Abstract
The unfolded protein response (UPR) is an important regulatory network that responds to perturbations in protein homeostasis in the endoplasmic reticulum (ER). In mammalian cells, the UPR features translational and transcriptional mechanisms of gene expression aimed at restoring proteostatic control. A central feature of the UPR is phosphorylation of the α subunit of eukaryotic initiation factor-2 (eIF2) by PERK (EIF2AK3/PEK), which reduces the influx of nascent proteins into the ER by lowering global protein synthesis, coincident with preferential translation of key transcription activators of genes that function to expand the processing capacity of this secretory organelle. Upon ER stress, the apicomplexan parasite Toxoplasma gondii is known to induce phosphorylation of Toxoplasma eIF2α and lower translation initiation. To characterize the nature of the ensuing UPR in this parasite, we carried out microarray analyses to measure the changes in the transcriptome and in translational control during ER stress. We determined that a collection of transcripts linked with the secretory process are induced in response to ER stress, supporting the idea that a transcriptional induction phase of the UPR occurs in Toxoplasma. Furthermore, we determined that about 500 gene transcripts showed enhanced association with translating ribosomes during ER stress. Many of these target genes are suggested to be involved in gene expression, including JmjC5, which continues to be actively translated during ER stress. This study indicates that Toxoplasma triggers a UPR during ER stress that features both translational and transcriptional regulatory mechanisms, which is likely to be important for parasite invasion and development.
INTRODUCTION
Perturbations in protein folding and assembly can induce the unfolded protein response (UPR), which consists of translational and transcriptional regulation of gene expression that is designed to expand the processing capacity of the endoplasmic reticulum (ER) and alleviate damage in eukaryotic organisms (1, 2). PERK (PEK/EIF2AK3) phosphorylation of the α subunit of eukaryotic initiation factor-2 (eIF2) rapidly represses protein synthesis, lowering the influx of nascent proteins into the ER. Concurrently, phosphorylation of eIF2α enhances the translation of select mRNAs, such as the basic zipper (bZIP) transcription factor ATF4, which serves to reprogram expression of genes involved in restoration of proteostatic control.
In mammalian cells, PERK functions in conjunction with ATF6 and IRE1 to detect the accumulation of malfolded proteins and signal transcriptional changes. ATF6 is a bZIP transcription factor localized to the ER that is subject to proteolytic cleavage in response to ER stress and the accumulation of malfolded secretory proteins. Proteolytic cleavage releases an active, cytosolic form of ATF6 that can translocate into the nucleus to activate expression of chaperones and other proteins to alleviate stress damage (3). IRE1 is also an ER stress sensor that is activated when protein folding conditions exceed ER capacity. The activated form of IRE1 induces the nonconventional splicing and translation of XBP1 mRNA, encoding another bZIP transcription factor that facilitates transcription of genes involved in regulating ER protein folding and homeostasis. While mammals possess all three ER stress sensors (PERK, ATF6, and IRE1), yeast only express IRE1 (1, 2). Among the genes induced by the UPR are those involved in amino acid transport, protein folding and assembly, vesicle transport, antioxidation, lipid biogenesis, and ER-associated protein degradation (ERAD), which can collectively serve to expand the processing capacity of the ER. If the ER stress is sustained, there is induced expression of genes associated with autophagy and, ultimately, programmed cell death.
Protozoan parasites have complex life cycles that can involve multiple hosts and oscillating periods of proliferation and latency. Toxoplasma gondii is an opportunistic intracellular parasite that represents a major threat to immunocompromised individuals, including AIDS patients and newborns with a congenital infection (4). Toxoplasma is suggested to tightly regulate ER homeostatic pathways to allow for the production of properly folded secretory proteins required for host cell adhesion and invasion, evasion of the host response, and conversion from a highly invasive and actively proliferating form (tachyzoite) to a latent cyst (bradyzoite) (5). These processes are central for parasite viability, and as a result, pharmacological agents that alter homeostasis within the ER are detrimental to apicomplexan parasites (6–9).
Phosphorylation of Toxoplasma eIF2α (TgIF2α) was previously suggested to repress translation initiation during parasite stress responses and developmental changes (10–12). Furthermore, we established a link between ER stress and the development of latent forms of Toxoplasma (10) and a PERK homologue in the kinetoplastid parasite Leishmania that has been suggested to be critical for proper developmental transitions (11). In this study, we address the nature of the gene expression networks in the UPR in Toxoplasma. We determine that while there is only a single identifiable UPR sensor related to the eIF2α kinase PERK (TgIF2K-A), both transcriptional and translational programs are elicited upon ER stress in Toxoplasma, and they affect many facets of parasite biology. Preferential translation, as judged by increased association of mRNAs with large polysomes during ER stress, includes about 500 genes, including a subset of AP2 transcription factors and other transcription modulators. These findings suggest that the translational phase of the UPR is pervasive in Toxoplasma and is a major regulatory mechanism contributing to gene expression.
MATERIALS AND METHODS
Measurement of TgIF2α phosphorylation.
Phosphorylation of TgIF2α was measured by Western blotting as previously described (10). Intracellular parasites were released from human foreskin fibroblast (HFF) host cells by scraping and syringe passage, followed by collection and purification by using 3-μm polycarbonate filters (13). Purified parasites were incubated in Dulbecco's modified essential medium (DMEM) containing 1% fetal bovine serum (FBS) at 37°C under 5% CO2 in the presence of 10 μM tunicamycin (T7765; Sigma-Aldrich) or dimethylsulfoxide (DMSO) (vehicle) for 1 h. During the course of this study, thapsigargin (0215899905; MP Biomedicals) was found to be a more reliable ER stress agent; hence, it was used in later experiments to elicit ER stress. In comparison, different lots and vendors for tunicamycin showed variations in TgIF2α phosphorylation. Tunicamycin is a nucleoside antibiotic comprised of at least 10 homologues with a single fatty acid side chain that differs in length and fatty acid component (14). The composition of different preparations of tunicamycin can vary significantly in amounts of each homologue, and the differences in TgIF2α phosphorylation may lie with chemical variations from different vendors and lots of the drug (T7765; Sigma-Aldrich). ER stress was also induced by the addition of the calcium ionophore A23187 (C7522; Sigma-Aldrich).
