Abstract
Naegleria fowleri, a pathogenic free-living amoeba, causes fatal primary amoebic meningoencephalitis (PAM) in humans and animals. The nfa1 gene (360 bp), cloned from a cDNA library of N. fowleri, produces a 13.1-kDa recombinant protein which is located on pseudopodia, particularly the food cup structure. The nfa1 gene plays an important role in the pathogenesis of N. fowleri infection. To examine the effect of nfa1 DNA vaccination against N. fowleri infection, we constructed a lentiviral vector (pCDH) expressing the nfa1 gene. For the in vivo mouse study, BALB/c mice were intranasally vaccinated with viral particles of a viral vector expressing the nfa1 gene. To evaluate the effect of vaccination and immune responses of mice, we analyzed the IgG levels (IgG, IgG1, and IgG2a), cytokine induction (interleukin-4 [IL-4] and gamma interferon [IFN-γ]), and survival rates of mice that developed PAM. The levels of both IgG and IgG subclasses (IgG1 and IgG2a) in vaccinated mice were significantly increased. The cytokine analysis showed that vaccinated mice exhibited greater IL-4 and IFN-γ production than the other control groups, suggesting a Th1/Th2 mixed-type immune response. In vaccinated mice, high levels of Nfa1-specific IgG antibodies continued until 12 weeks postvaccination. The mice vaccinated with viral vector expressing the nfa1 gene also exhibited significantly higher survival rates (90%) after challenge with N. fowleri trophozoites. Finally, the nfa1 vaccination effectively induced protective immunity by humoral and cellular immune responses in N. fowleri-infected mice. These results suggest that DNA vaccination using a viral vector may be a potential tool against N. fowleri infection.
INTRODUCTION
The pathogenic free-living amoeba Naegleria fowleri is commonly found in water, soil, and sediment (1). N. fowleri is the causal agent of primary amoebic meningoencephalitis (PAM) in animals and humans (2, 3). PAM is a fulminating disease, causing death within 1 to 2 weeks from the onset of symptoms (4). It occurs mainly in children and young adults and has been associated with swimming or water activities in contaminated waters (3, 5). The adherence of the amoeba is the critical initial step in the infection process, and N. fowleri enters the central nervous system (CNS) through the olfactory bulb (6). Amphotericin B is the only known agent for the treatment of N. fowleri infection (7–9). However, not all PAM patients treated with amphotericin B have survived, and amphotericin B has side effects (10, 11). Unfortunately, until now, there have been no satisfactory therapeutic agents for the treatment of PAM.
In a previous study, we cloned an antigenic gene, nfa1, from the N. fowleri cDNA library, which had a coding nucleotide sequence of 360 bp, producing a recombinant protein of 13.1 kDa (12). The nfa1 gene, which is involved with amoebic pseudopodial activity and especially with food cup formation, plays an important role in the pathogenicity of N. fowleri infection (13, 14). Moreover, an anti-Nfa1 antibody caused a decrease in the cytotoxicity of N. fowleri against target cells (15). Therefore, because the nfa1 gene is the key molecule concerned with cytotoxicity against host cells in regard to contact-dependent pathogenesis of N. fowleri, the nfa1 gene is an appropriate candidate for DNA vaccination.
In 1990, DNA vaccination was first introduced, and the induction of protein expression upon direct intramuscular injection of plasmid DNA into myocytes was demonstrated (16). DNA vaccination has been shown to be the most effective way of inducing specific humoral and cellular immune responses; this represents a promising strategy for protecting humans against pathogenic microorganisms, such as human immunodeficiency virus, mycobacteria, and parasites (17–19). Recently, lentiviral vectors have emerged as very promising vaccination tools. Lentiviral vectors have been widely used for the development of DNA vaccines to deliver genes effectively. Lentiviral vectors have been evaluated in various preclinical models of gene therapy and immunization because they can infect dividing and nondividing cells (20, 21). These vectors elicit both specific cytotoxic and strong humoral immune responses in animal models (22). Lentiviral vectors are regarded as promising vaccine vector candidates for the treatment of infectious disease and cancer (23).
Host protective immunity to N. fowleri infection has been studied in an in vivo model of PAM following administration of amoebic extracts, culture fluid, and amoebic trophozoites (24). Mice immunized with an intraperitoneal inoculation with live or killed trophozoites of N. fowleri showed variable levels of partial protective immunity (25). According to our previous studies, the nfa1 gene may be a proper candidate for DNA vaccination against N. fowleri infection (12–14, 26). Based on these findings, to evaluate the effect of our lentiviral vector systems expressing the nfa1 gene in the in vivo mouse model, vaccinated mice were tested for the development of specific immunity against N. fowleri infection, measured by humoral and cellular immune responses and by survival rates.
