Abstract
Biological motors are molecular nanomachines, which convert chemical energy into mechanical forces. The combination of mechanoenzymes with structural components, such as the cytoskeleton, enables eukaryotic cells to overcome entropy, generate molecular gradients, and establish polarity. Hyphae of filamentous fungi are among the most polarized cells, and polarity defects are most obvious. Here, we studied the role of the kinesin-3 motor, NKIN2, in Neurospora crassa. We found that NKIN2 localizes as fast-moving spots in the cytoplasm of mature hyphae. To test whether the spots represented early endosomes, the Rab5 GTPase YPT52 was used as an endosomal marker. NKIN2 colocalized with YPT52. Deletion of nkin2 caused strongly reduced endosomal movement. Combined, these results confirm the involvement of NKIN2 in early endosome transport. Introduction of a rigor mutation into NKIN2 labeled with green fluorescent protein (GFP) resulted in decoration of microtubules. Interestingly, NKIN2rigor was associated with a subpopulation of microtubules, as had been shown earlier for the Aspergillus nidulans orthologue UncA. Other kinesins did not show this specificity.
INTRODUCTION
Cell shape, polarity, intracellular movements, and differentiation in eukaryotic cells depend on a complex network of filaments and associated proteins that constitute the cytoskeleton. Microtubules (MTs), with a diameter of 25 nm, are characterized by dynamic instability. This inherent instability almost contradicts their structural function. Microtubules rather constitute a sophisticated network providing tracks for molecular motors, which in turn distribute many cellular components. Since biological motors are ATP-driven molecular nanomachines, they provide a great possibility to break the symmetry in a cell, generate molecular gradients, and give rise to polarity. Eukaryotic cells generally contain a number of different motor proteins: myosins for the transport along actin microfilaments, one cytoplasmic dynein for microtubule minus-end-directed movement, and several kinesins for microtubule plus-end-directed transportation. After the first kinesin had been discovered in squid axons, a large number of kinesins have been characterized in many organisms. Kinesins share a conserved motor domain, where ATP binding and hydrolysis as well as microtubule binding take place. They have meanwhile been grouped into 14 different families (1).
The first kinesin-3 family member discovered in Caenorhabditis elegans was Unc104, which transports synaptic vesicles (2). Kinesin-3 motor proteins contain two characteristic FHA (forkhead-associated) and PH (pleckstrin homology) domains, in addition to the strongly conserved N-terminal motor domain. FHA domains are involved in phosphorylation-dependent protein-protein interactions and signaling (3, 4), while the PH domain was shown to enable specific binding to phosphatidylinositol-4,5-bisphosphate in membranes (5).
In the basidiomycetous fungus Ustilago maydis, it was shown that kinesin-3 (Kin3) is required for anterograde endosome movement (6–8), whereas retrograde movement of endosomes is accomplished by dynein (7, 9). Deletion of the U. maydis kin-3 gene resulted in reduced endosome motility to 33% and abolished endosome clustering at the cell pole and at septa. Schuchardt et al. also presented evidence that Kin3 is involved in exocytosis, because acid phosphatase secretion was lowered to 50% in kin-3 deletion strains (10). An additional role of Kin3 in mRNA transport has been demonstrated recently (11, 12). Furthermore, deletion of the RNA-binding protein Rrm4 resulted in changes of the expression pattern of many genes. Among 5 identified mRNAs, three of them encode mitochondrial proteins, suggesting delivery of such mRNAs toward mitochondria (13). Koepke et al. suggested that mitochondrial protein import depends on proper mRNA localization. In agreement with the function in U. maydis, Aspergillus nidulans kinesin-3 (UncA) also transports endosomes anterograde toward the hyphal tip (14). The Neurospora crassa kinesin-3, named NKIN2, appeared to be associated with mitochondria and plays some role in mitochondrial distribution (15). In addition to the described kinesin-3 proteins, many fungi contain a truncated kinesin-3, which lacks the FHA and PH domains (16). Although the structure of the protein is very different, it is very interesting that in N. crassa this kinesin (named NKIN3) could rescue the lack of nkin2 (15).
Conventional kinesin, the first discovered kinesin, constitutes the kinesin-1 family. This motor is involved in a number of different transport processes ranging from vesicle transport in axons to nuclear migration and vacuole distribution in fungi (17–19). The continuous delivery of vesicles to the growing tip in filamentous fungi is very important for growth (20). The incorporation of the vesicles into the cytoplasmic membrane is necessary for the expansion. In addition, the vesicles deliver enzymes required for cell wall synthesis. Before the final delivery of the vesicles to the membrane, they accumulate in a structure called the Spitzenkörper, which functions as a vesicle supply center (20, 21). Deletion of nkin in N. crassa results in several morphological defects of the hyphae, such as a reduced growth rate and increased branching. Likewise, the anterograde transport of secretory vesicles to the hyphal tip is strongly reduced, and thus exoenzyme secretion is decreased (17). One remarkable property of conventional kinesin is the ability to transport dynein toward the microtubule's plus end (22). This links the function of conventional kinesin to endosome motility and distribution. If dynein is not delivered to the hyphal tip region, retrograde transport of endosomes is affected and they accumulate thus at the tip (22, 23).
Microtubule-dependent transport processes are thus numerous and highly sophisticated. The level of complexity however, is not only due to the high number of different motor proteins along with their specific cargos, but also to the regulation of the microtubules itself. Microtubule plus-end tracking proteins (+TIPs) at the tips together with many microtubule-associated proteins (MAPS) at the lattice of the microtubule filaments regulate the dynamics as well as the interaction with cortical proteins (24, 25). In addition, tubulin can be posttranslationally modified, and thus microtubules with specific functions may be generated. Such posttranslational modifications (PTMs) comprise, for instance, phosphorylation, acetylation, polyglutamylation, or the cleavage of the C-terminal tyrosine (26). Some of these modifications are already conserved in primitive eukaryotes, such as Giardia lamblia, suggesting that they arose early during eukaryotic evolution (27). There is good evidence that molecular motors may use PTMs as traffic signs. Recently it was shown that in rat neurons detyrosination of tubulin in axons navigates kinesin-1 specifically to axons and not to dendrites (28). In A. nidulans, we have evidence for the presence of detyrosinated microtubules and for a preference of kinesin-3 for detyrosinated microtubules (14). Recent data indicated that an 86-amino-acid (aa)-long stretch in the tail of UncA is required for the discrimination between different microtubules (29).
Here we show that the kinesin-3 motor functions and the microtubule specificity are evolutionarily conserved between A. nidulans and N. crassa.
MATERIALS AND METHODS
Strains and culture conditions.
The bacterial and N. crassa strains used or generated in this study are described in Table 1. The standard laboratory Escherichia coli strain Top 10 F′ was used. The N. crassa strains generated during this study were derived from FGSC 9717 (mat A his-3 Δmus-51::bar+), which is deficient in the nonhomologous end-joining process (30). The strains were routinely grown at 28°C in Vogel's minimal medium (VMM) supplemented with 1.5% sucrose and 1.5% agar. When needed, 200 μg/ml hygromycin B, 25 μg/ml nourseothricin, and/or 0.5 mg/ml histidine were added for selection. Transformations were performed by electroporation as described previously (31). Transformed conidia were plated on VMM-FGS (0.5% fructose, 0.5% glucose, 20% sorbose) and selected for histidine prototrophy or hygromycin B or nourseothricin resistance. The expression of the fusion proteins was examined in at least 10 transformants by wide-field fluorescence microscopy. For growth analyses, N. crassa strains were grown on VVM agar plates at 28°C. Microscopy of all N. crassa strains was performed at room temperature.
Table 1.
