxicity of type III phosphatidylinositol-4-kinase beta inhibitors. Antimicrob. Agents Chemother. 56:5149–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Décor A, Grand-Maître C, Hucke O, O'Meara J, Kuhn C, Constantineau-Forget L, Brochu C, Malenfant E, Bertrand-Laperle M, Bordeleau J, Ghiro E, Pesant M, Fazal G, Gorys V, Little M, Boucher C, Bordeleau S, Turcotte P, Guo T, Garneau M, Spickler C, Gauthier A. 7 May 2013, posting date Design, synthesis and biological evaluation of novel aminothiazoles as antiviral compounds acting against human rhinovirus. Bioorg. Med. Chem. Lett. 10.1016/j.bmcl.2013.04.077 [DOI] [PubMed] [Google Scholar]