Parasites were collected and lysed in a solution containing 50 mM Tris (pH 7.9), 150 mM NaCl, 2 mM EDTA, and 0.1% Nonidet P-40 that was supplemented with a phosphatase inhibitor (50 mM NaF) and protease inhibitors (100 μM phenylmethylsulfonyl fluoride, 0.15 μM aprotinin, 1 μM pepstatin, and 1 μM leupeptin). Forty micrograms of each of the protein lysates was separated by SDS-PAGE using a 10% Bis-Tris-acrylamide gel (Invitrogen). Proteins were transferred to nitrocellulose membranes and then probed with either rabbit anti-TgIF2α antibody (diluted 1:10,000) or phosphospecific (Ser71) TgIF2α antibody (diluted 1:500), followed by an anti-rabbit IgG-horseradish peroxidase conjugate (GE Healthcare) (10). Total and phospho-TgIF2α were visualized using an ECL Western blotting substrate (Pierce).
Polyribosome fractionation.
For polysome fractionations, ∼109 parasites were freshly harvested and purified from HFF monolayers as described above and then incubated for 1 h in the presence of 10 μM tunicamycin or DMSO supplemented in DMEM containing 1% FBS at 37°C under 5% CO2. Following treatment for 1 h, parasites then were exposed to 50 μg/ml cycloheximide for 10 min and collected by centrifugation at 4°C. Parasite pellets were washed twice in cold 1× phosphate-buffered saline (PBS) and resuspended in breaking solution (20 mM Tris-HCl [pH 7.9], 150 mM NaCl, 10 mM MgCl2, 0.1% Triton, 50 μg/ml cycloheximide, and 0.04 U/μl RNase Out) supplemented with protease inhibitors. To ensure lysis, parasites were briefly sonicated on ice three times and then clarified by centrifugation at 16,000 × g at 4°C. Cell lysates were layered onto 15 to 45% sucrose gradients prepared in breaking solution without Triton or RNase Out and then resolved by centrifugation using a Beckman SW41Ti rotor at 40,000 rpm at 4°C for 2 h as previously described (10). Gradients were fractionated using a BioComp Instruments gradient station. During fractionation, the absorbance was measured using an ISCO UA-6 absorbance monitor set at 254 nm, and 0.7-ml fractions were collected.
RNA purification.
Fractions from the polyribosome analysis were pooled into two groups: free and monoribosomal RNA (fractions 1 to 5) and polyribosomal RNA containing three or more associated ribosomes (fractions 7 to 13). For each 1-ml fraction pool, an equal amount of synthetic Affymetrix poly(A) RNA was added as previously described (15). The Affymetrix poly(A) RNA contains different amounts of four prokaryotic polyadenylated RNAs and serves as a control for variance in subsequent RNA isolation and purification steps. The Affymetrix-based ToxoGeneChip contains probes that specifically hybridize to the ∼8,000 predicted Toxoplasma genes as well as prokaryotic control RNA (16). RNA was precipitated with 2.5 volumes of 100% ethanol and purified using Qiagen RNeasy columns. The quality and quantity of the purified RNA was assessed using an Agilent Bioanalyzer and a NanoDrop NC-1000 UV/Vis spectrophotometer. In parallel, total RNA was prepared from parasites treated with 10 μM tunicamycin or DMSO for 1 h. The parasite pellets were lysed in buffer LRT (Qiagen RNeasy kit), and equal amounts of the Affymetrix poly(A) RNAs were added to each total RNA sample. The RNA was purified according to the manufacturer's protocol.
Microarray hybridization and data analysis.
Microarray hybridizations were carried out at the Center for Medical Genomics at the Indiana University School of Medicine. Preparation of cDNA, cRNA, and labeling were completed for three biological replicates of samples prepared from sucrose gradient fractionation and the unfractionated RNA according to the protocols recommended for the Affymetrix 3′ IVT express kit (Affymetrix, Santa Clara, CA), starting with 100 ng of RNA. Arrays were hybridized for 17 h at 42°C. The arrays were washed and hybridized by a fluidics station controlled by GeneChip operating software (GCOS) using the standard Affymetrix protocol. The microarrays were scanned using a dedicated Model 3000 scanner controlled by GCOS software. Data were extracted using the Affymetrix microarray suite 5 (MAS5) algorithm without scaling. The data were normalized for each array using a Mod factor, which is calculated by subtracting the mean log2 intensities for the poly(A) control RNAs from 10. The gene expression data for each data set was then normalized by adding the Mod factor to the log2-transformed data intensities from each array.
Results from three biological replicates, representing total RNA isolated from equal numbers of parasites cultured in the presence of tunicamycin or DMSO, were normalized and transformed into log2 intensities. Probe sets were normalized to the bacterial poly(A) control RNAs that were spiked into each lysate as discussed above. The change in mRNA abundance following tunicamycin treatment was calculated by determining the ratio of the intensities of the mRNA species from the tunicamycin-treated parasite samples to the vehicle-treated parasite population. Probe sets that were not significantly changed (P > 0.05) during tunicamycin treatment were filtered out so that the resulting data set would further represent ER stress-dependent genes. The Student t test (P ≤ 0.05) was used to determine the statistical significance. Key findings of the microarray analysis were confirmed by quantitative PCR (qPCR) with primers listed in Table S3 in the supplemental material.
Probe sets with fewer than 2 samples called present (above background as determined by using the Affymetrix MAS5 algorithm) in both treatment groups (DMSO/tunicamycin) were filtered out before calculating the false discovery rates (FDR) using the q value (17). FDR values are included in Data Sets S1 and S2 in the supplemental material to assess the quality of those genes showing changes in mRNA levels and preferential association with polysomes in response to ER stress. We filtered data sets to show statistically significant changes in mRNA abundance after tunicamycin treatment using P < 0.05 and FDR < 0.05. Analysis of changes in the polysome distribution following treatment with tunicamycin indicated that the 501 preferentially translated mRNAs (P < 0.02) have FDR of less than 0.063 (6.3% rate of false discovery).
In parallel, a microarray analysis was carried out using RNA prepared from the (i) free or monosomal RNA and (ii) polysomal RNA, as described above. Three biological replicates were analyzed from parasites treated with tunicamycin or DMSO. The mean signal intensities of the stressed and unstressed samples were calculated. The change in mRNA-polysome association following treatment with tunicamycin was calculating using the following equation:
Thus, a positive value (i.e., percent shift) would indicate enhanced abundance of polysome-associated gene transcripts following treatment with tunicamycin, whereas a negative value would indicate reduced mRNA association with polysomes in response to tunicamycin treatment. The mRNAs suggested to be preferentially translated were defined as having a statistically significant shift to the polyribosome fraction following exposure to tunicamycin (P ≤ 0.01). Selected gene transcripts subject to preferential association with polysomes were also analyzed by qPCR of the mRNA prepared from the sucrose gradient fractions using primers listed in Table S3 in the supplemental material.
Bioinformatic analyses.
