Abstract
The MAT1-1 and MAT1-2 idiomorphs associated with the MAT1 locus of Histoplasma capsulatum were identified by PCR. A total of 28 fungal isolates, 6 isolates from human clinical samples and 22 isolates from environmental (infected bat and contaminated soil) samples, were studied. Among the 14 isolates from Mexico, 71.4% (95% confidence interval [95% CI], 48.3% to 94.5%) were of the MAT1-2 genotype, whereas 100% of the isolates from Brazil were of the MAT1-1 genotype. Each MAT1 idiomorphic region was sequenced and aligned, using the sequences of the G-217B (+ mating type) and G-186AR (− mating type) strains as references. BLASTn analyses of the MAT1-1 and MAT1-2 sequences studied correlated with their respective + and − mating type genotypes. Trees were generated by the maximum likelihood (ML) method to search for similarity among isolates of each MAT1 idiomorph. All MAT1-1 isolates originated from Brazilian bats formed a well-defined group; three isolates from Mexico, the G-217B strain, and a subgroup encompassing all soil-derived isolates and two clinical isolates from Brazil formed a second group; last, one isolate (EH-696P) from a migratory bat captured in Mexico formed a third group of the MAT1-1 genotype. The MAT1-2 idiomorph formed two groups, one of which included two H. capsulatum isolates from infected bats that were closely related to the G-186AR strain. The other group was formed by two human isolates and six isolates from infected bats. Concatenated ML trees, with internal transcribed spacer 1 (ITS1) -5.8S-ITS2 and MAT1-1 or MAT1-2 sequences, support the relatedness of MAT1-1 or MAT1-2 isolates. H. capsulatum mating types were associated with the geographical origin of the isolates, and all isolates from Brazil correlated with their environmental sources.
INTRODUCTION
Histoplasma capsulatum is a heterothallic ascomycete that has an anamorphic or asexual stage with two types of sexual compatibility, + and −, represented at the mating locus (MAT1) by the idiomorphic regions MAT1-1 and MAT1-2, respectively. The teleomorphic (sexual) stage that results from + and − mating was first described as Emmonsiella capsulata by Kwon-Chung (1–4). Nowadays it is known as Ajellomyces capsulatus, which temporarily exhibits the dikaryotic and diploid phases that form haploid ascospores after two meiotic reductions. Thus, the species H. capsulatum and A. capsulatus constitute the same holomorphic organism. The classical studies of sexual compatibility in H. capsulatum were performed by mating fungal specimens in culture plates. However, this procedure is difficult because H. capsulatum isolates rapidly lose the ability to mate in vitro (5); therefore, molecular methods were developed to identify the mating type in this microorganism (6–8).
To date, there have been a few relevant studies in the United States about the use of genetic tools to determine sexual compatibility in H. capsulatum, in which the − mating type predominates (6–8). Recent findings have involved the product of the Velvet A gene (VeA), which belongs to the proteins of the Velvet family, in mating structure formation (cleistothecial) and virulence of H. capsulatum (9). H. capsulatum isolates exhibit a wide distribution and an important genetic diversity, as has been documented in Latin America (10–16). However, the frequency and genetic diversity of the + and − mating types are not well documented in most countries where H. capsulatum is found, and data reported in the United States are not necessarily representative of other geographical areas.
In the present work, we studied indigenous H. capsulatum isolates from Mexico (North America) and Brazil (South America) to determine the frequency and genetic diversity of the sexual compatibility types of H. capsulatum in these two distant geographical areas. Most of the isolates studied were obtained from naturally infected bats and contaminated soils, although some isolates from human clinical cases were also analyzed. PCR was used to identify and to perform genetic analyses of the MAT1-1 and MAT1-2 idiomorphs. The internal transcribed spacer 1 (ITS1)-5.8S-ITS2 region of the H. capsulatum isolates of each MAT1 idiomorph was used for concatenated phylogenetic analyses of both genomic regions to increase the genetic relatedness of H. capsulatum isolates from different mating types. Therefore, our contribution is original, mainly for its frequency data and because it is the first one to use the MAT1 locus as a geographical marker for H. capsulatum.
MATERIALS AND METHODS
Histoplasma capsulatum.
We studied 28 Histoplasma capsulatum isolates (14 from Mexico and 14 from Brazil) obtained from different infectious sources (Table 1). The isolates were maintained in the yeast phase by culture at 37°C in brain heart infusion broth (Bioxon; Becton, Dickinson, Mexico City, Mexico) supplemented with 0.1% l-cysteine and 1% glucose.
Table 1.