Plasmids and N. crassa strains used in this study
| Plasmid or strain | Description or genotype | Source or reference |
|---|---|---|
| Plasmids | ||
| pCCG::N-GFP | Pccg-1::c-Gly::GFP, 5′Δhis-3 flank, Ampr | 34 |
| pJV16-N | Pccg-1::mChFP, 5′Δhis-3 flank, Ampr | 33 |
| ve2gfp | Pgpd::natR | A. Dettmann |
| pCoS136 | Pccg-1::mChFP::nkin2, 5′Δhis-3 flank, Ampr | This study |
| pCoS139 | Pccg-1::mChFP::nkin2rigor, 5′Δhis-3 flank, Ampr | This study |
| pCoS172 | Pccg-1::c-Gly::GFP::nkin2, 5′Δhis-3 flank, Ampr | This study |
| pCoS173 | Pccg-1::c-Gly::GFP::nkin2rigor, 5′Δhis-3 flank, Ampr | This study |
| pCoS182 | Pccg-1::c-Gly::GFP::nkin, 5′Δhis-3 flank, Ampr | This study |
| pCoS183 | Pccg-1::c-Gly::GFP::nkinrigor, 5′Δhis-3 flank, Ampr | This study |
| pCoS204 | Pccg-1::c-Gly::GFP::ypt52, 5′Δhis-3 flank, Ampr | This study |
| pCoS212 | Pccg-1::c-Gly::GFP::nkin3, 5′Δhis-3 flank, Ampr | This study |
| pCoS113 | Pccg-1::c-Gly::GFP::nkin3rigor, 5′Δhis-3 flank, Ampr | This study |
| Strains | ||
| N9717 | mat A his-3; Δmus-51::bar+ | FGSC |
| N11722 | mat a Δnkin2 heterokaryon; hph+ | FGSC |
| N2524 | ridRIP4 mat a his-3+; Pccg-1::bml::GFP | 48 |
| N3618 | mat A ro-10 (mutant of dynactin subunit); hph+ | 43 |
| NCoS1 | mat A his-3+::Pccg-1::mChFP::nkin2rigor Δmus-51::bar+ | This study |
| NCoS2 | mat A his-3+::Pccg-1::mChFP::nkin2 Δmus-51::bar+ | This study |
| NCoS3 | mat A his-3+::Pccg-1::c-Gly::GFP::nkin2 Δmus-51::bar+ | This study |
| NCoS4 | mat A his-3+::Pccg-1::c-Gly::GFP::nkin2rigor Δmus-51::bar+ | This study |
| NCoS5 | mat A his-3+::Pccg-1::c-Gly::GFP::nkin Δmus-51::bar+ | This study |
| NCoS6 | mat A his-3+::Pccg-1::c-Gly::GFP::nkinrigor Δmus-51::bar+ | This study |
| NCoS7 | mat A his-3+::Pccg-1::c-Gly::GFP::ypt52 Δmus-51::bar+ | This study |
| NCoS8 | mat A his-3; Δnkin2::hph+ Δmus-51::bar+ | This study |
| NCoS10 | mat A his-3+::Pccg-1::c-Gly::GFP::nkin3 Δmus-51::bar+ | This study |
| NCoS11 | mat A his-3+::Pccg-1::c-Gly::GFP::nkin3rigor Δmus-51::bar+ | This study |
| NCoS12 | mat A his-3+::Pccg-1::c-Gly::GFP::ypt52 Δnkin2::hph+ Δmus-51::bar+ | This study |
| NCoS13 | mat A his-3+::Pccg-1::c-Gly::GFP::nkin2 Δnkin2::hph+ Δmus-51::bar+ | This study |
| NCoS14 | mat A his-3+::Pccg-1::c-Gly::GFP::nkin2rigor Δnkin2::hph+ Δmus-51::bar+ | This study |
| NCoS15 | mat A Pccg-1::c-Gly::GFP::ypt52::natR Δro-10::hph+ | This study |
Molecular techniques.
For PCR experiments, standard protocols were applied using a Biometra personal cycler (Biometra, Göttingen, Germany) for the reaction cycles. DNA sequencing was done commercially (Eurofins MWG Operon, Ebersberg, Germany). Genomic DNA was extracted from N. crassa with a DNeasy plant minikit (Qiagen, Hilden, Germany). Southern blots were performed as described in reference 32.
Plasmid construction and fusion PCR.
Standard PCR and cloning procedures were used to fuse green fluorescent protein (GFP) and mCherry fluorescent protein (mChFP) N terminally using plasmids pCCG::N-GFP and pJV16-N, respectively, for targeted integration into the his-3 locus of N. crassa strain FGSC 9717 (33, 34). The full-length 5.77-kb open reading frame (ORF) of nkin2 (kinesin-3; NCU06733) was amplified by PCR from FGSC 9717 genomic DNA using primers Nkin2_PacI_fwd (5′-CTTAATTAACATGGCTCCAGGCATGGC-3′) and Nkin2_XbaI_rev (5′-GCTCTAGATCAAAACCTCAACTGACTG-3′), which included PacI and XbaI restriction sites (which are underlined). The amplified PCR product was directly digested and subsequently cloned into the corresponding restriction sites of pCCG::N-GFP (for GFP tagging) and in pJV16-N (for mChFP tagging), yielding Pccg-1::c-Gly::GFP::nkin2 (pCoS 172) and Pccg-1::mChFP::nkin2 (pCoS 136). The same procedure was performed for the nkin full-length 2.99-kb ORF (kinesin-1; NCU09730) using the primers Nkin_PacI_fwd (5′-GTTAATTAACATGTCGTCAAGTGCGAATAG-3′) and Nkin_XbaI_rev (5′-CTCTAGATTACGACTTCTGGAAGAACC-3′), yielding plasmid pCoS182 [Pccg-1::C-Gly::GFP::nkin], and for the nkin3 2.02-kb ORF (kinesin-3; NCU03715) by using the primers Nkin3_PacI_fwd (5′-GTTAATTAACATGCCGAACTCCCTCGAC-3′) and Nkin3_XbaI_rev (5′-CTCTAGATCATATGATATTAGTCCTCAC-3′), yielding plasmid pCoS212. The ypt52 0.83-kb open reading frame (NCU06410) was amplified using the primers Ypt52_PacI_fwd (5′-GTTAATTAACATGGCCGATACGAACGCG-3′) and Ypt52_XbaI_rev (5′-CTCTAGATTAACAGGCGCAACCTTCC-3′); this yielded plasmid pCoS204 [Pccg-1::c-Gly::GFP::ypt52].
To transform the ro-10 mutant with a Pccg-1::GFP::ypt52::natR cassette, fusion PCR was performed. First the Pccg-1::GFP::ypt52 cassette was amplified using the primers ccg1(p)_fwd (5′-GCCAATGGTCTAATACACCG-3′) and Ypt52_linker_rev (5′-atccacttaacgttactgaaatcTTAACAGGCGCAACCTTCC-3′) with pCoS204 as the template (linker arms are indicated in lowercase letters). The natR cassette was amplified from the plasmid ve2gfp using the primers Nat_fwd (5′-gatttcagtaacgttaagtggatGGCGGCTCTGAGGTGCAGTGG-3′) and Nat_rev (5′-gacagaagatgatattgaaggagTCAGGGGCAGGGCATGCTCATG-3′). The final fusion PCR to fuse both cassettes was performed with TaKaRa LA Taq DNA polymerase (Clontech, Heidelberg, Germany) using the primers ccg-1_nested_fwd (5′-CTTGATTGACAGCGAACGAAACCC-3′) and Nat_nested_rev (5′-GCGCCTGCTCGCCGTCCG-3′). The purified PCR fragment (innuPREP PCRpure kit; Analytic Jena, Jena, Germany) was transformed into the ro-10 mutant by electroporation.
Insertion of mutations by site-directed mutagenesis.
To introduce the rigor mutation into the P-loop of the kinesins, a QuikChange XL mutagenesis kit (Stratagene, Heidelberg, Germany) was used. To generate the G115E mutation in NKIN2, G93E in NKIN and T146P and G149E in NKIN3, the primers Nkin2_rigor_p-loop_fwd (5′-GGTCAAACCGGTTCCGAGAAGTCCTACTCG-3′) and Nkin2_rigor_p-loop_rev (5′-CGAGTAGGACTTCTCGGAACCGGTTTGACC-3′), Nkin_rigor_p-loop_fwd (5′-CGGCCAGACGGGTGCCGAGAAATCATACAC-3′) and Nkin_rigor_p-loop_rev (5′-GTGTATGATTTCTCGGCACCCGTCTGGCCG-3′), and Nkin3_rigor_p-loop_fwd (5′-GGTCAGCCTGGCTCGGAGAAGTCTTATAC-3′) and Nkin3_rigor_p-loop_rev (5′-GTATAAGACTTCTCCGAGCCAGGCTGACC-3′) (the mutation sites are underlined) were used, yielding plasmids pCoS139, pCoS173, pCoS183, and pCoS213.
All plasmids were verified by sequencing, used for transformation, and incorporated by homologous integration into the his-3 locus of N. crassa strain FGSC 9717.
Deletion of nkin2.
The flanking regions of nkin2 were amplified by PCR with genomic DNA of N. crassa strain FGSC 9717 using the primers DNkin2_P1_fwd (5′-CGAGCTGTGATGTTGTCAGACATC-3′) and DNkin2_P2_rev (5′-ATCCACTTAACGTTACTGAAATCGGTGGCGCGAGATAGCTCTAACG-3′) for the upstream region of nkin2 and DNkin2_P3_fwd (5′-CTCCTTCAATATCATCTTCTGTCGGAAGGGATGGGATATGATGATG-3′) and DNkin2_P4_rev (5′-CGGAGATTATTATTACTCTATAGCGC-3′) for the downstream region (with linkers indicated by underlined nucleotides). The hph gene (hygromycin B phosphotransferase) was amplified with the primers hph_fwd (5′-GACAGAAGATGATATTGAAGGAGC-3′) and hph_rev (5′-GATTTCAGTAACGTTAAGTGG AT-3′) using genomic DNA of strain FGSC 11722 (FGSC heterokaryon of nkin2 deletion) as the template. The final deletion cassette was amplified by fusion PCR using the primers DNkin2_P7_fwd (5′-CAGAATCTAGCACAGGGCATGC-3′) and DNkin2_P8_rev (5′-CTAAGGAGTAGGACCTATATATAGG-3′); this product was directly transformed into N. crassa FGSC 9717.