MATERIALS AND METHODS
Cultivation of Naegleria fowleri.
Trophozoites of N. fowleri (Carter NF69 strain; American Type Culture Collection no. 30215) were cultured at 37°C in axenic Nelson's medium supplemented with 10% fetal bovine serum (FBS) (Gibco BRL, Gaithersburg, MD) (27).
Expression and purification of recombinant Nfa1 protein.
The recombinant Nfa1 (rNfa1) protein was produced according to the method previously described (12). Purified DNA (5 μg/μl) obtained from a PCR-T7/NT TOPO expression vector (Invitrogen, Groningen, Netherlands) containing the nfa1 gene was subsequently transferred to the BL21(DE3)-pLysS Escherichia coli strain using the heat shock method. Cells were cultured at 37°C in Luria-Bertani medium containing 100 μg/ml of ampicillin and 34 μg/ml of chloramphenicol (LAC) for selection. A transformed colony was selected and cultured in the LAC broth at 37°C. After 4 h of incubation with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the cells were harvested by centrifugation (6,000 × g for 15 min). Transformed and nontransformed BL21(DE3)pLysS E. coli cell extracts were analyzed by SDS-PAGE, and the presence of the expressed gene product (a His tag fusion rNfa1 protein) was confirmed by Western blotting using sera from both immunized and infected mice. The rNfa1 protein was purified by metal affinity chromatography using a nickel-nitrilotriacetic acid (Ni-NTA) agarose column (Qiagen, Hilden, Germany), as described previously (28). Elution was carried out using imidazole buffer (5 M urea, 20 mM Na2HPO4, 5 M NaCl, and 500 mM imidazole). The protein was dialyzed in phosphate-buffered saline (PBS) (pH 7.4), and the purity of the rNfa1 protein was evaluated by SDS-PAGE and by Western blotting.
Construction of the nfa1 lentiviral vector and collection of viral particles.
The nfa1 gene (GenBank accession no. AF230370) was previously cloned from an nfa1 gene-cloned vector, PCR-T7/NT TOPO (Invitrogen) (12). For the construction of the lentiviral vector expressing the nfa1 gene, the amplified egfp-nfa1 vector was digested with NheI and EcoRI restriction enzymes and was cloned into the lentiviral vector pCDH (System Biosciences, Mountain View, CA) according to the method previously described (26). The egfp-nfa1 lentiviral vector was transformed in a ligation mixture into E. coli DH5α. The egfp-nfa1 vector was purified from transformed E. coli DH5α using a plasmid kit (Qiagen, Hilden, Germany). The lentiviral vector pCDH/egfp-nfa1 was packaged to 293TN packaging cells with the pPACK packaging mix using a lentivector expression system (System Biosciences) according to the manufacturer's instructions. At 48 h after transfection to packaging cells, we collected the medium from packaging cells, centrifuged the medium at 3,000 × g for 15 min at room temperature to pellet cell debris, and filtered the medium through a 0.45-μm filter. Each viral particle (VP) was aliquoted in cryogenic vials and stored at −70°C until ready for use. The titer of the viral particles was determined with a QuickTiter kit (Cell Biolabs, Inc., San Diego, CA) according to the manufacturer's instructions.
DNA vaccination in mice.
For the DNA vaccination, 6-week-old female BALB/c mice (Samtaco, Suwon, South Korea) were divided into 5 experimental groups, each consisting of 10 animals. All mice were anesthetized for the intranasal infection with a mixture of 5 mg/kg of body weight of ketamine and 0.5 mg/ml xylazine. In the uninfected groups, 50 μl of either saline or Dulbecco's modified Eagle's medium (DMEM) was intranasally instilled into the nostril of each mouse. Mice in the empty vector group were infected intranasally with VP at concentrations of 5 × 1011. In the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups, mice were infected with VPs (lentiviral vectors expressing the nfa1 gene) at concentrations of 2.5 × 1011 and 5 × 1011, respectively. The second and third vaccinations were performed at 1-week intervals. Animal care and experimental procedures were carried out under approval from the Animal Care Committee of the Ajou University School of Medicine (AMC66).
IgG and IgG subclass analysis.