PFAM analysis was carried out to identify putative domains that belong to a protein domain family. TMHMM and SignalP informatics was used to predict the presence of a transmembrane domain and signal sequences in gene coding sequences (18, 19). Percentages of genes containing secretory protein properties were evaluated using the Search for Genes feature in the ToxoDB (20), and statistical significance between the biological properties of targeted UPR genes and the entire Toxoplasma genome were determined by using the chi-square test. Furthermore, classifications of molecular functions of genes whose transcripts showed increased association with polysomes upon ER stress were carried out using Gene Ontology (GO) terms by adding search steps, including the indicated classifications in the EuPathDB programming available in ToxoDB 8.1. The 3′- and 5′-untranslated region (UTR) sequences of annotated mRNAs suggested to be preferentially translated during ER stress was obtained from ToxoDB. Alternatively, RNA sequencing data along with the optimal translation start site based on ToxoDB gene predictions were used to manually annotate the 5′-UTR sequence (B. D. Gregory, unpublished data). The resulting 5′-UTR sequence was analyzed for the presence of upstream open reading frames (uORFs) as defined by an initiation codon (ATG).
Radiolabel incorporation and immunoprecipitation of newly synthesized JmjC5.
A Toxoplasma strain stably expressing TgJmjC5 tagged at its C terminus with 3× hemagglutinin (HA) epitopes was generated by targeting the endogenous JmjC5 (TGME49_061260) locus using homologous recombination in the RHΔKu80Δhxgprt background (21) (the strain was a gift of Vern Carruthers, University of Michigan). RHΔKu80Δhxgprt genomic DNA was used to PCR amplify a 0.9-kb fragment of the JmjC5 3′ end using forward primer C5HA_F (5′-TACTTCCAATCCAATTTAATGCGGAGGAACCAACAGACCAGC-3′) and reverse primer C5HA_R (5′-TCCTCCACTTCCAATTTTAGCTTCCGAGGTGAGAAGGCGTG-3′) that contained ligation-independent cloning (LIC) sequences (underlined). This DNA fragment was inserted into the pLIC_HAx3_HXGPRT endogenous tagging vector such that the TgJmjC5 coding sequence was fused in frame with the epitope coding region as described previously (21). A positive pLIC_C5HAx3_HXGPRT construct was identified and confirmed by Sanger sequencing. For transfection, 15 μg of the pLIC_C5HAx3_HXGPRT vector was linearized by overnight digestion with BaeI within the JmjC5 homologous region and ethanol precipitated. RHΔKu80Δhxgprt tachyzoites were transformed with the linearized construct by electroporation, and after overnight growth in HFF, parasite cultures were selected with MPA and xanthine for HXGPRT expression as described previously (22). Drug-resistant parasites were cloned by limiting dilution and screened by Western blotting and immunofluorescence for expression of JmjC5-HA.
JmjC5-HA parasites were released from HFF monolayers by syringe passage and purified using 3-μm polycarbonate filters. The parasites were washed three times in DMEM lacking l-methionine, l-cysteine, l-glutamine, or sodium pyruvate (21013-024; Invitrogen) supplemented with 5% FBS, 1 mM l-glutamine, and 0.5 mM sodium pyruvate. Express protein label mix (0.145 mCi) containing [35S]methionine and [35S]cysteine (PerkinElmer Life Sciences) was added to the sample and incubated for 90 min in the presence of 10 μM thapsigargin or DMSO. Samples were washed twice in PBS, and a portion was counted to determine the uptake of the radiolabeled amino acids. There was no difference in 35S uptake between parasites treated with thapsigargin or DMSO. The radiolabeled parasites were lysed in 20 mM Tris-HCl (pH 7.9), 150 mM NaCl, 1 mM dithiothreitol (DTT), 1 mM EDTA, and 0.1% Triton, supplemented with protease inhibitors. Two percent of the input protein sample was separated by PAGE and stained with Coomassie blue, and the radiolabeled proteins were visualized by autoradiography. JmjC5-HA was immunoprecipitated from each lysate using anti-HA-coupled agarose beads, resolved by SDS-PAGE, and visualized by autoradiography.
Microarray accession number.
Microarray data have been deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE43722.
RESULTS AND DISCUSSION
Tunicamycin induces an unfolded protein response in Toxoplasma.
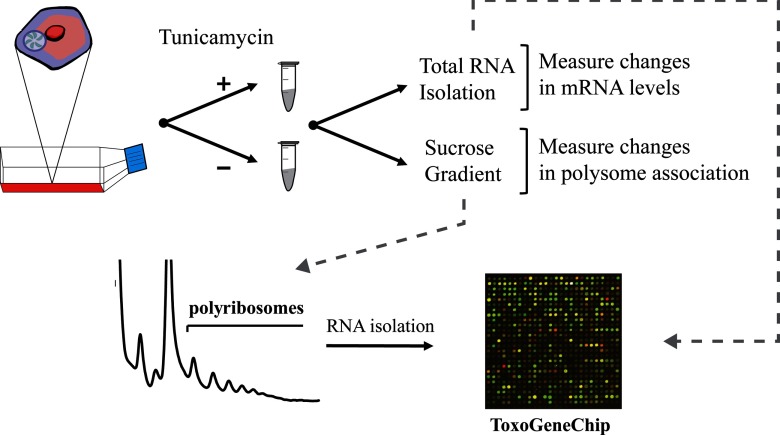
Tunicamycin is a potent inducer of ER stress through its inhibition of protein glycosylation in the ER, and it has been validated as an inhibitor of N-linked glycosylation in Toxoplasma (6). Furthermore, we showed that Toxoplasma TgIF2α is phosphorylated in response to ER stress, such as that elicited by tunicamycin treatment, leading to a dampening of global translation as measured by decreased polysomes accompanied by enhanced free ribosomes and monosomes (10). To measure the changes in the Toxoplasma transcriptome that occur upon ER stress, we purified tachyzoites (RH strain) from host cells and treated them with 10 μM tunicamycin or vehicle (DMSO) for 1 h at 37°C. Total RNA was isolated from each sample and analyzed on Affymetrix ToxoGeneChip microarrays, which contain probe sets for the predicted ∼8,000 Toxoplasma genes (16) (Fig. 1). The complete data sets can be accessed through GEO (GSE43722).
Fig 1.