In each case, putative transformants were screened by PCR for the homologous integration event. Single homologous integration of the construct and the homokaryon condition were confirmed by Southern blotting (see Fig. S1 in the supplemental material). Two Δnkin2 strains were selected from the transformants and named NCoS8a and NCoS8b. The coupling of the observed phenotypes with the gene deletion events was confirmed by complementation of the nkin2 deletion phenotype with GFP-NKIN2 (pCoS172).
Microscopy and image processing.
For live-cell imaging, conidia of 3- to 5-day-old N. crassa cultures were isolated and grown on petri dishes containing solid VMM plus supplements for 8 to 15 h at 28°C. Preparation of the samples for imaging was performed by using the “inverted agar block method” (35). To analyze the in vivo colocalization, we forced formation of heterokaryons on VMM agar plates; two different N. crassa strains of the same mating type were inoculated side by side and grown overnight at 28°C. Images were captured at room temperature with an Axio Imager Z1 microscope (Carl Zeiss, Jena, Germany) using 63× Plan Apochromat 1.4 oil and 63× Plan-Neofluar 100× 1.3 oil objectives. Exposure times ranged from 200 to 400 ms. Images were collected and analyzed with the AxioVision system (Carl Zeiss) and further processed with Adobe Photoshop CS (version 8.0) and ImageJ. Dynamic processes in the hyphae were quantified using the same software analyzing series of single pictures. Additionally, a Leica SP5 laser-scanning microscope (Leica, Wetzlar, Germany) was used for high-resolution imaging.
Fluorescent dyes.
For staining of endocytic organelles, the steryl dye N-3-triethylammoniumpropyl-4-p-diethylaminophenylhexatrienyl-pyridinium-dibromide (FM4-64; Invitrogen, Karlsruhe, Germany) was used at a final concentration of 25 μM. To stain the vacuoles, 7-amino-4-chloromethylcoumarin (CMAC) (CellTracker Blue, Life Technologies, Regensburg, Germany) was used at a final concentration of 10 μM. For staining of the cell wall, calcofluor white (Sigma-Aldrich, Taufkirchen, Germany) was used at a final concentration of 25 μM. MitoTracker Red (Life Technologies, Regensburg, Germany) was used at a final concentration of 20 μM to stain mitochondria. For each dye, the agar blocks with hyphae were inverted onto coverslips with 10 μl of the diluted working stock. Stained cells were imaged after recovery.
RESULTS
Localization of NKIN2 in different developmental stages of N. crassa.
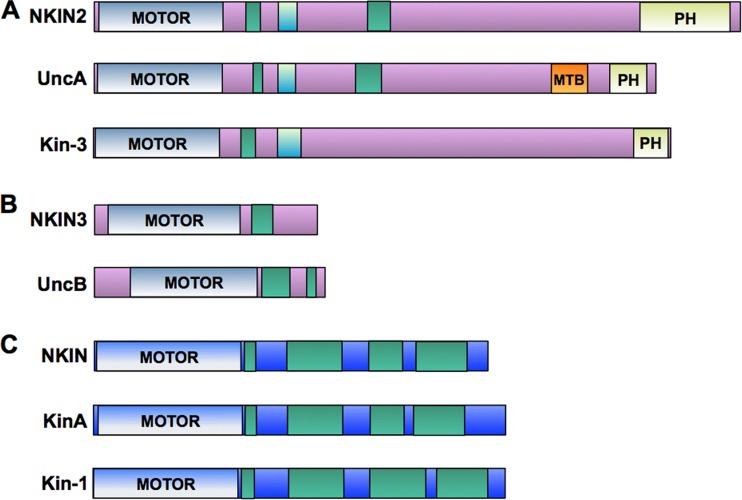
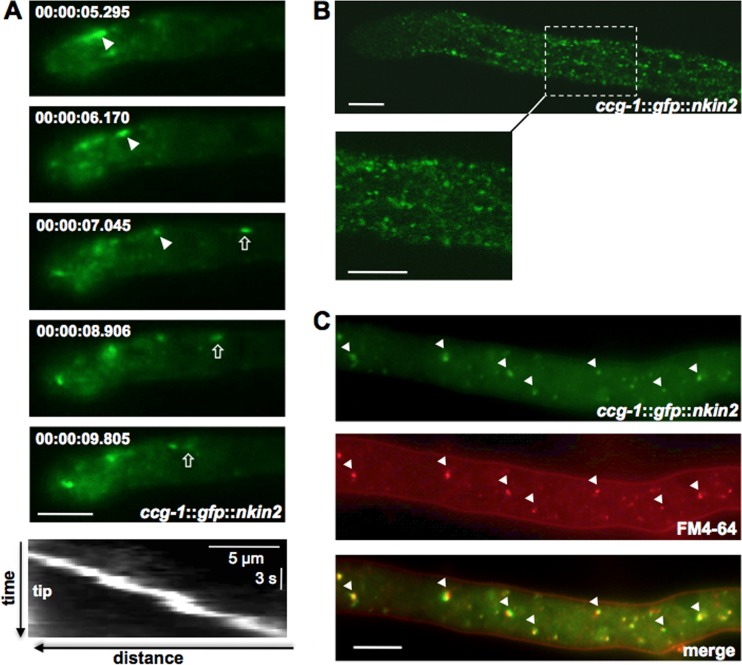
N. crassa contains two kinesin-3 motors (NKIN2 and NKIN3) and one conventional kinesin (NKIN). This resembles the situation in A. nidulans. In comparison, U. maydis does not contain a second kinesin-3 protein (Fig. 1). As a first step to analyze the molecular function of the N. crassa kinesin-3, NKIN2 (NCU06733), we visualized it by N-terminal tagging with GFP or mChFP through targeted homologous integration of the constructs into the his-3 locus of N. crassa strain FGSC 9717. Expression of the constructs was driven from the ccg-1 promoter. Although we cannot exclude that the use of this promoter does not reflect the natural expression level, the tagged motor expressed under the control of ccg-1 was sufficient to complement a Δnkin2 strain. The restored wild-type phenotype indicates the functionality of the construct (see Fig. 6A below). Because there was some earlier evidence that NKIN2 is associated with mitochondria, we expected colocalization of the motor with these organelles. Instead, we observed a spot-like distribution of NKIN2 (Fig. 2). The spots did not resemble mitochondria (see below) but rather vesicles decorated with the motor. The localization pattern was analyzed throughout mycelial development, starting from a germinating spore to differentiated, branched hyphae and to the different types of spores. In young germlings, NKIN2 appeared as bidirectionally moving spots with occasional accumulations at hyphal tips (Fig. 2A). A similar localization pattern was observed in lateral branches (see Movie S1 in the supplemental material). In leading hyphae, the behavior of NKIN2 was slightly different. Due to fast cytoplasmic flow and the high growth rate of these hyphae, a differentiation between active transport and passive movement was difficult, but retrograde movement was frequently observed (Fig. 2B; see Movie S2 in the supplemental material). A prominent accumulation at the tip, like in young germlings and lateral branches, was never observed. Motile spots, which accumulated to some extent at the tips, were also observed in arthrospores, whereas conidia did not show any accumulation of NKIN2 at the poles but also showed rapid movement of NKIN2 (data not shown). The speed of spots varied significantly, ranging from 0.7 to 2.6 μm/s (average, 1.3 ± 0.5 μm; n = 20). Differences in the velocities of anterograde- or retrograde-directed movement were not observed. Measurement of the top speed of NKIN2 at the tips of leading hyphae through tracking of individual vesicles was very difficult, due to the high number of spots. Pausing of the spots during long distance movement was frequently observed. The determined speed of NKIN2 is slightly lower than the corresponding values for the UncA motor in A. nidulans (up to 4 μm/s) and the Kin3 motor in U. maydis (2.87 ± 0.19 μm/s) (6, 14). Riquelme et al. observed that the vesicle traffic of chitin synthase CHS-3- and CHS-6-labeled vesicles (chitosomes) in N. crassa exhibits a velocity of 0.09 to 0.4 μm/s (36), while CHS-1-marked vesicles show speeds of 3.8 ± 0.9 μm/s (37).
Fig 1.
Scheme comparing the domains of kinesin-1 and kinesin-3 in N. crassa, A. nidulans, and U. maydis. (A and B) Kinesin-3 homologues of the two ascomycetes N. crassa (NKIN2 and NKIN3) and A. nidulans (UncA and UncB) and in the basidiomycete U. maydis (Kin-3). An NKIN3 homologue is missing in U. maydis. (C) Kinesin-1 homologues in N. crassa (NKIN), A. nidulans (KinA), and U. maydis (Kin-1).