One week after the final vaccination, blood samples from each group were analyzed for antibodies. Serum samples collected from the tail vein were centrifuged and analyzed for IgG, IgG1, and IgG2a antibodies by enzyme-linked immunosorbent assays (ELISAs). The rNfa1 protein (1 μg/ml) diluted in coating buffer (0.05 M carbonate-bicarbonate [pH 9.6]) was placed into 96-well ELISA plates (Nunc, Roskilde, Denmark) in 100-μl/well reactions. After incubation and washing, the reactions were blocked with 3% bovine serum albumin (BSA) (Sigma) in PBS (pH 7.4) at 4°C. After the serum samples were washed, they were diluted in PBS and added to each well. After being incubated and washed, 100 μl of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG, IgG1, or IgG2a antibodies (Bethyl Laboratories, Montgomery, TX) was added to each well. After the antibodies were incubated and washed, they were detected with 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) substrate solution (Kirkegaard & Perry Laboratories, Gaithersburg, MD). The plates were read at 405 nm in a microplate reader (Bio-Rad Laboratories, Hercules, CA).
Cytokine release assay.
Spleens were removed from the vaccinated mice under aseptic conditions 2 weeks after the final vaccination. Single-splenocyte suspensions were prepared by mashing the tissue through sterile metal screens in RPMI 1640 medium (Gibco BRL) supplemented with 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (complete RPMI 1640 medium). After lysing red blood cells with lysis buffer and washing them, we resuspended the splenocytes in complete RPMI 1640 medium containing 10% FBS. Splenocytes were adjusted at a density of 4 × 106 cells/ml and were seeded onto 24-well tissue culture plates (Nunc). For cytokine induction, splenocytes were stimulated in vitro with 10 μg/ml of rNfa1 protein.
After cultivation for 24 h (interleukin-4 [IL-4]) and 72 h (gamma interferon [IFN-γ]) at 37°C in a humidified atmosphere with 5% CO2, culture supernatants were harvested and stored at −80°C until they were assayed for cytokine production. Cytokines produced by activated splenocytes were measured by sandwich ELISAs. Monoclonal antibodies specific for mouse IFN-γ and IL-4 were used as the capture antibodies, and an enzyme-linked polyclonal antibody was used as the reporter antibody according to the manufacturer's instructions (R&D Systems, Inc., Minneapolis, MN). Tetramethylbenzidine microwell peroxidase was used as the substrate, and the reaction was terminated by 1 M H3PO4. The plate was read at 405 nm in a microplate reader (Bio-Rad Laboratories).
Duration of the Nfa1-specific IgG levels in vaccinated mice.
The Nfa1-specific IgG responses were evaluated by ELISAs in each group at 1, 4, 8, and 12 weeks after the last vaccination. The saline and rNfa1 groups were used as negative- and positive-control groups and received saline and recombinant Nfa1 (rNfa1) protein, respectively, under the same conditions.
N. fowleri infection and experimental PAM in mice.
N. fowleri trophozoites cultured for 3 days in Nelson's medium at 37°C were harvested and inoculated into mice. One week after the final vaccination, mice were infected with N. fowleri and monitored for the development of experimental PAM. According to a previous study (29), we decided that a lethal dose was 1 × 104 N. fowleri trophozoites. N. fowleri trophozoites (1 × 104) were inoculated intranasally into the mice under anesthesia. All the following experiments were carried out with 10 mice per group.
Survival rates and mean times to death of mice for evaluation of vaccination.
Mice were held for 1 month after inoculation with N. fowleri trophozoites, and the cumulative percentage of death was recorded on a daily basis. The mean time to death was also determined for each group. In order to verify the cause of death, brain tissue from dead mice was cultured at 37°C in Nelson's medium, and N. fowleri trophozoites were observed microscopically.
Statistical analysis.
Statistical differences between groups or samples were determined using the Student t test. The difference was considered statistically significant when the P value was <0.05.
RESULTS
Construction of a lentiviral vector expressing the nfa1 gene.
To establish viral vector systems for DNA vaccination, we used a pCDH lentiviral vector as a backbone. In order to construct the viral vectors, amplified egfp-nfa1 was digested with restriction enzymes and was inserted into the pCDH vector (Fig. 1A). The flowchart in Fig. 1B presents the schedule for vaccination, N. fowleri trophozoite inoculation, and immune responses of vaccinated mice. One week after the final vaccination, the mice were infected with N. fowleri trophozoites and monitored for the development of experimental PAM.
Fig 1.

The map of the lentiviral vector and flow chart of the in vivo study. (A) The pCDH/egfp-nfa1 vector was constructed. The egfp-nfa1 gene was inserted into the multiple cloning site (MCS) of the lentiviral vector (pCDH). LTR, long terminal repeat; CMV, cytomegalovirus; SV40, simian virus 40; RSV, respiratory syncytial virus. (B) The schedule for mouse vaccination, N. fowleri infection, and in vivo experiments in this study.
Specific immune responses in vaccinated mice.