Method used to measure changes in mRNA levels and polysome association in response to ER stress. Toxoplasma tachyzoites were purified from host cell monolayers and exposed to 10 μM tunicamycin (+) or DMSO vehicle (−) for 1 h. To measure changes in transcript abundance, total RNA was isolated from the treated parasites, and gene transcript levels were measured by microarray hybridization. Additionally, to measure translational changes as judged by mRNA association with polysomes, lysates were generated from parasite populations treated with tunicamycin or the DMSO vehicle control and subjected to sucrose gradient centrifugation. RNA was isolated from the sucrose fraction, namely, the free ribosomes and monosomes or the polysome fractions, and gene transcript levels were measured by microarray analysis.
Results from the microarray study show that 136 genes (∼1.7%) were significantly (P ≤ 0.05) upregulated following acute exposure to tunicamycin (Table 1). A similar study performed in the yeast Saccharomyces cerevisiae found that the UPR increases the levels of nearly 400 gene transcripts (∼7%) (23). Bioinformatic analysis of the predicted amino acid sequences for the differentially upregulated genes was performed to identify predicted protein domains and biological functions (see Data Set S1 in the supplemental material). The portion of the upregulated mRNAs encoding genes with a predicted signal peptide and/or a transmembrane domain was 40%, which was significantly larger than the 32% of genes in the entire Toxoplasma genome that were identified as having these properties (P < 0.05). We conclude that the gene products induced by ER stress are more frequently targeted to the secretory pathway (see Data Set S1). Consistent with ER stress remedy, many of the induced genes play an integral role in protein processing (folding, degradation, and vesicle transport), lipid biosynthesis, and oxidative stress (Table 2). Key findings include upregulation of glycosyl transferases (TGME49_007070 and TGME49_097720) and Derlin-1 (TGME49_017160), which is critical for the degradation of misfolded ER proteins (24, 25). Several protein chaperones were also induced, including a calreticulin family member (TGME49_077230); calreticulin is an ER-resident chaperone that prevents export of misfolded protein (26). Expression of amino acid transporters is also enhanced during UPR (27), and one predicted transporter (TGME49_026060) is induced in Toxoplasma. We note that this short period of ER stress only induced a subset of genes (including those encoding other chaperones or bradyzoite antigens) that are seen upon longer stress treatments (10).
Table 1.
ER stress induces changes in the Toxoplasma transcriptomea
| No. of Toxoplasma gene transcripts that are: |
P value | |||
|---|---|---|---|---|
| Activated |
Repressed |
|||
| >1-fold | ≥2.0-fold | <1.0-fold | ≤0.5-fold | |
| 710 | 305 | 7,347 | 3,493 | |
| 136 | 121 | 5,546 | 3,169 | ≤0.05 |
| 58 | 55 | 4,136 | 2,654 | ≤0.01 |
Shown are the number of Toxoplasma gene transcripts that are activated or repressed in parasites subjected to ER stress by treatment with tunicamycin for 1 h. Changes in transcript levels were assayed using three biological replicates and were analyzed for statistical significance as the means ± standard errors (SE). Data were statistically significant according to P value (<0.05 or 0.01, as indicated). A total of 8,057 probe sets were analyzed.
Table 2.
Tunicamycin induces expression of genes consistent with the unfolded protein response in other eukaryotesa
| ToxoDB Version 5 ID | Product | Fold induction | P value | TM (no.) | SP |
|---|---|---|---|---|---|
| Protein modification | |||||
| TGME49_007070 | Glycosyl transferase | 4.10 | 0.007 | No | No |
| TGME49_097720 | Glycosyl transferase domain | 6.84 | 0.044 | No | No |
| Protein degradation | |||||
| TGME49_017160 | Derlin-1 | 6.97 | 0.012 | 4 | Yes |
| TGME49_095670 | E3 ubiquitin ligase | 2.80 | 0.010 | No | No |
| TGME49_071490 | Endoprotease | 7.72 | 0.005 | 1 | Yes |
| Quality control and protein folding | |||||
| TGME49_063170 | Heat shock protein | 9.36 | 0.000 | No | No |
| TGME49_077230 | Calruticulin | 3.44 | 0.002 | No | No |
| TGME49_094870 | Universal stress protein/chaperone | 4.05 | 0.007 | No | No |
| Vesicle transport | |||||
| TGME49_073070 | GTPase activating protein for Arf domain-containing protein | 3.55 | 0.044 | No | Yes |
| TGME49_014170 | Prenyltransferase | 4.14 | 0.041 | No | Yes |
| Lipid and membrane metabolism | |||||
| TGME49_063740 | ATP-binding cassette, lipid transporter | 5.31 | 0.005 | 5 | No |
| TGME49_078110 | 1,3-Beta-glucan synthase | 4.34 | 0.038 | 11 | No |
| TGME49_046010 | Esterase/lipase | 10.76 | 0.008 | No | No |
| TGME49_050360 | Esterase/lipase | 1.42 | 0.043 | No | No |
Shown are Toxoplasma genes whose expression is induced by tunicamycin treatment. Like other eukaryotes, the Toxoplasma UPR contains factors involved in protein modification, folding, degradation, vesicle transport, and lipid/membrane metabolism. Statistical analysis was performed as described for Table 1. TMHMM and SignalP were used to predict the presence or absence of transmembrane (TM) domains and signal peptide (SP) sequences.
To confirm the fidelity of the microarray results, we carried out qPCR for several induced genes which supported our key findings (see Table S1 the supplemental material). To address whether another ER stress agent also induces the expression of the Toxoplasma UPR genes, we carried out qPCR measurements of mRNA changes that occurred in response to 5 μM A23187, a calcium ionophore that induces ER stress and is a potent inducer of TgIF2α phosphorylation (10). Expression of mRNAs for both the calreticulin family member (TGME49_077230) and E3 ubiquitin ligase (TGME49_095670) were strongly induced following exposure to tunicamycin or A23187 (Fig. 2). Our findings suggest that mRNA changes containing hallmark features of the UPR occur when Toxoplasma is exposed to ER stress agents.
Fig 2.