Fig 6.
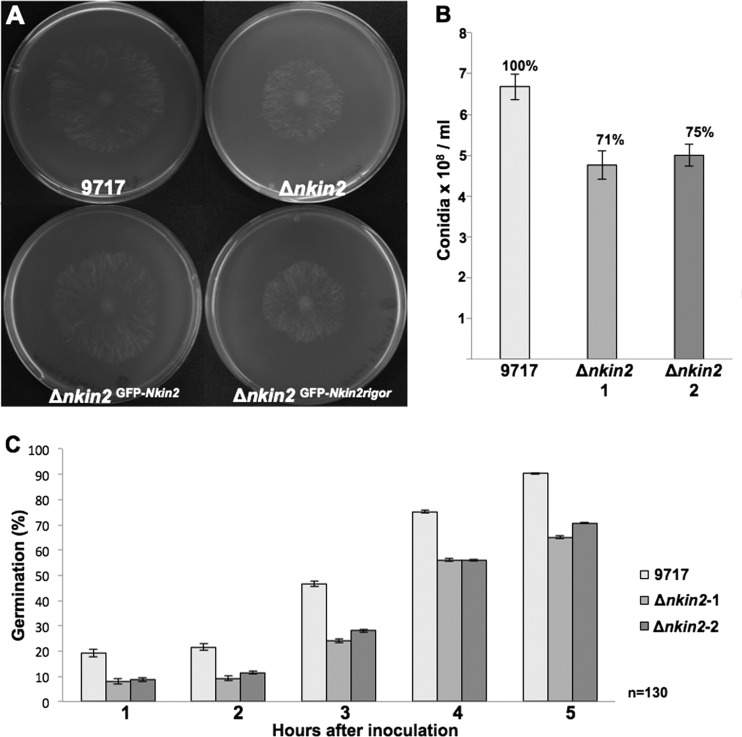
Deletion of nkin2. (A) Growth comparison of the wild-type strain and Δnkin2 and Δnkin2 strains complemented with Pccg-1::GFP::nkin2 and Pccg-1::GFP::nkin2rigor. (B) Quantification of conidial production in both nkin2 deletion strains. Ten microliters of a cell suspension of 3.6 × 106 conidia/ml was plated on VMM plus histidine, the mixture was incubated for 72 h in light at 28°C, and the conidia were counted. (C) Comparison of germination rates. A total of 1.5 × 106 conidia were inoculated on VMM plus histidine, and 130 cells per time point and strain were counted.
Fig 2.
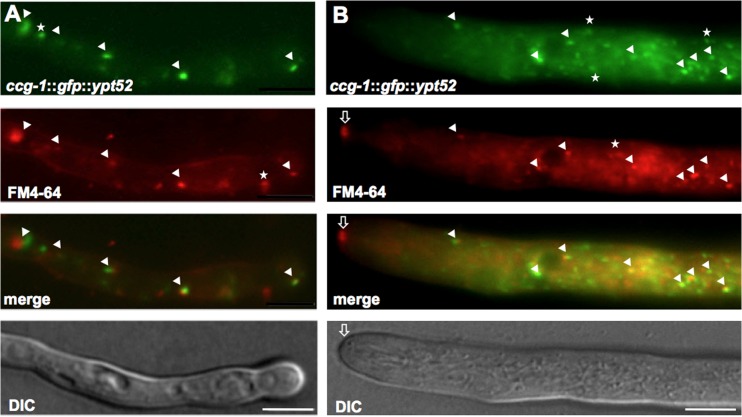
Movement of NKIN2 in germlings, lateral branches, and leading hyphae of N. crassa. (A) GFP-NKIN2 localizes in germlings to small, fast anterograde (arrows)- and retrograde (arrowheads)-moving spots, which sometimes accumulate at the hyphal tip. A kymograph (bottom panel) shows the movement of one mChFP-NKIN2 spot toward the tip of a branch. (B) Leading hyphae show an increased number of spots that moved faster. The movement is so fast that single spots sometimes appear as short rods. Accumulations at the apex were never observed. (C) Colocalization of GFP-NKIN2 and FM4-64 in a leading hypha confirms that NKIN2 is associated with endosomes (arrowheads). Scale bars, 5 μm.
To investigate if NKIN2 is associated with endosomes and the observed spots thus represent NKIN2-decorated endosomes, colocalization studies with the lipophilic dye FM4-64 were performed. Because of the high velocities of GFP-NKIN2 at the tip of the leading hyphae, the colocalization experiments were performed slightly distal to the apex. Indeed, NKIN2 colocalized with the FM4-64-stained organelles (Fig. 2C). Due to the rapid movement of the spots, a small shift of the two fluorescent signals was commonly observed. To get further evidence that these spots represent endosomes, an endosomal marker was chosen for further analysis.
NKIN2 transports early endosomes.
The S. cerevisiae small Rab5 GTPase Ypt52 is a well-accepted endosomal marker involved in clathrin-dependent endocytosis (38–40). Therefore, Ypt52 was used to analyze the N. crassa genome for an orthologue. The predicted protein NCU06410 displayed 48% identity to Saccharomyces cerevisiae Ypt52 and groups together with A. nidulans RabA (see Fig. S2 in the supplemental material). N. crassa YPT52 is a Rab5 GTPase composed of 237 amino acids with a conserved Rab domain. In S. cerevisiae, it was shown that Ypt52 and the two other closely related Rab5s, Ypt51 and Ypt53, are involved in endocytosis and vacuolar protein sorting (41). RabA, the A. nidulans Rab5 homologue, was shown to bind to early endosomes (38). We named the gene ypt52.
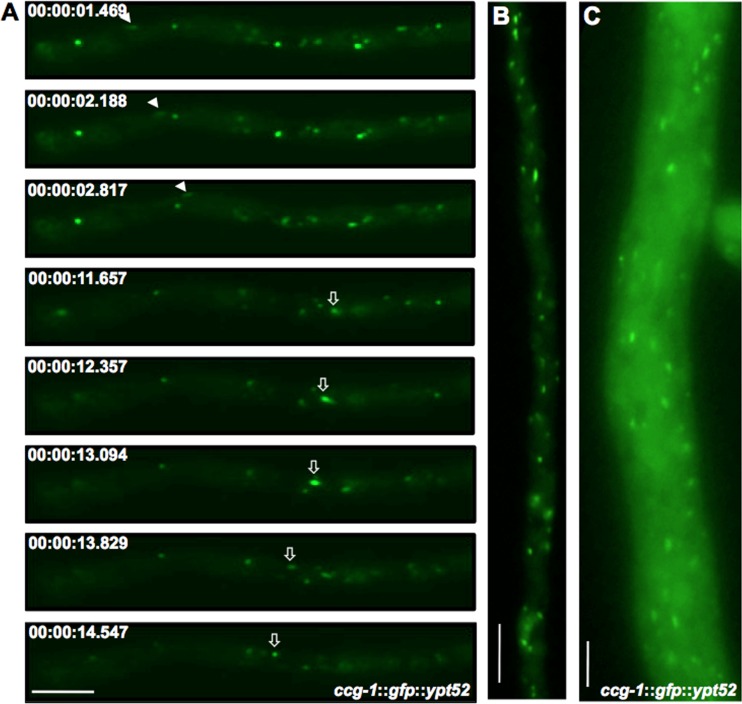
YPT52 was N-terminally tagged with GFP and expressed from the ccg-1 promoter. YPT52 localized to bidirectionally moving spots in germlings and young hyphae (Fig. 3A and B; see Movie S3 in the supplemental material). In leading hyphae, a stronger cytoplasmic background of GFP signals and considerably more GFP-YPT52-labeled spots were observed, which also moved bidirectionally within the hyphal compartments (Fig. 3C). FM4-64 stained the Ypt52-labeled spots, although 31% of the vesicles were stained with either FM4-64 or GFP-YPT52 (Fig. 4, asterisks). These results indicate that YPT52 does not bind to all endosomes and may also specifically bind to early endosomes as was shown for its homologues RabA in A. nidulans and Ypt51 in S. cerevisiae (38, 41). In order to show that NKIN2 is involved in endosome transport, a strategy applied earlier in A. nidulans was followed, namely, the inactivation of the NKIN2 motor through the introduction of a rigor mutation into the motor domain. This causes strong binding of the motor to the microtubules and leads to decoration of the tracks with a bound cargo. We introduced the rigor mutation into the P-loop of the NKIN2 motor domain and fused the full-length motor protein N terminally with mChFP, placed it under the control of the ccg-1 promoter, and integrated it into the his-3 locus of FGSC 9717. In order to coexpress mChFP-NKIN2rigor and GFP-YPT52, two strains that harbor the corresponding single modifications were cocultured to allow hyphal fusions. Heterokaryotic hyphae showed both GFP-YPT52- and mChFP-NKIN2rigor-decorated microtubules (Fig. 5). Such localization was observed in all developmental stages of N. crassa. The result was somewhat surprising, given the fact that the fused hyphae contain in addition to the NKIN2rigor protein nonmodified versions of NKIN2 derived from both parental strains. This indicates that the modified motor acts as a sink for all early endosomes and that binding of the motor to the endosome is very strong or even irreversible. One possibility for the dominant effect of the modified NKIN2 motor could be that it causes a steric block of wild-type NKIN2 or other motor proteins and thereby prevents endosomal movement.