To determine specific immune responses in mice vaccinated with the lentiviral vector expressing the nfa1 gene, all serum samples were tested using ELISAs. In the ELISAs, anti-Nfa1 monoclonal antibodies and PBS were used as the positive and negative controls, respectively. The uninfected (saline), uninfected (DMEM), and empty vector (5 × 1011 VP) groups, representing mice vaccinated with saline, DMEM, or viral particles (lentiviral vector only), respectively, were used as controls for the in vivo mouse experiments. The pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups showed significantly higher levels of IgG than the control groups (uninfected [saline], uninfected [DMEM], and empty vector [5 × 1011 VP]) (P < 0.001). In the case of IgG, the optical density (OD) values for the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups were 1.287 and 1.442, while the OD values for the uninfected (saline), uninfected (DMEM), and empty vector (5 × 1011 VP) groups were 0.114, 0.127, and 0.154, respectively (Fig. 2). To determine whether Th1-type or Th2-type immune responses were elicited in the vaccinated mice, we also measured the levels of IgG subclasses (IgG1 and IgG2a). The levels of IgG1 as well as IgG2a in the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups were significantly higher than those of the other control groups (P < 0.001). These data show that the nfa1 vaccination using a lentiviral vector efficiently induced Th1-type and Th2-type immune responses in the mice.
Fig 2.
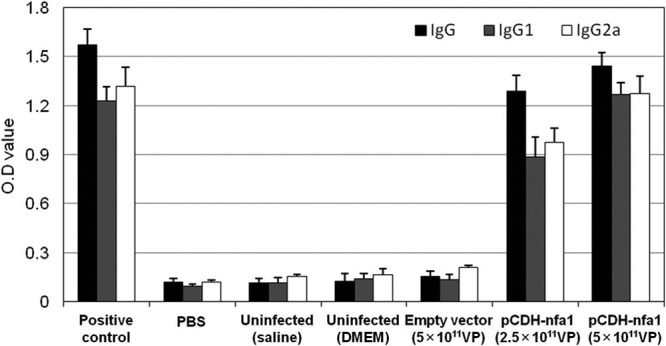
Specific immune responses in mice vaccinated with the lentiviral vector expressing the nfa1 gene. The OD values of specific anti-Nfa1 IgG, IgG1, and IgG2a antibodies were measured at 405 nm using indirect ELISAs. Serum samples were collected from the mice 1 week after the last vaccination. The OD values are expressed as means ± SD from 3 independent experiments. Positive control, anti-Nfa1 monoclonal antibody; PBS, phosphate-buffered saline.
Duration of IgG in vaccinated mice.
To evaluate the duration of the Nfa1-specific IgG antibody responses, Nfa1-specific IgG antibodies were evaluated by ELISAs in each group at 1, 4, 8, and 12 weeks after the last vaccination. As shown in Fig. 3, in the pCDH-nfa1 (5 × 1011 VP) group, higher levels of Nfa1-specific IgG antibodies were detected at 1 week and continued until 12 weeks postvaccination than those in the uninfected (saline) and empty vector (5 × 1011 VP) groups. Moreover, the level of IgG in the pCDH-nfa1 (5 × 1011 VP) group was higher than that of the mice immunized with recombinant Nfa1 protein (rNfa1 group) used as a positive control (P < 0.01) (Fig. 3).
Fig 3.

Duration of the Nfa1-specific IgG levels in the sera of mice vaccinated with the lentiviral vector. The Nfa1-specific IgG responses were evaluated by ELISAs in each group at 1, 4, 8, and 12 weeks after the last vaccination. The uninfected (saline) and rNfa1 groups were used as negative- and positive-control groups, respectively.
Cytokine responses of vaccinated mice.
To elucidate the T-cell responses in mice vaccinated with a viral vector expressing the nfa1 gene, the levels of a type 1 cytokine (IFN-γ) and a type 2 cytokine (IL-4) in splenocytes stimulated with the Nfa1 protein were analyzed by ELISAs (Fig. 4). In the case of IFN-γ, the levels in the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups were markedly enhanced compared with those of the other groups (uninfected [saline], uninfected [DMEM], and empty vector [5 × 1011 VP]) (P < 0.01) (Fig. 4). The levels of IFN-γ in the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups were increased in a dose-dependent manner (P < 0.01). Similarly, increased IL-4 levels were detected in splenocytes from mice vaccinated with the nfa1 gene (Fig. 4). The levels of IL-4 in the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups were increased on a dose-dependent basis (P < 0.01). These data show that vaccination with a lentiviral vector expressing the nfa1 gene leads to Th1/Th2 mixed-type immune responses in mice.
Fig 4.