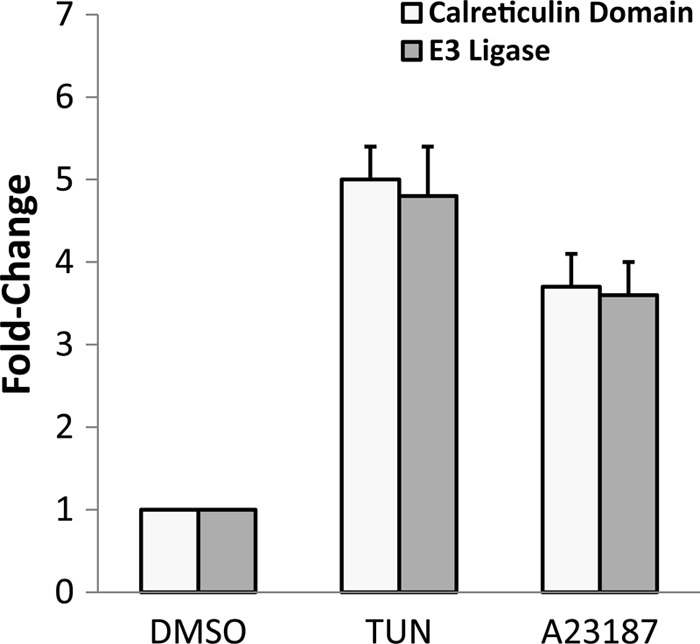
UPR genes are induced by multiple ER stress agents. Freshly purified tachyzoites were exposed to 10 μM tunicamycin (TUN), 5 μM A23187, or vehicle (DMSO) for 1 h. Levels for two UPR gene transcripts, a calreticulin family domain member (TGME49_077230) and a putative E3 ubiquitin ligase (TGME49_095670), were measured by qPCR. The histograms represent statistically significant changes (P < 0.01) between ER-stressed samples and the DMSO control, with standard errors (SE) indicated by the error bars.
The transcriptome analysis of the Toxoplasma ER stress response indicates several new features associated with this parasite UPR. Apicomplexan parasites lack most conventional transcription factors and appear to employ an expanded lineage of plant-like DNA-binding proteins harboring Apetela-2 (AP2) domains (28–30). Two such AP2 factors were found to be transcriptionally upregulated: AP2IX-3 (TGME49_064480) and AP2VIII-7 (TGME49_069010). The levels of bradyzoite-specific surface antigen SAG2C (SRS49D; TGME49_007160) mRNA is also increased upon ER stress, consistent with a stress-induced developmental shift to bradyzoites. Seventy gene transcripts are of hypothetical genes of unknown function, underscoring the great potential for novel features of the UPR in apicomplexan parasites such as Toxoplasma.
Treatment with tunicamycin also caused a significant reduction (P ≤ 0.05) in almost 70% of the measured gene transcripts (Table 1; also see Fig. S1 in the supplemental material). The brief 1-h insult with tunicamycin suggests that mRNA decay plays a role in the mRNA reduction. In Drosophila melanogaster and mammals, ER-stressed cells appear to have widespread IRE1-dependent degradation of ER-associated mRNAs (31, 32). However, as we describe in the following section, Toxoplasma does not appear to possess an IRE1 homologue. An alternative possibility for mRNA degradation is through enhanced expression of a PUF gene (TGME49_060600) during tunicamycin exposure (see Fig. S1). PUF proteins bind to 3′-UTRs of specific mRNAs to repress their translation and/or to facilitate their decay (33). Recently it was found in Plasmodium that Puf2 participates in the developmental transition from sporozoite to liver stages (34, 35).
Apicomplexa lack clear homologues of IRE1 and ATF6.
In higher eukaryotes, the UPR is mediated by three ER stress sensors, IRE1, ATF6, and PERK. Bioinformatic analyses performed using Toxoplasma and other apicomplexan databases indicate there is no homologue of IRE1. Furthermore, apicomplexan parasites lack bZIP transcription regulators (28), such as the sensor ATF6, and affiliated UPR bZIP transcriptional regulators XBP1 and ATF4. As noted above, Apicomplexa express an expanded group of proteins containing a plant-like DNA-binding domain called AP2. We surveyed all of the predicted AP2 domain proteins in Toxoplasma for transmembrane domains, which would make it a candidate that is functionally analogous to ATF6. However, none of the predicted Toxoplasma AP2 domain proteins contain a transmembrane domain. Searches for PERK homologues among apicomplexan parasites revealed a number of predicted eIF2α kinases containing transmembrane domains (Table 3). Only Toxoplasma TgIF2K-A has been verified to be present in the ER to date (10). Furthermore, TgIF2K-A associates with the ER-resident chaperone BiP, and this binding is released in an ER stress-dependent fashion, as reported for activation of mammalian PERK (10, 36). None of the other eIF2α kinases, designated TgIF2K-B, TgIF2K-C, and TgIF2K-D, possess predicted signal sequences or transmembrane segments, suggesting that they are not localized to the ER, as described for the known UPR sensors. Interestingly, the transmembrane domain is localized to the extreme N terminus of putative PERK homologues in Cryptosporidium species. This is not likely due to problematic gene annotation, since it occurs in multiple related species, suggesting that these eIF2α kinases have a distinct mechanism for stress sensing. The results from this analysis suggest that Apicomplexa lack two major branches of the standard eukaryotic UPR but possess transmembrane-containing eIF2α kinases that localize to the ER to mediate UPR through translational control. The absence of IRE1 and presence of PERK homologues in apicomplexan protozoa challenges proposed models that IRE1 arose early in evolution as the sole modulator of UPR (23, 37).
Table 3.
Parasite eIF2α kinases with transmembrane domain(s)a
| Species | Accession no. | TM domain(s) |
|---|---|---|
| Toxoplasma gondii | TGME49_029630 | 2915–2937; 2942–2964 |
| Neospora caninum | NCLIV_030460 | 2977–2999; 3004–3026 |
| Plasmodium falciparum | PFF1370w | 108–130 |
| Cryptosporidium hominis | Chro.10109 | 20–42 |
| Cryptosporidium parvum | cgd1_890 | 20–39 |
| Theileria parva | TP01_1167 | 175–197; 674–696 |
| Theileria annulata | TA16695 | 183–205; 552–569; 701–723 |
List of putative parasite eIF2α kinases with a predicted transmembrane (TM) domain. The numbers presented under the TM domain represent the encoded amino acid residue positions for each transmembrane segment.
ER stress induced by tunicamycin reduces global protein synthesis.
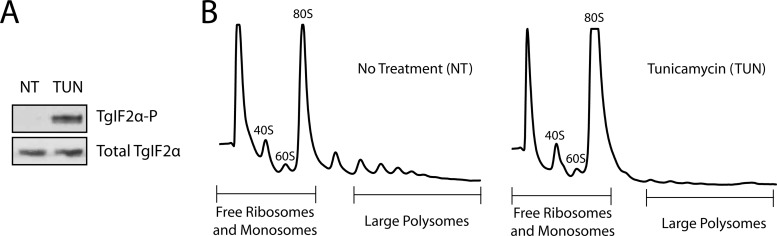
Given the presence of a potential PERK orthologue in TgIF2K-A, we assessed if tunicamycin treatment induced TgIF2α phosphorylation and a subsequent decrease in general protein synthesis using the conditions for ER stress outlined above (10 μM tunicamycin or vehicle for 1 h at 37°C). Parasites treated with tunicamycin showed a pronounced increase in TgIF2α phosphorylation relative to those treated with the vehicle control (Fig. 3A), consistent with our previous results (10). To evaluate the effect of tunicamycin on mRNA translation, parasites exposed to tunicamycin or vehicle were subjected to polysome fractionation by using sucrose gradient centrifugation. Following exposure to tunicamycin, parasites showed a significant reduction in mRNA-bound polyribosomes (two or more ribosomes) and a concomitant increase in free ribosomes and monoribosome-bound transcripts (Fig. 3B). These results support the idea that translational control is a central feature of the UPR in Toxoplasma.