General data and sexual compatibility of the H. capsulatum isolates studied
| Isolate | Associated clinical forma | Sourceb | Geographical originc | Mating typed |
|---|---|---|---|---|
| EH-46 | D | Human (liver) | GR-Mexico | − |
| EH-53 | D | Human (blood) | HG-Mexico | − |
| EH-317 | D/HIV+ | Human (blood) | MS-Mexico | + |
| EH-315 | NA | Mormoops megalophylla (gut) | GR-Mexico | + |
| EH-373 | NA | Artibeus hirsutus (lung) | MS-Mexico | − |
| EH-374 | NA | Artibeus hirsutus (spleen) | MS-Mexico | − |
| EH-376 | NA | Artibeus hirsutus (lung) | MS-Mexico | − |
| EH-378 | NA | Artibeus hirsutus (lung) | MS-Mexico | − |
| EH-391 | NA | Leptonycteris nivalis (liver) | MS-Mexico | − |
| EH-405 | NA | Leptonycteris nivalis (lung) | PL-Mexico | − |
| EH-449B | NA | Leptonycteris nivalis (spleen) | MS-Mexico | − |
| EH-521 | NA | Artibeus hirsutus (lung) | MS-Mexico | + |
| EH-672H | NA | Tadarida brasiliensis (liver) | HG-Mexico | − |
| EH-696P | NA | Tadarida brasiliensis (lung) | NL-Mexico | + |
| 18H | D/HIV+ | Human (blood) | RJ-Brazil | + |
| 37307 | D/HIV+ | Human (bone marrow) | RJ-Brazil | + |
| 247BL | ND | Human (ND) | MS-Brazil | + |
| M396/08 | NA | Molossus molossus (NR) | SP-Brazil | + |
| M1084/08 | NA | Molossus molossus (NR) | SP-Brazil | + |
| M487/08 | NA | Molossus molossus (NR) | SP-Brazil | + |
| M975/08 | NA | Molossus molossus (NR) | SP-Brazil | + |
| AC05 | NA | Soil | RJ-Brazil | + |
| TI01 | NA | Soil | RJ-Brazil | + |
| IGS19 | NA | Soil | RJ-Brazil | + |
| RPS51 | NA | Soil | RJ-Brazil | + |
| CO2 | NA | Soil | RJ-Brazil | + |
| CO4 | NA | Soil | RJ-Brazil | + |
| IgS4/5 | NA | Soil | RJ-Brazil | + |
Abbreviations: D, Disseminated histoplasmosis; HIV+, human immunodeficiency virus positive; NA, not applicable; ND, not determined.
ND, not determined; NR, not registered.
The state and country are shown. The country is shown after the hyphen, and the state is shown before the hyphen as follows: in Mexico, GR, Guerrero; HG, Hidalgo; MS-Mexico, Morelos, Mexico; PL, Puebla; NL, Nuevo León; in Brazil, RJ, Rio de Janeiro; MS-Brazil, Mato Grosso do Sul, Brazil; SP, São Paulo.
The + mating type has the MAT1-1 idiomorphic region, and the − mating type has the MAT1-2 idiomorphic region.
Identification of sexual compatibility types.
We followed the protocols for yeast DNA extraction and the processing of PCR products of Bubnick and Smulian (6) with minor modifications. We used two sets of primers designed for the MAT1 locus: (i) for the MAT1-1 sequence, MAT1-1S (5′-CGTGGTTAGTTACGGAGGCA-3′) and MAT1-1AS (5′-TGAGGATGCGAGTGATGGGA-3′), which generated an amplicon of 440 bp; and (ii) for the MAT1-2 sequence, MAT1-2S (5′-ACACAGTAGCCCAACCTCTC-3′) and MAT1-2AS (5′-TCGACAATCCCATCCAATACCG-3′), which generated an amplicon of 528 bp. The PCR was performed in a 25-μl reaction mixture, containing 200 μM each deoxynucleoside triphosphate (dNTP) (Applied Biosystems Inc., Foster City, CA, USA), 1.5 mM MgCl2, 50 ng/μl of each primer, 2.5 U Taq DNA polymerase (New England BioLabs Inc., MA, USA), 1× Taq commercial buffer, and 75 ng/μl of each DNA sample.
PCR assays were performed in a Thermal iCycler (Bio-Rad Laboratories Inc., Hercules, CA, USA) programmed as follows: (i) 3 min at 95°C; (ii) 35 cycles, consisting of 30 s at 95°C, 30 s at 58°C, and 1 min 30 s at 72°C; and (iii) 10 min at 72°C. The PCR products were resolved by 1.5% agarose gel electrophoresis under the same conditions described by Bubnick and Smulian (6). The 100-bp DNA ladder was used as a molecular marker.