Fig 3.
GFP-YPT52 labels putative early endosomes in hyphae of N. crassa. (A) Time-lapse series of GFP-YPT52 displays bidirectional movement of small spots, whereby a retrograde-moving spot is indicated by arrowheads and an anterograde one by arrows. (B) GFP-YPT52 shows several motile vesicles in a young hypha. In a leading hypha (C), the cytoplasmic background is stronger and the number of spots is higher. Scale bars, 10 μm.
Fig 4.
Colocalization of GFP-YPT52- and FM4-64-labeled putative endosomes. (A and B) Most of the FM4-64-stained vesicles colocalize with the GFP-YPT52-labeled spot in a young hypha (A) as well as in leading hyphae (B) (arrowheads). A total of 31% of the spots do not colocalize (asterisks). DIC, differential interference contrast. Scale bars, 5 μm.
Fig 5.
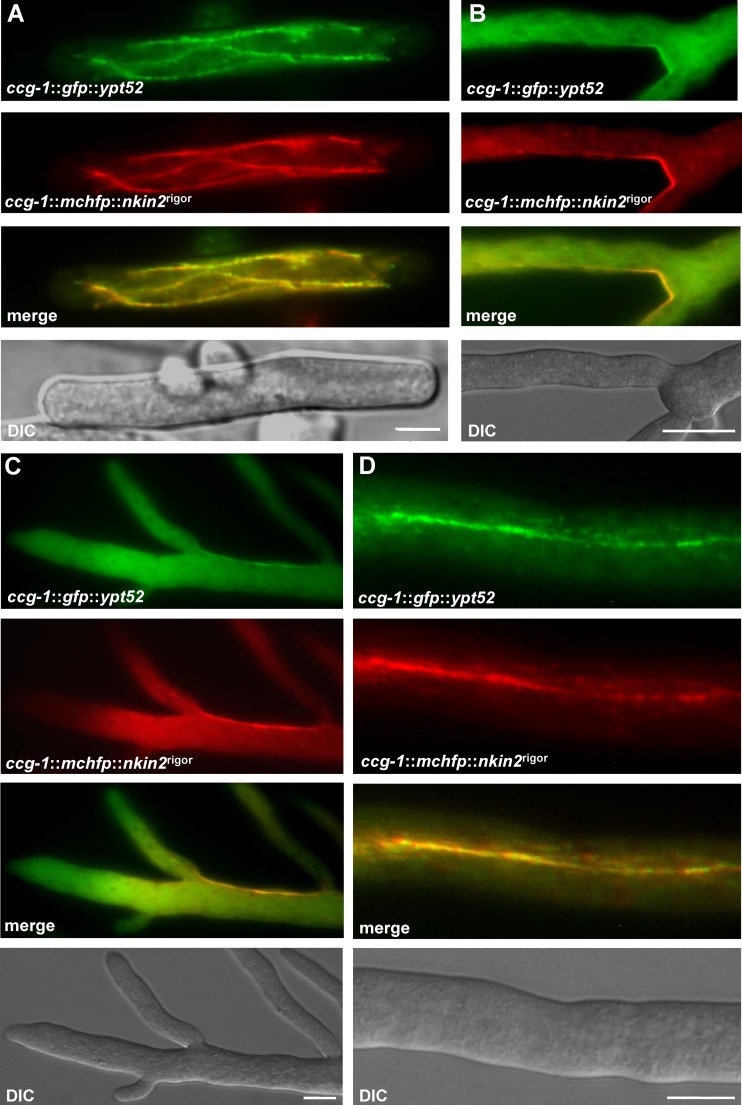
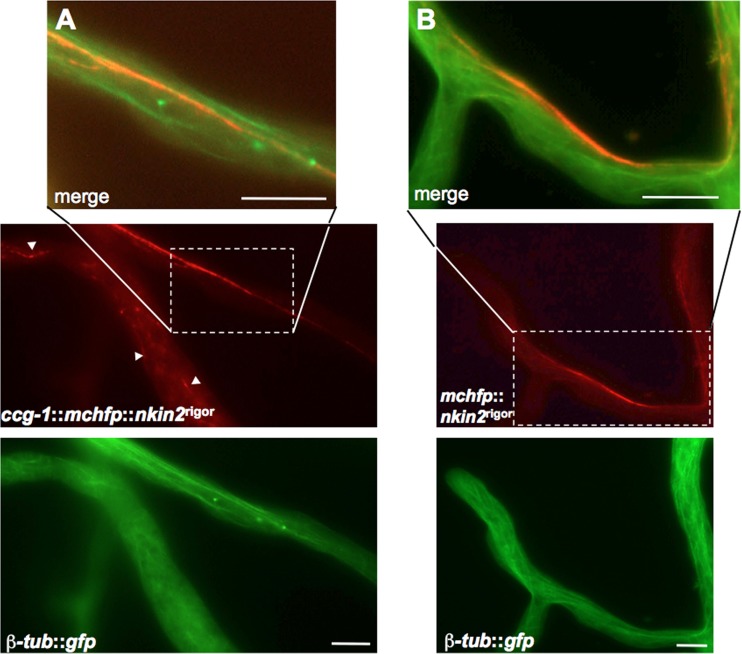
Colocalization of GFP-YPT52-labeled early endosomes and mChFP-NKIN2rigor. (A) In an arthrospore, several microtubules are decorated with mChFP-NKIN2rigor. GFP-YPT52 vesicles do not show any movement. (B) Similar observation in an apical branch of leading hyphae, where NKIN2rigor shows a preference for certain microtubules close to the cell wall. (C) An apical region of leading hyphae also displays no motile GFP-YPT52-labeled early endosomes but does show decoration of the same microtubule subpopulation, which is colabeled by mChFP-NKIN2rigor. (D) In lateral branches, where mChFP-NKIN2rigor prefers a certain microtubule bundle, GFP-YPT52 also sticks to the rigor-mutated kinesin. Scale bars, 5 μm in panel A and 10 μm in panels B to D.
Deletion of nkin2 causes endosomal motility defects.
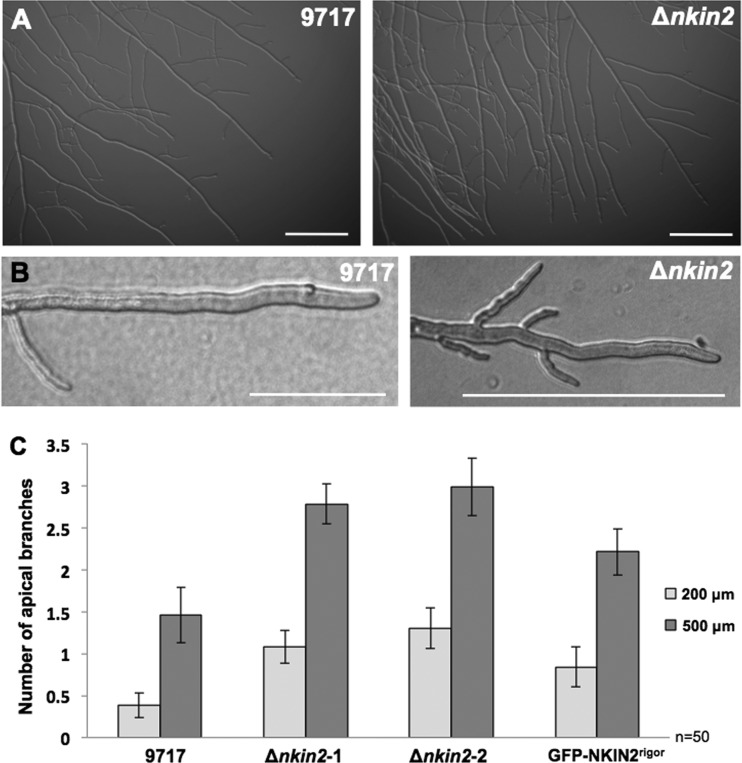
To investigate if NKIN2 is the only motor involved in the transport of early endosomes, an nkin2 deletion strain was constructed. A corresponding homokaryotic deletion strain was not available through the Fungal Genetics Stock Center. Therefore, we deleted the nkin2 open reading frame in N. crassa strain FGSC 9717 with hph as a selection marker. Single integration of the deletion construct in the N. crassa genome was confirmed by Southern blotting (see Fig. S2 in the supplemental material). Two independent nkin2 deletion strains were isolated and analyzed. Deletion of nkin2 resulted in a reduced colony expansion rate (32.06 μm/min and 34.83 μm/min, respectively) in comparison to that of the wild-type strain FGSC 9717 (39.83 μm/min) (Fig. 6A). However, the effect was not as strong as the effect of the deletion of conventional kinesin, Δnkin (17). In order to prove that the observed reduction of the growth rate was due to nkin2 deletion, the mutation was complemented. To this end, ccg-1::GFP::nkin2 and ccg-1::GFP::nkin2rigor constructs were integrated into the his-3 locus of Δnkin2. The wild-type version of NKIN2 was able to restore growth, whereas integration of NKIN2rigor did not rescue the Δnkin2 phenotype (Fig. 6A). Localization analysis of GFP-NKIN2rigor in the Δnkin2 mutant showed the same preference for one microtubule (MT) subpopulation as in wild type, indicating that an interaction between wild-type kinesin-3 and its mutated rigor version does not affect the specificity of GFP-NKIN2rigor which traverse the hyphal compartments even through septa (see Fig. S3D, arrowheads, in the supplemental material).