Production of cytokines by splenocytes from mice vaccinated with the lentiviral vector. Splenocytes were collected 2 weeks after the last vaccination. Cell-free supernatants were harvested and assayed for IFN-γ activity at 96 h and for IL-4 activity at 24 h after stimulation with 10 μg/ml Nfa1 protein. The values are expressed as means ± SD with 3 mice per group.
Effective protection of vaccination in mice.
To evaluate whether nfa1 vaccination could induce protection against N. fowleri infection, nfa1-vaccinated mice were challenged intranasally with 5 × 104 trophozoites of N. fowleri at 1 week after the last vaccination. We observed the mortality rate of the mice, and the cumulative percentage was recorded on a daily basis. The cause of death was confirmed by culturing the brain tissue of dead mice prior to examination under a light microscope to identify N. fowleri (data not shown). Survival rates of the different groups of mice are shown in Fig. 5 and Table 1. Substantially increased survival rates were found in the mice vaccinated with the viral vector expressing the nfa1 gene. Mice in the control groups (uninfected [saline], uninfected [DMEM], and empty vector [5 × 1011 VP]) began dying on day 8, and all mice were dead by day 16 postinfection (Fig. 5). The mean times to death (MTDs) were 12.3, 12.3, and 12.7 days for the uninfected (saline), uninfected (DMEM), and empty vector (5 × 1011 VP) groups, respectively (Table 1). On the other hand, the survival rates of the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups were stably maintained at 70% and 90% throughout the experimental periods (1 month). The MTDs of the pCDH-nfa1 (2.5 × 1011 VP) and pCDH-nfa1 (5 × 1011 VP) groups were 15 and 20 days, respectively.
Fig 5.
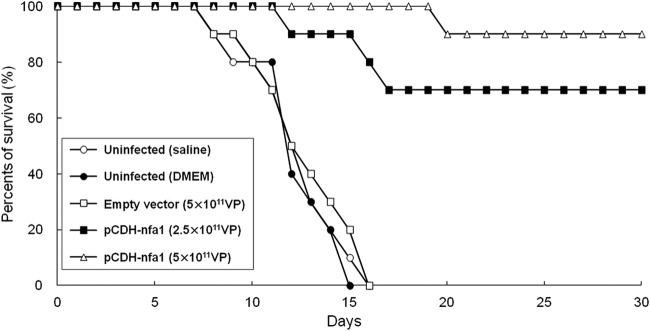
Survival curves of vaccinated mice after N. fowleri infection. The mice were intranasally challenged with 5 × 104 N. fowleri trophozoites at 1 week after the last vaccination and observed for mortality. Each group had 10 mice. The uninfected (saline or DMEM) groups were used as control groups and received saline or DMEM, respectively, under the same conditions.
Table 1.
Survival rates and mean times to death of N. fowleri-infected mice vaccinated with the lentiviral vector expressing the nfa1 gene
| Groupa | No. of mice | No. (%) of surviving miceb | MTD (days)c |
|---|---|---|---|
| Uninfected (saline) | 10 | 0 (0) | 12.3 |
| Uninfected (DMEM) | 10 | 0 (0) | 12.3 |
| Empty vector (5 × 1011 VP) | 10 | 0 (0) | 12.7 |
| pCDH-nfa1 (2.5 × 1011 VP) | 10 | 7 (70) | 15 |
| pCDH-nfa1 (5 × 1011 VP) | 10 | 9 (90) | 20 |
The uninfected groups were used as control groups and received saline or DMEM under the same conditions. All mice were intranasally inoculated with 1 × 104 N. fowleri trophozoites.
Mice were held for 30 days after inoculation, and the cumulative survival percentage was recorded on a daily basis.
MTD, mean time to death calculated by dead mice only.
DISCUSSION
Infection with N. fowleri occurs by inhalation into the nasal cavity, and subsequently, the invasive amoeba enters the CNS through the olfactory apparatus (2, 3, 5, 6). In the infection process of the amoeba, the adherence to host cells is the critical step (6). Previously, we reported that the nfa1 gene cloned from the N. fowleri cDNA library is a key molecule that mediates contact between the amoeba and host cells (13, 14, 29). Recently, we constructed lentiviral vector systems expressing the nfa1 gene and showed that our vector system delivered the nfa1 gene effectively into the target cells (26). Therefore, to find a potentially effective DNA vaccination for N. fowleri infection, the in vivo efficacies of our lentiviral vector system expressing the nfa1 gene should be evaluated.