ITS1-5.8S-ITS2 PCR.
The PCR assay was performed by the method of Muniz et al. (13), using the following primers: ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′), which generated an amplicon of 607 bp.
Sequencing.
The resolved PCR products were purified using the Montage PCR centrifugal filter devices kit (Millipore Corporation, Bedford, MA, USA). The purified products were sent to the Molecular Biology Unit of the Cellular Physiology Institute, Universidad Nacional Autónoma de México (UNAM)-Mexico, for sequencing in an ABI automated apparatus (Applied Biosystems). Sequencing was performed for both DNA strands. The generated consensus MAT1-1 and MAT1-2 or ITS1-5.8S-ITS2 sequences for each isolate are deposited in GenBank or in the Fungi Barcode of Life Database (Bold System) and their available accession numbers are shown in Tables 2 and 3.
Table 2.
Database accession numbers of H. capsulatum MAT1 sequences analyzed
| Isolate | GenBank accession no. |
|---|---|
| MAT1-1 isolates | |
| EH-696P | KC282441 |
| EH-315 | KC282442 |
| EH-317 | KC282443 |
| EH-521 | KC282444 |
| 37307 | KC282445 |
| 18H | KC282446 |
| RPS51 | KC282447 |
| IGS19 | KC282448 |
| AC05 | KC282449 |
| IgS4/5 | KC282450 |
| TI01 | KC282451 |
| CO2 | KC282452 |
| CO4 | KC282453 |
| 247BL | KC282454 |
| M396/08 | KC282455 |
| M975/08 | KC282456 |
| M487/08 | KC282457 |
| M1084/08 | KC282458 |
| MAT1-2 isolates | |
| EH-46 | KC282431 |
| EH-53 | KC282432 |
| EH-373 | KC282433 |
| EH-374 | KC282434 |
| EH-376 | KC282435 |
| EH-378 | KC282436 |
| EH-672H | KC282437 |
| EH-391 | KC282438 |
| EH-449B | KC282439 |
| EH-405 | KC282440 |
Table 3.
Database accession numbers of H. capsulatum ITS1-5.8S-ITS2 sequences analyzed
| Isolate | Accession no.a |
|---|---|
| Mexican isolates | |
| EH-46 | HIST019-13 |
| EH-53 | HIST001-13 |
| EH-315 | HIST002-13 |
| EH-317 | HIST003-13 |
| EH-373 | HIST004-13 |
| EH-374 | HIST020-13 |
| EH-376 | HIST021-13 |
| EH-378 | HIST022-13 |
| EH-391 | HIST006-13 |
| EH-449B | HIST025-13 |
| EH-521 | HIST026-13 |
| EH-672H | HIST029-13 |
| EH-696P | HIST018-13 |
| Brazilian isolates | |
| IgS4/5 | GU320945.1 |
| TI01 | GU320964.1 |
| CO2 | KF114466 |
| CO4 | KF114465 |
| 37307 | KF114464 |
| 247BL | KF114463 |
| 18H | KF114471 |
| M396/08 | KF114467 |
| RPS51 | GU320962.1 |
| M975/08 | KF114469 |
| IGS19 | GU320944.1 |
| M487/08 | KF114468 |
| AC05 | GU320980.1 |
| M1084/08 | KF114470 |
The Mexican isolates were deposited in the Fungi Barcode of Life Database (Bold System), and the Brazilian isolates were deposited in GenBank.
Genetic analyses.
Sequences were aligned with Clustal-W in MEGA version 5.0 (MEGA-5) (http://www.megasoftware.net) and edited manually.
The sequences of the MAT1-1 and MAT1-2 idiomorphs from all the H. capsulatum isolates studied were compared by searching the GenBank database for homologous nucleotide sequences with the BLASTn algorithm. The sequence of the G-217B strain from Louisiana-United States, ATCC 22636 (GenBank accession number EF433757), was used as a reference for the MAT1-1 idiomorphic region. The sequence of the G-186AR strain from Panama, ATCC 22635 (GenBank accession number EF433756), was used as a reference for the MAT1-2 idiomorphic region.
The aligned sequences were submitted to evolutionary analyses to assume the similarity or divergence among isolates of each mating type, using the maximum likelihood (ML) method. Concatenated ML trees, with ITS1-5.8S-ITS2 and MAT1-1 or MAT1-2 sequences, were processed to provide more-robust phylogenetic data. Unrooted ML trees were constructed in MEGA-5, based on the Hasegawa-Kishino-Yano (HKY) model (17) for MAT1 idiomorphs and Tamura-Nei (TrN) model for concatenated analyses (18). Gaps and missing data were eliminated. A bootstrapping algorithm was implemented on the data set for 1,000 replicates. The highest bootstrap values were registered in each node of each ML tree.