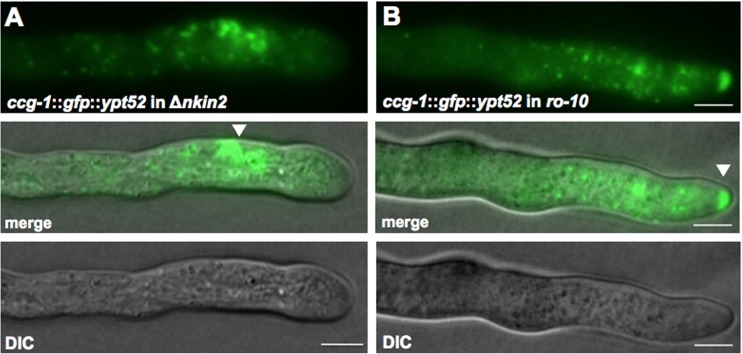
In addition to the reduced growth rate, the nkin2 deletion strains produced 27% less conidia than the wild type (Fig. 6B). Furthermore, germination of conidia was largely delayed in the Δnkin2 strains. While after 5 h, 90% of the conidia of the wild type had produced a germ tube, only 65 to 70% of the conidia of both nkin2 deletion strains had germinated (Fig. 6C). Microscopic analyses of the hyphae revealed increased branching at apical and subapical regions of leading hyphae of both Δnkin2 strains in comparison to the wild type (Fig. 7A and B). The number of branches observed in the first 200 μm and 500 μm behind the tip of the leading hyphae was more than doubled in the nkin2 mutants (Fig. 7C). Likewise, branching was also increased in the heterokaryotic NKIN2/GFP-NKIN2rigor strain, which confirmed the dominant effect of the rigor version (Fig. 7C). Septation and vacuolar distribution did not show any obvious abnormality (see Fig. S3 in the supplemental material). Interestingly, staining with FM4-64 revealed that the Spitzenkörper is less robust in Δnkin2 strains than in the wild type. A similar behavior of the Spitzenkörper was observed in the nkin deletion strain (42). However, movement of FM4-64-stained organelles was not completely abolished, only reduced. On the other hand, GFP-YPT52-stained early endosomes showed nearly no movement in Δnkin2 strains and accumulated frequently close to the apex, most likely the subapical endocytotic ring (Fig. 8A; see Movie S4 in the supplemental material). The still motile spots stained by FM4-64 could be the population of vesicles, which did not colocalize with GFP-YPT52. This suggests that NKIN2 may not be the only motor transporting different endosome populations along the hyphae of N. crassa.
Fig 7.
Analysis of the branching patterns of different NKIN2 mutants. (A) The Δnkin2 strain displays more branches. Scale bar, 200 μm. (B) Examples of the apical hyphal region with branches. (C) Quantification of branching within the first 200 μm and 500 μm away from the tip of leading hyphae of the wild-type strain (N9717), two Δnkin2 strains, and GFP-NKIN2rigor integrated into the his-3 locus of N9717.
Fig 8.
Analysis of endosomal motility associated with Δnkin2 and ro-10. (A) GFP-YPT52 targeted into the his-3 locus of Δnkin2 shows a highly reduced motility of the labeled early endosomes and accumulation of these endosomes close to the tip of young hyphae (arrowhead). (B) GFP-Ypt52-marked early endosomes accumulate at the hyphal tip (arrowhead) and show strongly reduced movement. Scale bars, 5 μm.
Furthermore, the dynamics of early endosomes was also strongly affected in a ro-10 mutant. ro-10 encodes a dynactin subunit and is required for assembly of the dynactin complex and thereby for proper dynein function (43). An N. crassa ro-10 strain expressing GFP-YPT52 displayed fluorescence accumulations at the hyphal tip and strongly reduced early endosome movements (Fig. 8B). Sometimes anterograde movement was still observed, most likely driven by kinesin-3 (see Movie S5 in the supplemental material). Dynein was shown to be necessary for retrograde transport of vesicles and endosomes (6, 23, 42). These observations suggest that the bidirectional transport of early endosomes in N. crassa is similar to those of A. nidulans and U. maydis (6, 14).
Fuchs and Westermann reported that a Δnkin2 strain displays mitochondrial motility and distribution defects in young germlings (15). Using MitoTracker Red and confocal imaging, we were unable to observe any differences in mitochondrial distributions and motilities between wild-type and nkin2 deletion strains (see Fig. S4 in the supplemental material). Coimaging of MitoTracker Red- and GFP-labeled microtubules showed some association, but also some mitochondria were observed, which were not associated with any microtubule (see Fig. S5 in the supplemental material).
NKIN2rigor localizes to a subset of microtubules.
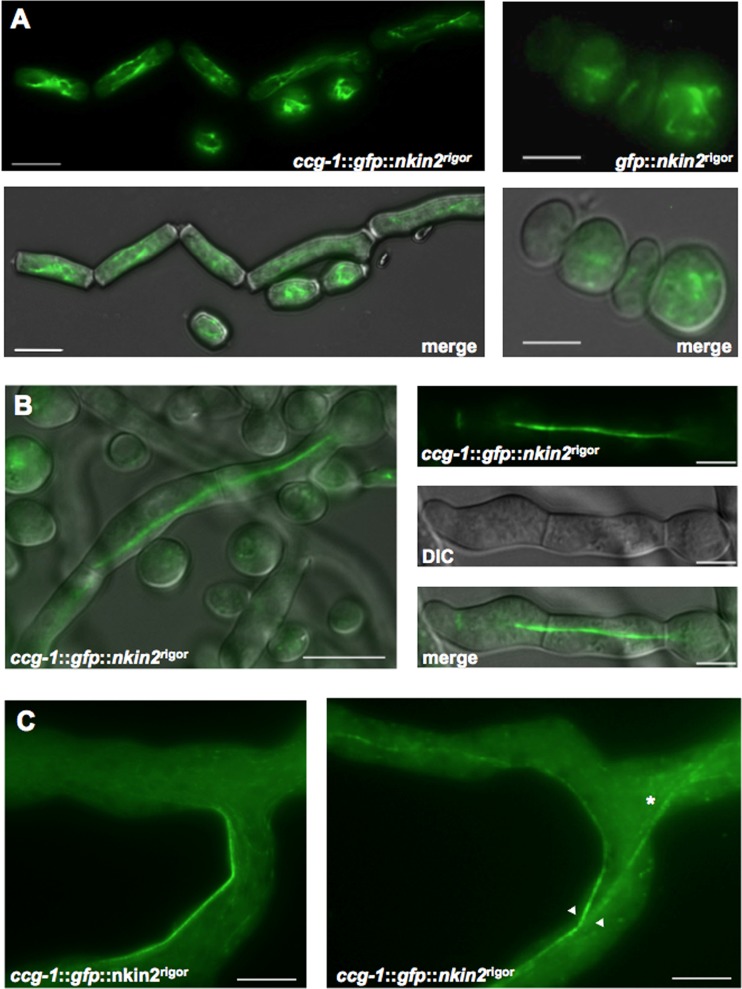
In the experiments presented above, it was observed that the rigor version of NKIN2 caused decoration of microtubules with GPF-YPT52. However, it appeared that not all microtubules were labeled, but rather a subpopulation of them. This is reminiscent of the behavior of UncA in A. nidulans. In order to test this hypothesis, microtubules and NKIN2rigor were both visualized. Heterokaryons coexpressing GFP-labeled microtubules and mChFP-NKIN2rigor were obtained (Fig. 9). The GFP-labeled microtubular cytoskeleton resembled the wild-type cytoskeleton, showing that the rigor mutation does not interfere with the cytoskeletal architecture. Interestingly, the localization pattern of GFP-NKIN2rigor was dependent on the developmental stage of N. crassa. Whereas in conidia and arthrospores, GFP-NKIN2rigor localized to several microtubules in the cell, it already showed in young germlings a preference for one microtubule or one microtubule bundle, as observed in mature hyphae (Fig. 10A and B). At the edge of the colony, GFP-NKIN2rigor localized in apical branches of leading hyphae cortically as a bar very close to the cell surface and often as a rigid rod, which crossed the branches and sometimes fused with each other and formed bundles (Fig. 10C, arrowheads). However, in lateral branches mChFP-NKIN2rigor bound preferentially to a microtubule, which traverses the hyphae very straight in the middle, whereas the other microtubules, labeled with β-tubulin–GFP, were not decorated (Fig. 9A). These observations suggest that NKIN2 performs different tasks during hyphal differentiation and development of N. crassa.