In this study, to determine specific immune responses in vaccinated mice, we showed Nfa1-specific IgG, IgG1, and IgG2a production. The levels of IgG in nfa1-vaccinated mice were increased significantly in comparison with those in the control groups. Concerning the humoral response, nfa1-vaccinated mice had significant levels of Nfa1-specific IgG2a and IgG1 that are characteristic of Th1 and Th2 immunity, respectively. Moreover, the duration of Nfa1-specific IgG antibodies in the vaccinated mice was prolonged during the 12 weeks postvaccination in comparison with that in the control groups. Consequently, these results suggest that our nfa1 lentiviral vector system delivered and expressed the nfa1 gene and protein effectively in the target cells and, subsequently, induced specific immune responses in in vivo mouse models. The mice exhibited high levels of both IgG1 and IgG2a, suggesting that the immune response was a Th1/Th2 mixed type, which was confirmed by analysis of the cytokine (IFN-γ and IL-4) production. The result shows that the levels of both IFN-γ and IL-4 in splenocytes of nfa1-vaccinated mice were increased significantly in comparison with those of the control groups. Moreover, we previously reported that the intraperitoneal or intranasal immunization of recombinant Nfa1 protein triggers a Th1/Th2/regulatory T cell (Treg) mixed type of immune response in a mouse model (29).
To investigate the potential of nfa1 vaccination against N. fowleri infection, we observed the mean times to death and mortality rates of the vaccinated mice after infection. Previous studies have shown that intranasal or intraperitoneal immunization with recombinant Nfa1 protein can induce immune responses and partial protective immunity against N. fowleri infection and prolong the survival times of infected mice in comparison with those of control mice, but all the mice still eventually died (29). In this study, an effective and remarkable protection was obtained in nfa1-vaccinated mice that was not seen in the control groups. Mice in the control groups (saline, DMEM, or empty vector) died within 16 days after N. fowleri infection. However, nfa1-vaccinated mice had longer mean times to death and higher survival rates (90%) after N. fowleri infection. The present study demonstrates that nfa1 vaccination was able to elicit a significant immune response and increase the mean times to death and survival rates in N. fowleri-infected mice. Consequently, we suggest that the nfa1 gene is an appropriate candidate for DNA vaccination against N. fowleri infection.
DNA vaccines have been used as novel delivery systems to achieve both humoral and cell-mediated immune responses. In addition, DNA vaccines are attractive because of their ease of production, low cost, and potential for long-lasting immunity (30). Recently, the effects of DNA vaccines against parasitic diseases, such as amebiasis, leishmaniasis, schistosomiasis, and toxoplasmosis, were reported (31–34). Viral vectors also have been widely used for gene therapy and DNA vaccination. Lentiviral vectors have emerged as very promising vaccination tools. Lentiviral vectors have been evaluated in various preclinical models for gene therapy and vaccination and are regarded as promising vaccine vector candidates. Our study is the first to report an effect of DNA vaccination using a lentiviral vector in N. fowleri infection. The present study shows that nfa1 vaccination using a lentiviral vector effectively induces protective immunity in N. fowleri-infected mice.
Vaccination via the intranasal route is efficient in in vivo models, inducing mucosal immune responses, and it requires comparatively lower doses of antigens, inducing a stronger systemic immune response than does vaccination via the oral route (35). Previously, we reported that Nfa1-specific IgE antibodies were not detected in the mouse model, suggesting that intranasal Nfa1 protein immunization does not induce immune hypersensitivity (29). Furthermore, our results show that nfa1 vaccination through the intranasal route effectively induced mucosal immune responses and significantly increased the survival rate of N. fowleri-infected mice. Collectively, the intranasal route appears to be suitable in comparison with other routes for vaccination against N. fowleri infection because it effectively inhibits the adhesion of amoebae via a contact-dependent pathway.
A successful vaccine requires an efficient adjuvant system as well as a vector system. The suitable adjuvant is one that induces a more effective immune response. Furthermore, improvements in the vaccine strategy are required to determine factors such as the adjuvant combination, vector selection, antigen presentation pathways, immune status of the mice, and protection against infection. In conclusion, these results show that nfa1 vaccination is able to strongly enhance specific IgG and IgG subclass antibody production, cytokine production, and survival rates of N. fowleri-infected mice. This is the first report to evaluate in vivo immune responses of DNA vaccination against N. fowleri infection. Finally, our results should be helpful for future investigations into the detailed pathogenesis of N. fowleri infection. Furthermore, we strongly suggest that nfa1 vaccination may be a new potential tool for the treatment of N. fowleri infection.
ACKNOWLEDGMENT
This work was supported by the Mid-career Researcher Program through an NRF grant (2011-0015429) funded by the MEST.