Statistics.
The MAT1-1 and MAT1-2 idiomorph frequencies were estimated in relation to each mating genotype, taking into account all H. capsulatum isolates from Mexico or Brazil studied. In regard to frequency data, the 95% confidence interval (95% CI) was calculated by normal distribution.
RESULTS AND DISCUSSION
Previous studies of the clinical and environmental H. capsulatum strains in the United States using conventional mating tests with the fungal mycelial phase reported a higher frequency of the − mating type in clinical strains than in strains isolated from soil samples (5, 19). However, the predominance of the − genotype in clinical strains in the United States, even if supported by a large number of H. capsulatum samples from different sources as indicated by Kwon-Chung et al. (5, 19), is not necessarily true in other geographical areas. Although the above-published data have emphasized interesting findings about the disequilibrium of the − and + mating types in human clinical isolates associated with the clinical form of the disease (5, 6, 19), these findings may not apply to clinical isolates from other geographical areas, since H. capsulatum isolates exhibit a wide distribution and a significant genetic diversity as has been documented in Latin America (11). On the basis of our present results, it was impossible to infer a disequilibrium of − and + mating types due to the small number of human clinical isolates studied, and in regard to the environmental isolates, we did not find any evidence of disequilibrium.
Currently, DNA-based mating studies have been used to evaluate the distribution of the sexual compatibility types of Ophiostoma quercus in different geographical areas (20). According to this antecedent, in the present paper, by PCR of the MAT1 locus, we determined the mating types of 6 autochthonous clinical isolates and 22 native environmental isolates (infected bats and contaminated soil) of H. capsulatum from two distant regions within the Americas (Mexico and Brazil) (Table 1). The MAT1-2 genotype was predominant in Mexico, representing 71.4% (95% CI, 48.3 to 94.5%) of the isolates, whereas 100% of the isolates originating in Brazil were of the MAT1-1 genotype.
Hence, the frequencies of H. capsulatum mating types were mainly associated with the geographical origin of the isolates and most likely correlate with their environmental sources.
Given that the − mating type is more widely distributed in the H. capsulatum isolates from the United States and Mexico (North America) and the + mating type is more frequent in Brazil (South America), as revealed by the present results, we suggest that the different mating types of H. capsulatum are distinctively spread across the American continent. In addition, the presence of two sexual compatibility types in the same geographical region, albeit unequally distributed, suggest that genetic dispersion of the MAT1 locus in the environment could be associated with natural reservoirs of H. capsulatum.
For the MAT1-1 idiomorph, 18 sequences from nucleotide (nt) 778 to 1171 were analyzed, whereas 10 sequences from nt 3038 to 3,584 were analyzed for the MAT1-2 idiomorph. The sequences obtained for each idiomorph were aligned with the matching sequences of the H. capsulatum reference strains obtained from GenBank, G-217B (MAT1-1) or G-186AR (MAT1-2).
Based on the known sequence of reference strain G-217B, the sequences of the MAT1-1 idiomorph region (14 from Brazilian H. capsulatum isolates and four from Mexican H. capsulatum isolates) were aligned by using MEGA-5, showing four noninformative sites common to all isolates (Table 4). In the MAT1-1 genotype, four informative sites stand out, these sites were shared by nine isolates (seven isolates from soil samples and two isolates from human clinical samples) from Rio de Janeiro, Brazil. In addition, one informative site was shared by four isolates recovered from Molossus molossus bats from São Paulo State and one human isolate from Mato Grosso do Sul State, Brazil (Table 4).
Table 4.
Variable sites within the MAT1-1 idiomorph sequences of H. capsulatum
| Variable site in the MAT1-1 idiomorph sequence |
| Noninformative sites common to all MAT1-1 isolates |
| Transitions |
| Guanine to adenine at nt 807, 813, and 822 |
| Cytosine to thymine at nt 856 |
| Informative sites for nine isolates from RJ, Brazila |
| Transitions |
| Cytosine to thymine at nt 782 and 1127 |
| Thymine to cytosine at nt 865 |
| Guanine to adenine at nt 1023 |
| Informative sites for five isolates (four from SP, Brazil, and one from MS, Brazil)a |
| Transition |
| Guanine to adenine at nt 1000 |
RJ, Rio de Janeiro; SP, São Paulo; MS, Mato Grosso do Sul.
Two isolates from Mexico (EH-317 from a clinical case and EH-315 from an infected bat) with the MAT1-1 genotype exhibited the same mutations, a transition (cytosine to thymine at nt 940) and a transversion (guanine to thymine at nt 955). The Mexican H. capsulatum isolate EH-521 was the most similar to reference strain G-217B, whereas the H. capsulatum isolate EH-696P, from the migratory Tadarida brasiliensis bat captured in Mexico, was the most divergent from all MAT1-1 H. capsulatum isolates studied (data not shown).