Fig 9.
Colocalization of mChFP-NKIN2rigor and β-tubulin-GFP. (A) Whereas in distal parts of primary hyphae, often only short pieces of the MT network are labeled (arrowheads) by mChFP NKIN2rigor, in lateral branches, one MT bundle appears to be preferred (magnification). (B) In apical branches of leading hyphae, GFP-NKIN2rigor decorates an MT subpopulation close to the edge of the hyphae, whereas the other MTs are not affected (magnification). Scale bars, 10 μm.
Fig 10.
Analysis of microtubule preference of NKIN2rigor at different developmental stages. (A) In arthrospores (left) and conidia (right), several microtubules are labeled by GFP-NKIN2rigor. Scale bars, 5 μm. (B) Already in young germlings, NKIN2rigor specifically binds to a subset of microtubules. Scale bars, 10 μm. (C) In apical branches of primary hyphae, GFP-NKIN2rigor sticks to an MT bundle close to the cell wall (arrowheads). This rigid rod also sometimes transverses branches of leading hyphae (asterisk). Scale bar, 10 μm.
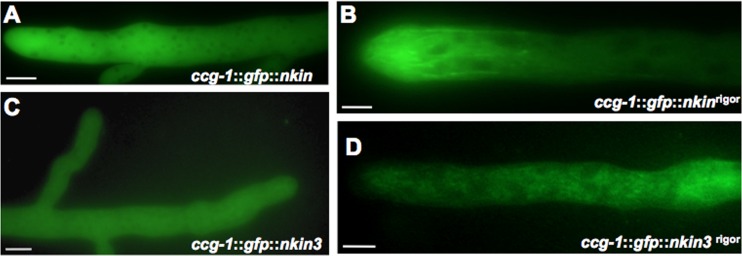
In order to emphasize the specificity of NKIN2, the localizations of kinesin-1, NKIN, and kinesin-3, NKIN3, were also studied in N. crassa. Both kinesins displayed cytoplasmic localization, which was different from that of NKIN2 (Fig. 11A and C). GFP-NKIN3 also frequently showed additional staining of the vacuoles in distal parts of primary hyphae (see Fig. S6A in the supplemental material). Immunolocalization studies of NKIN have previously revealed weak punctate structures on a strongly uniform background (42, 44). By introducing the rigor mutation into the P-loop of both kinesins, we investigated if they also exhibit any microtubule specificity. While GFP-NKIN3rigor still localized to the cytoplasm and vacuoles (probably for degradation) (Fig. 11D; see Fig. S6B), GFP-NKINrigor bound to several microtubules, but surprisingly only very close to the tip of the hyphae (Fig. 11B). Such a gradient of NKIN has been observed before with modified NKIN proteins unable to bind their cargoes (44). Specificity for a subpopulation of microtubules was not observed.
Fig 11.
Localization of conventional kinesin, NKIN, and NKIN3 (kinesin-3) and the corresponding rigor versions. (A) GFP-NKIN localizes in the cytoplasm but not in nuclei (dark spots). (B) Several microtubules are decorated by GFP-NKINrigor, most prominently at the hyphal tip. (C) GFP-NKIN3 localizes throughout the whole hypha. (D) The rigor version of NKIN3 does not bind to microtubules. Scale bar, 10 μm.
DISCUSSION
NKIN2 transports early endosomes but is required for mitochondrial function.
In higher eukaryotes, it has been shown that kinesin-3 motor proteins are neuron specific and transport synaptic vesicle precursors from the neuronal cell body along the axons (2, 45). In the filamentous fungi U. maydis and A. nidulans, kinesin-3 motors have been shown to be involved in the anterograde transport of endosomes (6, 14). In this work, we were able to unify the picture and showed by colocalization with an early endosomal marker, YPT52, and the lipophilic dye FM4-64 that in N. crassa, kinesin-3 also transports endosomes. nkin2 deletion strains showed early endosomal motility defects and reduction of polar growth. This function for this motor protein has not been described in the original report of the characterization of the motor protein (15). In contrast, the authors proposed a role for NKIN2 in mitochondrial movement along microtubules in young germlings (15). These conclusions were mainly drawn from cell fractionation experiments in which NKIN2 was shown to be associated with the mitochondrial outer membrane. However, in vivo colocalization experiments with microtubules and mitochondria in this work did not show strong association of the organelles with the cytoskeleton. Recent data obtained from U. maydis may explain the somewhat contradictory results. It was shown recently that Kin3 of U. maydis is involved in mRNA transport (11, 13). mRNA molecules are attached to endosomes and thus hitchhike these organelles (12). Most interestingly, the transport mechanism appears to be required for the proper localization of several mitochondrial proteins (13). This suggests transient association of kinesin-3 with mitochondria for delivery of mRNA encoding mitochondrial proteins. Thus, NKIN2 is apparently also necessary for proper mitochondrial function, but only indirectly through the endosomal transport.
During the analysis of endosome transportation, we noticed striking differences in different developmental hyphal stages, suggesting distinct functions of NKIN2. In young hyphae and branches, accumulations at the apex were observed frequently, which is consistent with the observations in A. nidulans and U. maydis. Such accumulations could never be observed in fast-growing leading hyphae. This can be easily explained if we consider the orientation of microtubules in hyphae and the fact that kinesin-3 moves toward the positive end of microtubules. In A. nidulans, nuclei are nicely aligned along the hyphae (46) and microtubules in the hyphal tip are almost exclusively oriented with their positive end facing the tip. In young germlings of N. crassa microtubules must be oriented similarly as in A. nidulans. However, older hyphae contain large numbers of nuclei, and the orientation of the microtubules is mixed (47).
Evidence for modified microtubules in N. crassa.
One interesting property of A. nidulans kinesin-3 is the ability to distinguish between different microtubules in one hyphal compartment. The nature of this special microtubule subpopulation has not been determined yet, but there is good evidence that A. nidulans contains detyrosinated α-tubulin. In order to show that this feature might be a general property of fungal kinesin-3 motors, it was very important to analyze this motor in a second fungus, N. crassa. Indeed, introduction of a rigor mutation into the motor domain of NKIN2 resulted in labeling a subpopulation of microtubules. Very interestingly, the labeling pattern varied depending on the developmental stage of the hyphae. This observation leads to the question how NKIN2 is able to discriminate between different microtubule populations. In A. nidulans UncA, we determined an 86-amino-acid-long stretch in the tail of the motor protein, named the microtubule binding (MTB) domain, as being required for the recognition (29). The N. crassa sequence of this MTB region is well conserved, especially in the C-terminal part of the MTB sequence and thus could also mediate the recognition of the modified microtubules. Another possibility to explain the observed rod-like MT structure in the hyphae could be that the motor cross-links several MTs and forms a bundle. It was indeed shown that the tail of A. nidulans UncA is able to interact with tubulin (29). However, given that NKIN2 appears to bind to many MTs under our experimental conditions (see Movie S1 in the supplemental material), one would expect several MT bundles in NKIN2rigor hyphae. This is not the case, and the majority of the MT cytoskeleton appears normal (Fig. 9).
The fact that NKIN2rigor specifically binds to a microtubule subpopulation in young hyphae and branches, which straight traverses the hyphae like a highway, similar to UncArigor in A. nidulans, suggests a similar function of NKIN2 in N. crassa. However, in primary hyphae NKIN2rigor associates with a microtubule rod close to the cell surface, which is most obvious during apical branching, indicating rather a mechanical function specifically during branching than transportation of early endosomes. This hypothesis is consistent with the observation that the nkin2 deletion strain shows the strongest defects during germination and colony establishment. This suggests different roles of KIN2 during N. crassa development.
Supplementary Material
ACKNOWLEDGMENTS
The work was funded through the German Science Foundation (DFG) Research Group 1334 and the CONACyT (Mexico). C. Seidel was partly supported by the Landesgraduiertenförderung of the state Baden-Württemberg.
We thank Stephan Seiler, Anne Dettmann, and Rosa Mouriño-Pérez for providing some strains and plasmids. We give special thanks to Eddy Sánchez-León, Rosa Fajardo-Somera, and Alexander Lichius for technical advice.
Footnotes
Published ahead of print 17 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00081-13.