Fig 3.
ER stress represses global translation initiation in Toxoplasma. (A) Equal amounts of lysate from parasites treated with tunicamycin (TUN) or vehicle (DMSO) were separated in a 4 to 12% polyacrylamide gel and transferred for Western blotting with antibodies that specifically recognize TgIF2α phosphorylated at Ser-71 (TgIF2α-P) or total TgIF2α protein. (B) Polysome profiles were generated from parasites exposed to 10 μM tunicamycin (TUN) or no treatment (NT) for 1 h. Fractions containing free RNA, ribosomal subunits, and the 80S monoribosomes were collected and pooled into a single tube (termed free ribosomes and monosomes). A second pool of fractions containing mRNAs engaged with three or more ribosomes (large polysomes) were collected for subsequent microarray analysis.
Preferential translation is suggested to facilitate the Toxoplasma UPR.
It has been established in other species that eIF2α phosphorylation induces preferential translation of a subset of mRNAs. To identify mRNAs that are suggested to be preferentially translated during ER stress in Toxoplasma, polysome profiles were generated from tachyzoites subjected to tunicamycin versus vehicle (Fig. 3B). An increase in mRNA associating with polysomes during ER stress would strongly suggest enhanced translation of the gene transcript. RNA was purified from fractions containing (i) free ribosomes and monosomes or (ii) actively translating polyribosomes. The purified RNA samples were processed for hybridization to ToxoGeneChip microarrays as discussed above (Fig. 1). Polysome association was calculated for each gene transcript as the percentage of the mRNA in the polyribosome fraction from parasites cultured in the presence of tunicamycin or DMSO (see Materials and Methods). To evaluate the change in global translation, a histogram plot was generated to illustrate the percentage of polysome mRNA for each probe set (percent polysomal) following exposure to tunicamycin or the DMSO vehicle control (Fig. 4A). Parasites cultured in the absence of ER stress had a significantly larger population of mRNAs that were primarily engaged with polysomes (≥60% of gene transcripts associated with polysomes) relative to parasites exposed to tunicamycin, further indicating that the ER stress agent causes a pronounced reduction in translation initiation (Fig. 4A). Furthermore, these results are consistent with those shown in Fig. 3, confirming the fidelity of the polysome microarray strategy to evaluate changes in global translation in response to ER stress. Interestingly, a population of mRNAs was suggested to be poorly translated (∼10% of the gene transcripts associated with polysomes) in the absence of stress (Fig. 4A, arrow). In mammals and yeast, preferentially translated mRNAs, such as ATF4 and its counterpart in yeast, the bZIP transcriptional activator GCN4, are each poorly translated in the absence of stress (38, 39).
Fig 4.
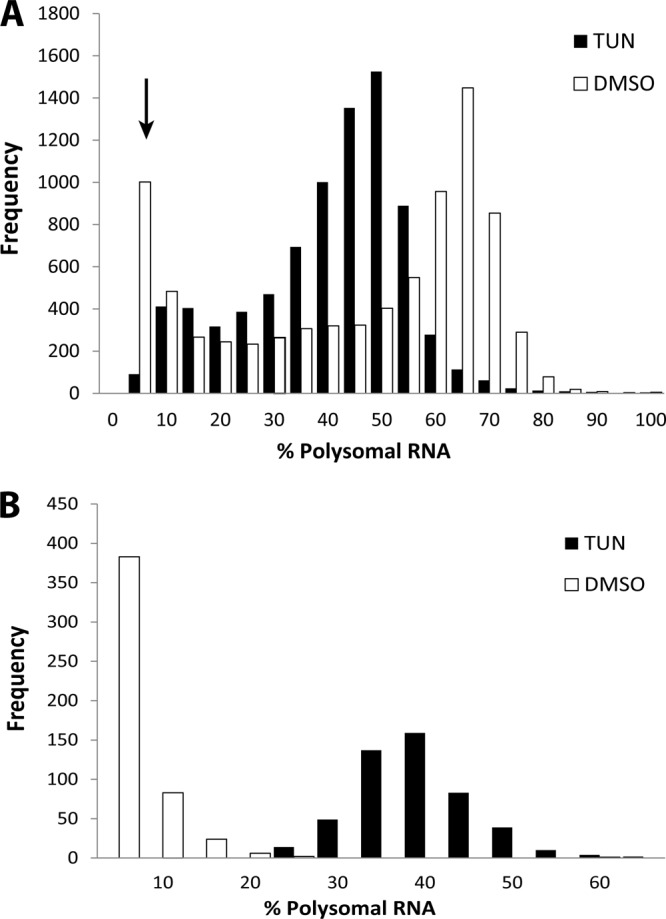
Polysome association of Toxoplasma gene transcripts is suggested to be reduced in response to ER stress. (A) Histogram representing the number of different gene transcripts (y axis) associated with polysomes (% polysomal RNA) from parasites cultured in the presence of tunicamycin (TUN) or vehicle (black and white bars, respectively). The arrow denotes a subset of mRNAs that are poorly translated in the absence of stress. (B) The percentage of polysomal RNA of the preferentially translated (i.e., enhanced) transcripts (following exposure to tunicamycin) represented in a histogram in the presence of tunicamycin or vehicle (black and white bars, respectively).
To identify the Toxoplasma mRNAs that are suggested to be preferentially translated in response to tunicamycin treatment, we filtered our data for probe sets that showed a statistically significant (P ≤ 0.01) increase in polysome abundance following exposure to ER stress. Interestingly, a large subset of gene transcripts (501 genes) was significantly enriched in the polyribosome fractions following exposure to tunicamycin, with 499 of these mRNAs showing a ≥20% increase in association with polysomes in response to tunicamycin treatment (see Data Set S2 in the supplemental material). A second histogram was generated for gene transcripts suggested to be preferentially translated under conditions of ER stress (Fig. 4B). As expected, in the absence of ER stress, a majority of these gene transcripts were found predominantly in the free ribosomes and monosome fractions (only ∼10% polysome association). Following treatment with tunicamycin, there was a pronounced shift in these mRNAs to the polyribosome fraction, consistent with higher translation efficiency during ER stress. The fidelity of our microarray data was confirmed using qPCR analysis of mRNAs subject to enhanced or repressed polysome association following exposure to tunicamycin (see Table S2). Interestingly, the collection of gene transcripts showing enhanced polysome association shows no significant overlap with mRNAs whose levels are induced upon ER stress (Table 2; also see Data Set S1).