The MAT1-2 sequences of the 10 Mexican isolates were compared to the MAT1-2 sequence of reference strain G-186AR (GenBank) using MEGA-5, showing six noninformative sites common to all isolates (Table 5). In addition, eight isolates from Morelos State, Mexico, shared three transitions and four transversions (Table 5). However, four out of these eight H. capsulatum isolates, which were recovered from bats captured in the same cave of Morelos, diverge from this group by the absence of mutations between nt 3510 and 3513 (data not shown).
Table 5.
Variable sites within the MAT1-2 idiomorph sequences of H. capsulatum
| Variable site in the MAT1-2 idiomorph sequence |
| Noninformative sites common to all MAT1-2 isolates |
| Transitions |
| Cytosine to thymine at nt 3098 and 3248 |
| Adenine to guanine at nt 3125 and 3285 |
| Thymine to cytosine at nt 3200 |
| Transversion |
| Thymine to adenine at nt 3068 |
| Informative sites for eight isolates from MS, Mexicoa |
| Transitions |
| Thymine to cytosine at nt 3156 |
| Guanine to adenine at nt 3284 |
| Adenine to guanine at nt 3449 |
| Transversions |
| Thymine to adenine at nt 3130 and 3212 |
| Cytosine to adenine at nt 3163 and 3188 |
MS, Morelos.
Two informative sites (both transversions, adenine to thymine at nt 3052 and guanine to thymine at nt 3480) were found only in isolates EH-374 and EH-672H (data not shown).
Alignment analysis showed that isolate EH-696P from Mexico, which had been recovered from a bat captured at the northeastern Mexican border, exhibited the greatest number of point mutations in its MAT1-1 sequence, whereas the EH-374 and EH-672H isolates showed fewer mutations among the MAT1-2 sequences (data not shown).
BLASTn analysis of nucleotide sequences of the MAT1-1 idiomorph region of 18 H. capsulatum isolates demonstrated that five of the Brazilian isolates exhibited 99% similarity to the sequence of reference strain G-217B. The fact that these isolates came from a circumscribed geographical region in Brazil, that four of the isolates (M396/08, M487/08, M975/08, and M1084/08) were recovered from M. molossus bats from São Paulo and one isolate (247BL) came from a human clinical sample from Mato Grosso do Sul State, which borders São Paulo State, suggest that an unusual MAT1-1 genotype is prevalent in this particular region. Likewise, nine Brazilian isolates from Rio de Janeiro (seven isolates from soil, isolates AC05, CO2, CO4, IgS4/5, IGS19, RPS51, and TI01, and two isolates from human samples, isolates 18H and 37307) exhibited 98% similarity. Three Mexican isolates (EH-315, EH-317, and EH-521) showed 98% similarity and the Mexican EH-696P isolate from the migratory bat T. brasiliensis was detected to have 96% similarity to the G-217B reference strain. The BLASTn algorithm search for similarities among sequences of the MAT1-1 genotype revealed only a very low similarity of 6.8% with the gene sequence of a hypothetical protein of the ascomycete Pyrenophora teres f. sp. teres, which supports the close relationship of all MAT1-1 H. capsulatum isolates studied.
BLASTn analysis of 10 sequences from Mexican H. capsulatum isolates of the MAT1-2 idiomorph region revealed their high similarity with the sequence of reference strain G-186AR. This reference sequence shared 98% similarity with sequences of two isolates from different bat species (EH-374 and EH-672H). The other eight Mexican isolates from the central zone of the country (EH-46 and EH-53 from human clinical samples; EH-373, EH-376, EH-378, EH-391, EH-405, and EH-449B from different bat species) showed 97% similarity with the sequence of the G-186AR reference strain. The BLASTn algorithm search for similarities among MAT1-2 genotype sequences showed 49.1% similarity with the gene sequence of a predicted protein, with a high-mobility-group (HMG) DNA binding domain, of the fungal pathogen Paracoccidioides brasiliensis (clone 60855 isolate C4-PS3) and 44.4% similarity with the gene sequence of a protein, with an HMG DNA binding domain, of Ajellomyces dermatitidis (strain SLH14081). These data also confirm the relationship among all MAT1-2 H. capsulatum isolates studied.