REFERENCES
- 1. Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. 2004. A standardized kinesin nomenclature. J. Cell Biol. 167:19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hall DH, Hedgecock EM. 1991. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65:837–847 [DOI] [PubMed] [Google Scholar]
- 3. Lee J, Shin H, Choi J, Ko J, Kim S, Lee H, Kim K, Rho S, Lee J, Song H, Eom S, Kim E. 2004. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 23:1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Westerholm-Parvinen A, Vernos I, Serrano L. 2000. Kinesin subfamily UNC104 contains a FHA domain: boundaries and physicochemical characterization. FEBS Lett. 486:285–290 [DOI] [PubMed] [Google Scholar]
- 5. Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. 2002. Role of phosphatidylinositol(4,5)bisphosphate organisation in membrane transport by the Unc104 kinesin motor. Cell 109:347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wedlich-Söldner R, Schulz I, Straube A, Steinberg G. 2002. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell 13:965–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuster M, Lipowsky R, Assmann MA, Lenz P, Steinberg G. 2011. Transient binding of dynein controls bidirectional long-range motility in early endosomes. Proc. Natl. Acad. Sci. U. S. A. 108:3618–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schuster M, Kilaru S, Fink G, Collemare J, Roger Y, Steinberg G. 2011. Kinesin-3 and dynein cooperate in long-range retrograde endosome motility along a nonuniform microtubule array. Mol. Biol. Cell 22:3645–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steinberg G. 2012. The transport machinery for motility of fungal endosomes. Fungal Genet. Biol. 49:675–776 [DOI] [PubMed] [Google Scholar]
- 10. Schuchardt I, Aßmann D, Thines E, Schuberth C, Steinberg G. 2005. Myosin-V, kinesin-1, and kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol. Biol. Cell 16:5191–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. König J, Baumann S, Koepke J, Pohlmann T, Zarnack K, Feldbrügge M. 2009. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 28:1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrügge M. 2012. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci. 125:5191–5201 [DOI] [PubMed] [Google Scholar]
- 13. Koepke J, Kaffarnik F, Haag C, Zarnack K, Luscombe NM, König J, Ule J, Kellner R, Begerow D, Feldbrügge M. 2011. The RNA-binding protein Rrm4 is essential for efficient secretion of endochitinase Cts1. Mol. Cell Proteomics 10:M111.011213. 10.1074/mcp.M111.011213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zekert N, Fischer R. 2009. The Aspergillus nidulans kinesin-3 UncA motor moves vesicles along a subpopulation of microtubules. Mol. Biol. Cell 20:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuchs F, Westermann B. 2005. Role of Unc104/KIF1-related motor proteins in mitochondrial transport in Neurospora crassa. Mol. Biol. Cell 16:153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schoch CL, Aist JR, Yoder OC, Turgeon BG. 2003. A complete inventory of fungal kinesins in representative filamentous ascomycetes. Fungal Genet. Biol. 39:1–15 [DOI] [PubMed] [Google Scholar]
- 17. Seiler S, Nargang FE, Steinberg G, Schliwa M. 1997. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16:3025–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Requena N, Alberti-Segui C, Winzenburg E, Horn C, Schliwa M, Philippsen P, Liese R, Fischer R. 2001. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 42:121–132 [DOI] [PubMed] [Google Scholar]
- 19. Lehmler C, Steinberg G, Snetselaar KM, Schliwa M, Kahmann R, Bölker M. 1997. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 16:3464–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riquelme M, Yarden O, Bartnicki-Garcia S, Bowman B, Castro-Longoria E, Free SJ, Fleissner A, Freitag M, Lew RR, Mouriño-Pérez R, Plamann M, Rasmussen C, Richthammer C, Roberson RW, Sanchez-Leon E, Seiler S, Watters MK. 2011. Architecture and development of the Neurospora crassa hypha—a model cell for polarized growth. Fungal Genet. Biol. 115:446–474 [DOI] [PubMed] [Google Scholar]
- 21. Bartnicki-Garcia S, Bartnicki DD, Gierz G, Lopez-Franco R, Bracker CE. 1995. Evidence that Spitzenkörper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp. Mycol. 19:153–159 [DOI] [PubMed] [Google Scholar]
- 22. Zhang J, Li S, Fischer R, Xiang X. 2003. Accumulation of cytoplasmic dynein and dynactin at microtubule plus-ends is kinesin dependent in Aspergillus nidulans. Mol. Biol. Cell 14:1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lenz JH, Schuchardt I, Straube A, Steinberg G. 2006. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 25:2275–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiang X. 2006. A +TIP for a smooth trip. J. Cell Biol. 172:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fischer R, Zekert N, Takeshita N. 2008. Polarized growth in fungi—interplay between the cytosekelton, positional markers and membrane domains. Mol. Microbiol. 68:813–826 [DOI] [PubMed] [Google Scholar]
- 26. Janke C, Bulinski JC. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12:773–786 [DOI] [PubMed] [Google Scholar]
- 27. Weber K, Schneider A, Westermann S, Muller N, Plessmann U. 1997. Posttranslational modifications of α- and β-tubulin in Giardia lamblia, an ancient eukaryote. FEBS Lett. 419:87–91 [DOI] [PubMed] [Google Scholar]
- 28. Konishi Y, Setou M. 2009. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 12:559–567 [DOI] [PubMed] [Google Scholar]
- 29. Seidel C, Zekert N, Fischer R. 2012. The Aspergillus nidulans kinesin-3 tail is necessary and sufficient to recognize modified microtubules. PLoS One 7:e30976. 10.1371/journal.pone.0030976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ninomiya Y, Suzuki K, Ishii C, Inoue H. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101:12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sachs MS, Selker EU, Lin B, Roberts CJ, Luo Z, Vaught-Alexander D, Margolin BS. 1997. Expression of herpes virus thymidine kinase in Neurospora crassa. Nucleic Acids Res. 25:2389–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Verdín J, Bartnicki-Garcia S, Riquelme M. 2009. Functional stratification of the Spitzenkörper of Neurospora crassa. Mol. Microbiol. 74:1044–1053 [DOI] [PubMed] [Google Scholar]
- 34. Honda S, Selker EU. 2009. Tools for fungal proteomics: multifunctional neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182:11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hickey PC, Swift SR, Roca G, Read ND. 2005. Live-cell imaging of filamentous fungi using vital dyes and confocal microscopy. Methods Microbiol. 34:63–87 [Google Scholar]
- 36. Riquelme M, Bartnicki-Garcia S, González-Prieto JM, Sánchez-León E, Verdín-Ramos JA, Beltrán-Aguilar A, Freitag M. 2007. Spitzenkörper localization and intracellular traffic of green fluorescent protein-labeled CHS-3 and CHS-6 chitin synthases in living hyphae of Neurospora crassa. Eukaryot. Cell 6:1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sánchez-León E, Verdín J, Freitag M, Roberson RW, Bartnicki-Garcia S, Riquelme M. 2011. Traffic of chitin synthase 1 (CHS-1) to the Spitzenkörper and developing septa in hyphae of Neurospora crassa: actin dependence and evidence of distinct microvesicle populations. Eukaryot. Cell 10:683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abenza JF, Pantazopoulou A, Rodríguez JM, Galindo A, Peñalva MA. 2009. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic 10:57–75 [DOI] [PubMed] [Google Scholar]
- 39. Abenza JF, Galindo A, Pantazopoulou A, Gil C, de los Ríos V, Peñalva MA. 2010. Aspergillus RabB Rab5 integrates acquisition of degradative identity with the long distance movement of early endosomes. Mol. Biol. Cell 21:2756–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grosshans BL, Andreeva A, Gangar A, Niessen S, Yates JR, Brennwald P, Novick P. 2006. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J. Cell Biol. 172:55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singer-Krüger B, Stenmark H, Düsterhöft A, Philippsen P, Yoo JS, Gallwitz D, Zerial M. 1994. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125:283–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seiler S, Plamann M, Schliwa M. 1999. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9:779–785 [DOI] [PubMed] [Google Scholar]
- 43. Minke PF, Lee IH, Tinsley JH, Bruno KS, Plamann M. 1999. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 32:1065–1076 [DOI] [PubMed] [Google Scholar]
- 44. Seiler S, Kirchner J, Horn C, Kallipolitou A, Schliwa GWM. 2000. Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat. Cell Biol. 2:333–338 [DOI] [PubMed] [Google Scholar]
- 45. Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81:769–780 [DOI] [PubMed] [Google Scholar]
- 46. Suelmann R, Sievers N, Fischer R. 1997. Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol. Microbiol. 25:757–769 [DOI] [PubMed] [Google Scholar]
- 47. Mouriño-Pérez RR, Roberson RW, Bartnicki-Garcia S. 2006. Microtubule dynamics and organization during hyphal growth and branching in Neurospora crassa. Fungal Genet. Biol. 43:389–400 [DOI] [PubMed] [Google Scholar]
- 48. Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897–910 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.