Footnotes
Published ahead of print 15 May 2013
REFERENCES
- 1. Culbertson CG. 1970. Pathogenic Naegleria and Hartmannella (Acanthamoeba). Ann. N. Y. Acad. Sci. 74:1018–1022 [DOI] [PubMed] [Google Scholar]
- 2. Carter RF. 1968. Primary amoebic meningoencephalitis: clinical, pathological and epidemiological features of six fatal cases. J. Pathol. Bacteriol. 96:1–25 [DOI] [PubMed] [Google Scholar]
- 3. Marciano-Cabral F, Cabral GA. 2007. The immune response to Naegleria fowleri amebae and pathogenesis of infection. FEMS Immunol. Med. Microbiol. 51:243–259 [DOI] [PubMed] [Google Scholar]
- 4. Visvesvara GS, Moura H, Schuster FL. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 50:1–26 [DOI] [PubMed] [Google Scholar]
- 5. Carter RF. 1972. Primary amoebic meningo-encephalitis. An appraisal of present knowledge. Trans. R. Soc. Trop. Med. Hyg. 66:193–213 [DOI] [PubMed] [Google Scholar]
- 6. Ma P, Visvesvara GS, Martinez AJ, Theodore FH, Daggett PM, Sawyer TK. 1990. Naegleria and Acanthamoeba infections: review. Rev. Infect. Dis. 12:490–513 [DOI] [PubMed] [Google Scholar]
- 7. Anderson K, Jamieson A. 1972. Primary amoebic meningoencephalitis. Lancet ii:379. [DOI] [PubMed] [Google Scholar]
- 8. Seidel JS, Harmatz P, Visvesvara GS, Cohen A, Edwards J, Turner J. 1982. Successful treatment of primary amebic meningoencephalitis. N. Engl. J. Med. 306:346–348 [DOI] [PubMed] [Google Scholar]
- 9. Wang A, Kay R, Poon WS, Ng HK. 1993. Successful treatment of amoebic meningoencephalitis in a Chinese living in Hong Kong. Clin. Neurol. Neurosurg. 95:249–252 [DOI] [PubMed] [Google Scholar]
- 10. Proffitt RT, Satorius A, Chiang SM, Sullivan L, Adler-Moore JP. 1991. Pharmacology and toxicology of a liposomal formulation of amphotericin B (AmBisome) in rodents. J. Antimicrob. Chemother. 28:49–61 [DOI] [PubMed] [Google Scholar]
- 11. Stevens AR, Shulman ST, Lansen TA, Cichon MJ, Willaert E. 1981. Primary amoebic meningoencephalitis: a report of two cases and antibiotic and immunologic studies. J. Infect. Dis. 143:193–199 [DOI] [PubMed] [Google Scholar]
- 12. Shin HJ, Cho MS, Jung SU, Kim HI, Park S, Kim HJ, Im KI. 2001. Molecular cloning and characterization of a gene encoding a 13.1 kDa antigenic protein of Naegleria fowleri. J. Eukaryot. Microbiol. 48:713–717 [DOI] [PubMed] [Google Scholar]
- 13. Cho MS, Jung SY, Park S, Kim KH, Kim HI, Sohn S, Kim HJ, Im KI, Shin HJ. 2003. Immunological characterizations of a cloned 13.1-kilodalton protein from pathogenic Naegleria fowleri. Clin. Diagn. Lab. Immunol. 10:954–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang SY, Song KJ, Jeong SR, Kim JH, Park S, Kim K, Kwon MH, Shin HJ. 2005. Role of the Nfa1 protein in pathogenic Naegleria fowleri co-cultured with CHO target cells. Clin. Diagn. Lab. Immunol. 12:873–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeong SY, Kang SY, Lee SC, Song KJ, Im KI, Shin HJ. 2004. Decreasing effect of an anti-Nfa1 polyclonal antibody on the in vitro cytotoxicity of pathogenic Naegleria fowleri. Korean J. Parasitol. 42:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. 1990. Direct gene transfer into mouse muscle in vivo. Science 247:1465–1468 [DOI] [PubMed] [Google Scholar]
- 17. Mascola JR, Nabel GJ. 2001. Vaccines for the prevention of HIV-1 disease. Curr. Opin. Immunol. 13:489–495 [DOI] [PubMed] [Google Scholar]
- 18. Mustafa AS. 2001. Biotechnology in the development of new vaccines and diagnostic reagents against tuberculosis. Curr. Pharm. Biotechnol. 2:157–173 [DOI] [PubMed] [Google Scholar]
- 19. Robinson HL. 1997. Nucleic acid vaccines: an overview. Vaccine 15:785–787 [DOI] [PubMed] [Google Scholar]