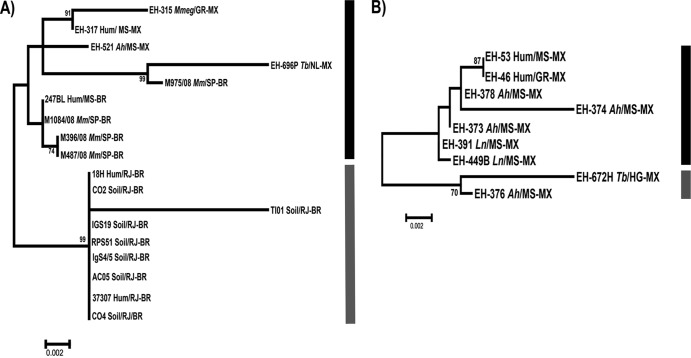
The ML analysis of the sequences of the MAT1-1 idiomorph demonstrated that our Brazilian isolates from infected bats captured in São Paulo State (Table 1) and the human clinical isolate from Mato Grosso do Sul, Brazil, constitute a well-defined group, representing a probable clonal population of H. capsulatum. In addition, seven H. capsulatum isolates from contaminated soil and two clinical isolates, all from Rio de Janeiro, Brazil (Table 1), formed a distinct subgroup that shared a probably clonal MAT1-1 genotype, which is in agreement with previous data using different molecular markers published by Muniz et al. (12, 13). This subgroup clustered with three MAT1-1 isolates from Mexico and the G-217B reference strain. The EH-696P isolate from a migratory bat captured in Mexico constituted a third group of the MAT1-1 genotype and showed the largest genetic distance to all the H. capsulatum isolates with the MAT1-1 genotype studied (Fig. 1A).
Fig 1.
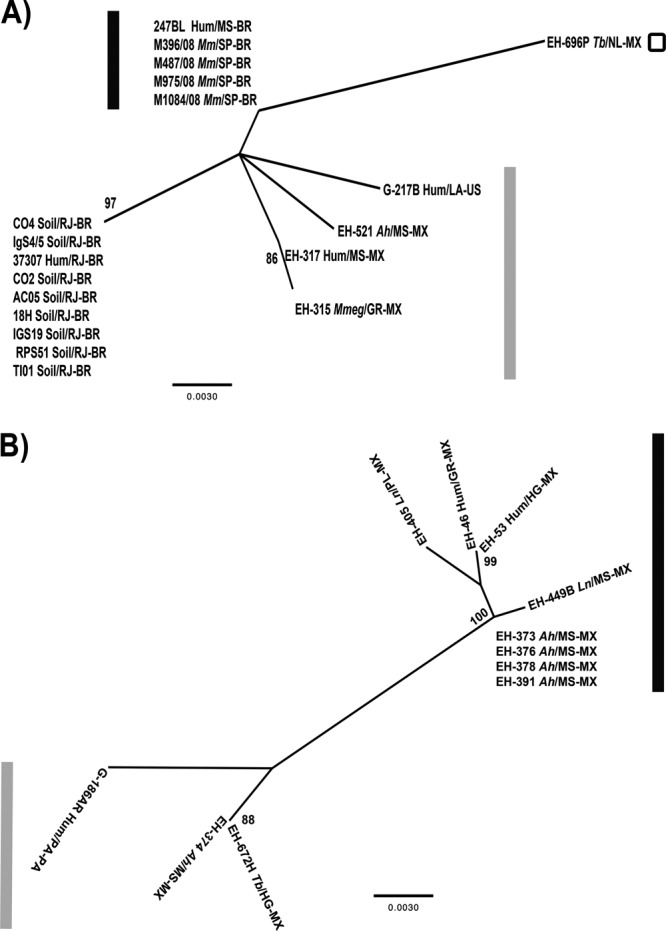
Maximum likelihood trees for the MAT1 locus of the H. capsulatum isolates studied. (A) MAT1-1 tree. (B) MAT1-2 tree. The ML analysis was based on the HKY model. The trees were generated by 1,000 replications, as outlined in Materials and Methods. The bootstrap values that were ≥70% are shown at the nodes. The G-217B (MAT1-1) and G-186AR (MAT1-2) sequences were obtained from the GenBank database and were used as reference strains. The black and gray bars indicate the different isolate groups. The isolates are named by their biological and geographical sources. The source (soil or biological) of the isolate is shown before the slash as follows: Hum, human; Ah, Artibeus hirsutus; Ln, Leptonycteris nivalis; Mm, Molossus molossus; Mmeg, Mormoops megalophylla; Tb, Tadarida brasiliensis. The geographical source of the isolate is shown after the slash. The state is shown after the slash and before the hyphen as follows: GR, Guerrero; HG, Hidalgo; LA, Louisiana; MS, Morelos (Mexico); MS, Mato Grosso do Sul (Brazil); NL, Nuevo León; PL, Puebla; PA, Panama; RJ, Rio de Janeiro; SP, São Paulo. The country is shown after the hyphen (Brazil [BR], Mexico [MX], United States [US]).