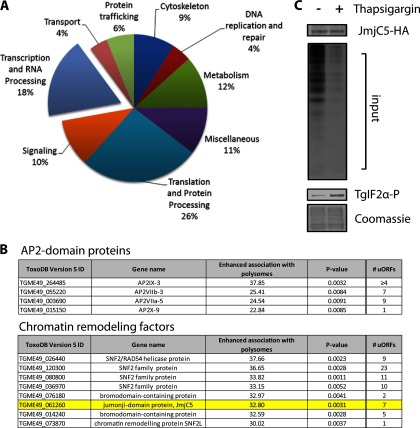
The genes suggested to be subject to preferential translation encode proteins present in all major GO categories (Fig. 5A). The greatest representation was found in two categories: translation and protein processing (26%) and transcription and RNA processing (18%), both of which were significantly greater (P < 0.001) than the 6 and 4% representation estimated for the entire Toxoplasma genome. In comparison, 12% of the gene transcripts preferentially associated with polysomes upon ER stress involved metabolism, which was significantly lower than the 18% predicted genome-wide representation (P < 0.001). These results suggest that genes subject to preferential translation in response to ER stress are enriched for functions in mRNA and protein expression.
Fig 5.
Subset of transcriptional regulators is suggested to be preferentially translated in response to ER stress. (A) Pie chart displays GO categories of genes that were suggested to be preferentially translated following treatment with tunicamycin (P < 0.02). (B) A list of preferentially translated AP2 factors and chromatin remodeling factors that are suggested to be preferentially translated during ER stress. The chart indicates enhanced association with polysomes (percent shift to polysome fraction) following tunicamycin treatment and the predicted number of uORFs within the 5′-UTR of each preferentially translated mRNA. (C) Parasites were subjected to ER stress by treatment with 10 μM thapsigargin or control vehicle (DMSO) and incubated with [35S]Met/Cys to radiolabel the synthesized proteins in each sample for 90 min. Two percent of each sample (input) was resolved by SDS-PAGE, followed by autoradiography (second panel from top). Coomassie blue staining indicated equal loading of input protein between the DMSO- and thapsigargin-treated samples (bottom panel). JmjC5-HA was immunoprecipitated from each lysate using anti-HA-coupled agarose beads and subjected to autoradiography after being resolved by SDS-PAGE (top panel). Immunoblotting with antibodies that specifically recognize TgIF2α phosphorylated at Ser-71 (TgIF2α-P) was performed to confirm induction of ER stress by thapsigargin treatment (third panel from the top).
Interestingly, we identified several parasite-specific AP2 factors that are suggested to be subject to preferential translation in response to ER stress (Fig. 5B). It is inviting to speculate that preferentially translated AP2 factors serve as the parasite counterpart to bZIP transcription factors that coordinate gene expression for the UPR in other species (2). Consistent with this idea, a recent report has demonstrated that preferential translation of AP2 transcription factors occurs in Arabidopsis following exposure to heat shock or high-salt conditions (40). Additionally, a number of genes involving transcription through chromatin remodeling were also suggested to be subject to preferential translation during ER stress (Fig. 5B). These included members of the SNF2 family of DNA-dependent ATPases and JmjC5, a jumonji C domain-containing lysine demethylase (TGME49_061260). In other species, these proteins have roles in chromatin regulation and development (41, 42).
Potential regulatory features of UPR genes suggested to be preferentially translated.
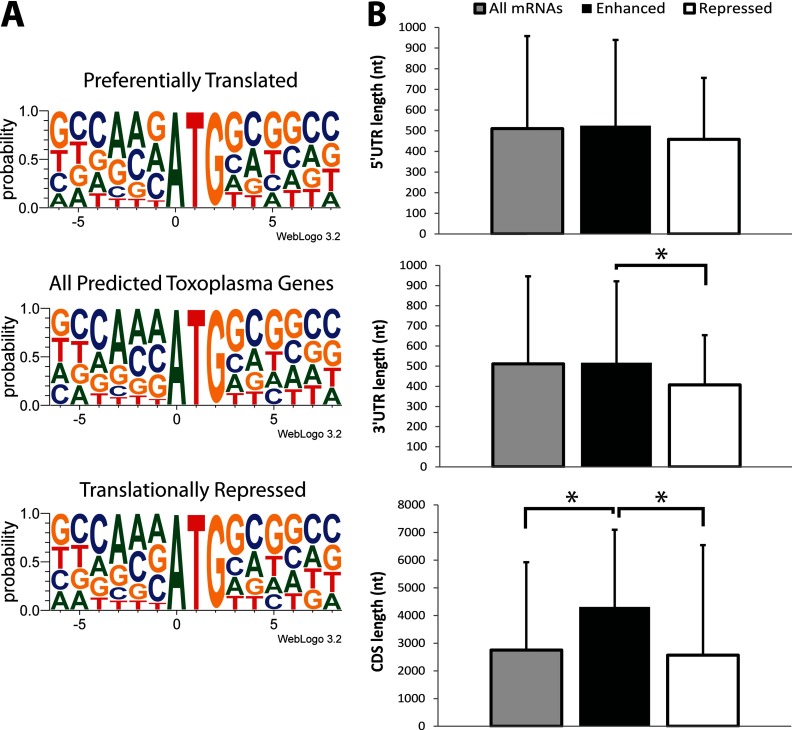
We wished to address whether the mRNAs that are preferentially associated with polysomes in response to ER stress share sequence or structural features. Bioinformatics analyses were performed to determine if the Kozak sequence context for the initiation codons or UTR length were associated with the suggested translational regulation of mRNAs in response to ER stress (Fig. 6A). The predicted Kozak sequences for initiation codons of coding sequences in Toxoplasma genes were divided into three groups: (i) all Toxoplasma genes present on the ToxoGeneChip, (ii) those suggested to be preferentially translated genes in response to ER stress, and (iii) genes that are suggested to be translationally repressed. The Kozak sequence for the predicted initiation codons for all three groups was similar (Fig. 6A), suggesting that the changes in polysome abundance of the gene transcripts in response to ER stress was not due to poor Kozak context. We note that our analysis of the initiation codon context indicates a prevalence for AAAATGG (boldface indicates translational start codon), which is consistent with a previous report by Seeber (43).