- 20. Arce F, Rowe HM, Chain B, Lopes L, Collins MK. 2009. Lentiviral vectors transduce proliferating dendritic cell precursors leading to persistent antigen presentation and immunization. Mol. Ther. 17:1643–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 22. Buffa V, Negri DR, Leone P, Borghi M, Bona R, Michelini Z, Compagnoni D, Sgadari C, Ensoli B, Cara A. 2006. Evaluation of a self inactivating lentiviral vector expressing simian immunodeficiency virus gag for induction of specific immune responses in vitro and in vivo. Viral Immunol. 19:690–701 [DOI] [PubMed] [Google Scholar]
- 23. Breckpot K, Emeagi PU, Thielemans K. 2008. Lentiviral vectors for anti-tumor immunotherapy. Curr. Gene Ther. 8:438–448 [DOI] [PubMed] [Google Scholar]
- 24. Thong YH, Ferrante A, Rowan-Kelly B, O'Keefe D. 1980. Immunization with live amoebae, amoebic lysate and culture supernatant in experimental Naegleria meningoencephalitis. Trans. R. Soc. Trop. Med. Hyg. 74:570–576 [DOI] [PubMed] [Google Scholar]
- 25. Thong YH, Shepherd C, Ferrante A, Rowan-Kelly B. 1978. Protective immunity to Naegleria fowleri in experimental amebic meningoencephalitis. Am. J. Trop. Med. Hyg. 27:238–240 [DOI] [PubMed] [Google Scholar]
- 26. Kim JH, Lee SH, Sohn HJ, Lee J, Chwae YJ, Park S, Kim K, Shin HJ. 2012. The immune response induced by DNA vaccine expressing nfa1 gene against Naegleria fowleri. Parasitol. Res. 111:2377–2384 [DOI] [PubMed] [Google Scholar]
- 27. Willaert E. 1971. Isolation and in vitro culture of the amoeba of the genus Naegleria. Ann. Soc. Belges Med. Trop. Parasitol. Mycol. 51:701–708 [PubMed] [Google Scholar]
- 28. Lee YJ, Kim JH, Jeong SR, Song KJ, Kim K, Park S, Park MS, Shin HJ. 2007. Production of Nfa1-specific monoclonal antibodies that influences the in vitro cytotoxicity of Naegleria fowleri trophozoites on microglial cells. Parasitol. Res. 101:1191–1196 [DOI] [PubMed] [Google Scholar]
- 29. Lee YJ, Kim JH, Sohn HJ, Lee J, Jung SY, Chwae YJ, Kim K, Park S, Shin HJ. 2011. Effects of immunization with the rNfa1 protein on experimental Naegleria fowleri-PAM mice. Parasite Immunol. 33:382–389 [DOI] [PubMed] [Google Scholar]
- 30. Gurunathan S, Wu C, Freidag BL, Seder RA. 2000. DNA vaccines: a key for inducing long-term cellular immunity. Curr. Opin. Immunol. 12:442–447 [DOI] [PubMed] [Google Scholar]
- 31. Ahmed SB, Touihri L, Chtourou Y, Dellagi K, Bahloul C. 2009. DNA based vaccination with a cocktail of plasmids encoding immunodominant Leishmania (Leishmania) major antigens confers full protection in BALB/c mice. Vaccine 27:99–106 [DOI] [PubMed] [Google Scholar]
- 32. Dai Y, Zhu Y, Harn DA, Wang X, Tang J, Zhao S, Lu F, Guan X. 2009. DNA vaccination by electroporation and boosting with recombinant proteins enhances the efficacy of DNA vaccines for Schistosomiasis japonica. Clin. Vaccine Immunol. 16:1796–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoseinian Khosroshahi K, Ghaffarifar F, D'Souza S, Sharifi Z, Dalimi A. 2011. Evaluation of the immune response induced by DNA vaccine cocktail expressing complete SAG1 and ROP2 genes against toxoplasmosis. Vaccine 29:778–783 [DOI] [PubMed] [Google Scholar]
- 34. Martínez MB, Rodríguez MA, García-Rivera G, Sánchez T, Hernández-Pando R, Aguilar D, Orozco E. 2009. A pcDNA-Ehcpadh vaccine against Entamoeba histolytica elicits a protective Th1-like response in hamster liver. Vaccine 27:4176–4186 [DOI] [PubMed] [Google Scholar]
- 35. Negri DR, Michelini Z, Bona R, Blasi M, Filati P, Leone P, Rossi A, Franco M, Cara A. 2011. Integrase-defective lentiviral-vector-based vaccine: a new vector for induction of T cell immunity. Expert Opin. Biol. Ther. 11:739–750 [DOI] [PubMed] [Google Scholar]