In contrast, ML analysis of the MAT1-2 idiomorph, which included most sequences from Mexican isolates, demonstrated that these sequences could be categorized into two major groups. The first group was formed by two isolates from bats (EH-374 and EH-672H) and the G-186AR reference strain (Fig. 1B). The other group was formed by two human clinical isolates and six isolates from infected bats, four of which (EH-373, EH-376, EH-378, and EH-391) belong to a probably clonal population of H. capsulatum, as reported by Kasuga et al. (11).
Our results reveal a preferential distribution of H. capsulatum isolates with respect to the sequences of the idiomorphic regions of the MAT1 locus, encompassing two distant geographical areas of the Americas, Mexico and Brazil. Moreover, our findings demonstrated a close relationship between the fungal isolation source and geographical origin within Brazil and also in the central states of Mexico (Guerrero, Morelos, Puebla, and Hidalgo), where most of the Mexican samples were isolated.
Regarding the population structure of the H. capsulatum isolates studied, some of the Mexican isolates classified by Kasuga et al. (11) were characterized as possibly recombinant, and others, such as environmental isolates from the Morelos State of Mexico, were characterized as a clonal population (11, 14–16).
Undoubtedly, understanding the H. capsulatum mating type distribution in areas of the Americas with a high prevalence of histoplasmosis would contribute to our knowledge of the H. capsulatum genetic plasticity generated by sexual recombination events, which could eventually occur under environmental conditions.
Results of concatenated analyses, using ITS1-5.8S-ITS2 region and each MAT1 idiomorph, support the relatedness of MAT1-1 or MAT1-2 isolates with robust data (Fig. 2A and B, respectively). In addition, ML concatenated analyses generated similar tree topologies as Fig. 1A and B, despite the fact that only two major groups were found. In these analyses, G-217B and G-186AR reference strains were not included because their ITS1-5.8S-ITS2 sequences were not available in different databases. The ITS1-5.8S-ITS2 sequence of the Mexican EH-405 isolate was not obtained due to the loss of this isolate and its DNA.
Fig 2.
Concatenated maximum likelihood trees for the ITS1-5.8S-ITS2 region and each MAT1 locus of the H. capsulatum isolates studied. (A) ITS1-5.8S-ITS2 and MAT1-1 concatenated tree. (B) ITS1-5.8S-ITS2 and MAT1-2 concatenated tree. The ML analysis was based on the TrN model. The trees were generated by 1,000 replications, as outlined in Materials and Methods. The bootstrap values that were ≥70% are shown at the nodes. The black and dark gray bars indicate the different isolate groups. For abbreviations, see the legend to Fig. 1.
In conclusion, knowledge regarding the distribution of the MAT1 locus in H. capsulatum and its genetic diversity should contribute to a better understanding of the biology of this fungus and the actual impact of its sexual compatibility genes distributed in natural conditions. We emphasize that in this paper we incorporated two completely new aspects. (i) We studied isolates from two very distant geographical areas to use the MAT1 locus as a geographical marker. (ii) Most of the H. capsulatum isolates studied came from natural sources (wild infected bats), which makes our contribution unique as a result of its geographical frequency data.
ACKNOWLEDGMENTS
This study was funded by the PAPIIT-DGAPA project from UNAM (IN204210), the Bilateral Collaboration Agreement between CONACYT-Mexico (B330/099/11) and CNPq-Brazil (490110/2009-6), the Bilateral Collaboration Agreement between UNAM-Mexico (15090-563-24-V-04) and UNESP-Brazil (000528/04/01/2005), and the FAPERJ E-26/111.532/2010. R.M.Z.-O. is supported in part by CNPq 350338/2000-0. C.N.D.S. is supported by INOVATEC FIOCRUZ/FAPERJ. T.V.-G. thanks CONACYT for scholarship 231985.
M.-L.T. thanks Ingrid Mascher for editorial assistance.
Rosely M. Zancopé-Oliveira and Maria-Lucia Taylor participated in the design and coordination of the study.
Footnotes
Published ahead of print 24 May 2013
REFERENCES
- 1. Kwon-Chung KJ. 1972. Sexual stage of Histoplasma capsulatum. Science 175: 326. [DOI] [PubMed] [Google Scholar]
- 2. Kwon-Chung KJ. 1972. Emmonsiella capsulata: perfect state of Histoplasma capsulatum. Science 177:368– 369 [DOI] [PubMed] [Google Scholar]
- 3. Kwon-Chung KJ. 1973. Studies on Emmonsiella capsulata. I. Heterothallism and development of the ascocarp. Mycologia 65: 109– 121 [PubMed] [Google Scholar]
- 4. Kwon-Chung KJ, Bennett JE. 1992. Medical mycology. Lea and Febiger, Philadelphia, PA [Google Scholar]
- 5. Kwon-Chung KJ, Weeks RJ, Larsh HW. 1974. Studies on Emmonsiella capsulata (Histoplasma capsulatum). II. Distribution of the two mating types in 13 endemic states of the United States. Am. J. Epidemiol. 99: 44– 49 [DOI] [PubMed] [Google Scholar]
- 6. Bubnick M, Smulian AG. 2007. The MAT1 locus of Histoplasma capsulatum is responsive in a mating type-specific manner. Eukaryot. Cell 6: 616– 621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fraser JA, Stajich JE, Tarcha EJ, Cole GT, Inglis DO, Sil A, Heitman J. 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6: 622– 629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laskowski MC, Smulian AG. 2010. Insertional mutagenesis enables cleistothecial formation in a non-mating strain of Histoplasma capsulatum. BMC Microbiol. 10: 49– 64. 10.1186/1471-2180-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laskowski-Peak MC, Calvo AM, Rohrssen J, Smulian AG. 2012. VEA1 is required for cleistothecial formation and virulence in Histoplasma capsulatum. Fungal Genet. Biol. 49: 838– 846 [DOI] [PubMed] [Google Scholar]
- 10. Kasuga T, Taylor JW, White TJ. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 37: 653– 663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kasuga T, White TJ, Koenig G, McEwen J, Restrepo A, Castañeda E, Da Silva-Lacaz C, Heins-Vaccari EM, De Freitas RS, Zancopé-Oliveira RM, Qin Z, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW. 2003. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol. Ecol. 12: 3383– 3401 [DOI] [PubMed] [Google Scholar]
- 12. Muniz MM, Pizzini CV, Peralta JM, Reiss E, Zancope-Oliveira RM. 2001. Genetic diversity of Histoplasma capsulatum strains isolated from soil, animals, and clinical specimens in Rio de Janeiro State, Brazil, by a PCR-based random amplified polymorphic DNA assay. J. Clin. Microbiol. 39: 4487– 4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muniz MM, Morais e Silva Tavares P, Meyer W, Nosanchuk JD, Zancope-Oliveira RM. 2010. Comparison of different DNA-based methods for molecular typing of Histoplasma capsulatum. Appl. Environ. Microbiol. 76: 4438– 4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taylor ML, Chávez-Tapia CB, Reyes-Montes MR. 2000. Molecular typing of Histoplasma capsulatum isolated from infected bats, captured in Mexico. Fungal Genet. Biol. 30: 207– 212 [DOI] [PubMed] [Google Scholar]
- 15. Taylor ML, Chávez-Tapia CB, Rojas-Martínez A, Reyes-Montes MR, Bobadilla del Valle M, Zuñiga G. 2005. Geographical distribution of genetic polymorphism of the pathogen Histoplasma capsulatum isolated from infected bats, captured in a central zone of Mexico. FEMS Immunol. Med. Microbiol. 45: 451– 458 [DOI] [PubMed] [Google Scholar]
- 16. Taylor ML, Hernández-García L, Estrada-Bárcenas D, Salas-Lizana R, Zancopé-Oliveira RM, García de La Cruz S, Galvão-Dias MA, Curiel-Quesada E, Canteros CE, Bojórquez-Torres G, Bogard-Fuentes CA, Zamora-Tehozol E. 2012. Genetic diversity of Histoplasma capsulatum isolated from infected bats randomly captured in Mexico, Brazil and Argentina, using the polymorphism of (GA)n microsatellite and its flanking regions. Fungal Biol. 1l6: 308– 317 [DOI] [PubMed] [Google Scholar]
- 17. Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160– 174 [DOI] [PubMed] [Google Scholar]
- 18. Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512– 526 [DOI] [PubMed] [Google Scholar]
- 19. Kwon-Chung KJ, Bartlett MS, Wheat LJ. 1984. Distribution of the two mating types among Histoplasma capsulatum isolates obtained from an urban histoplasmosis outbreak. Sabouraudia 22: 155– 157 [PubMed] [Google Scholar]
- 20. Wilken PM, Steenkamp ET, Hall TA, De Beer ZW, Wingfield MJ, Wingfield BD. 2012. Both mating types in the heterothallic fungus Ophiostoma quercus contain MAT1-1 and MAT1-2 genes. Fungal Biol. 116: 427– 437 [DOI] [PubMed] [Google Scholar]