Fig 6.
Bioinformatic analyses of Toxoplasma gene transcripts suggested to be preferentially translated in response to ER stress. (A) Comparison of the Kozak sequences for predicted initiation codons from all Toxoplasma genes, as well as those subject to preferential translation or translational repression in response to tunicamycin exposure. (B) Length of the annotated 5′-UTRs (top), 3′-UTRs (middle), and ORF (bottom) for all annotated gene transcripts (gray bar), those suggested to be subject to preferential translation (enhanced) (black bar), and those that are translationally repressed (white bar). An asterisk denotes statistically significant difference (P ≤ 0.05). CDS, coding DNA sequence.
We next addressed whether the preferentially translated mRNAs had longer UTRs, which may function to mediate translational control in a stress-dependent manner. We observed no significant differences in 5′-UTR or 3′-UTR length between the genes suggested to be preferentially translated compared to all predicted Toxoplasma genes (Fig. 6A). However, the translationally repressed mRNAs had a modestly smaller 3′-UTR relative to mRNAs that were preferentially translated in response to tunicamycin (Fig. 6B). It is noted that presently only ∼30% of Toxoplasma genes in the ToxoDB have annotated UTRs, many of which have yet to be validated; therefore, this analysis will be strengthened with more complete annotation of the Toxoplasma genome database. Furthermore, the coding regions for mRNAs that were preferentially associated with polysomes upon ER stress were significantly longer than those of all Toxoplasma mRNAs, as well as mRNAs that were suggested to be translationally repressed (Fig. 6B). This suggests that the gene transcripts identified to be preferentially associated with polysomes were enriched for those mRNAs that can accommodate a larger number of elongating ribosomes distributed along extended coding regions.
The preferential translation of GCN4 and ATF4 in yeast and mammals, respectively, is regulated through the presence of multiple upstream open reading frames (uORFs) in the 5′ leaders of these mRNAs. Under nonstressed conditions, the ribosome initiates translation at a 5′-proximal short uORF that facilitates reinitiation at a downstream inhibitory uORF(s), preventing translation of the ATF4 and GCN4 coding sequences. During stress, eIF2α phosphorylation inhibits its guanine nucleotide exchange factor, eIF2B, leading to a decrease in eIF2-GTP levels. As a result, the time required for ribosomes to reinitiate following translation of the 5′-proximal uORF is extended. Only after scanning through the inhibitory uORFs are the ribosomes thought to reacquire the eIF2/GTP/Met-tRNAiMet ternary complex, facilitating translation of the ATF4 and GCN4 coding sequences. Therefore, we analyzed the annotated 5′-UTRs in ToxoDB, as well as RNA sequencing data of the Toxoplasma genome, to identify uORFs in the predicted 5′ leader sequence in the preferentially translated AP2 mRNAs (44). These 5′-UTR sequences suggest that mRNAs encoding each of these AP2 factors and the chromatin remodeling factors mentioned above contain one or more uORFs adjacent to the protein-coding ORF (Fig. 5B), supporting the idea that their mRNAs are preferentially translated through a mechanism analogous to those of ATF4 and GCN4 (39, 45, 46). We also note that Toxoplasma contains predicted homologues for eIF2A (TGME49_258740) and eIF2D (TGME49_211410), which have been reported to facilitate delivery of Met-tRNAiMet to the P site of ribosomes independently of the canonical translation initiation factor eIF2 (47–49). eIF2A was reported to facilitate translation initiation by internal ribosome entry sites (IRESs), and this mode of translational control is suggested to facilitate preferential translation of mRNAs independently of eIF2 and uORFs (50, 51).
JmjC5 is actively translated during ER stress.
To address whether ER stress is capable of inducing preferential translation of a transcriptional regulator identified in our polysome microarray studies, we monitored the synthesis of JmjC5, which we endogenously tagged with a triple HA epitope at the C terminus. Parasites treated with thapsigargin, a potent SERCA pump inhibitor and a widely described ER stressor (52), led to robust phosphorylation of TgIF2α and a global reduction in protein synthesis as measured by [35S]Met/Cys incorporation (Fig. 5C). Equivalent amounts of input protein were confirmed by Coomassie staining of the thapsigargin-treated and DMSO control samples. In comparison, the tagged JmjC5 protein continued to be synthesized, as visualized by [35S]Met/Cys radiolabeling during treatment with thapsigargin (Fig. 5C, top). The undiminished presence of JmjC5 protein in the stressed sample strongly supports the idea that mRNAs identified in the polysomal fraction continue to be translated during TgIF2α phosphorylation and global repression of protein synthesis.
Based on phylogenetic analyses, the human homologue of TgJmjC5 is jumonji domain-containing 6 (JMJD6), which is reported to have histone arginine demethylation and lysyl hydroxylation activity (53–56), with potential biological functions in gene expression (53, 57, 58). Preliminary knockout studies in Toxoplasma suggest that the TgJmjC5 gene results in a growth defect in tachyzoites (Z. Tampaki and K. Kim, unpublished results), and an important question for the future is the precise role of TgJmjC5 in parasitic stress responses, including those affecting the ER.
Conclusions.
Bioinformatic analysis of protozoan parasites suggests that these early-branching eukaryotes have both unique and conserved features associated with the UPR (59). While Toxoplasma appears to lack ATF6 and IRE1 homologues, it possesses an ER-localized eIF2α kinase, TgIF2K-A. These data suggest that PERK is an ancient regulator of the UPR, which may differ from the view that IRE1 is the most evolutionarily ancient ER stress sensor (60, 61). ER stresses cause robust phosphorylation of TgIF2α and a concomitant reduction in translation initiation (Fig. 3 and 5C). RNA purified from total lysates as well as sucrose gradient fractions provided insight into the regulation of coordinated changes in the transcriptome and gene-specific translation during ER stress. Interestingly, the results indicate that a dynamic set of changes take place to conserve resources by repressing global translation while enhancing translation and transcription of a core set of genes to remedy the underlying cellular stress. Most notably, the acute stress appears to enhance translation of transcriptional regulators driving parasite adaptation.
Supplementary Material
ACKNOWLEDGMENTS
Support for this research was provided by the National Institutes of Health R21 grant AI084031 (R.C.W. and W.J.S.), R01 GM049164 (R.C.W.), R01 AI077502 (W.J.S.), R01 AI087625 (K.K.), RC4AI092801 (K.K. and W.J.S.), and F32 AI093271 (B.R.J.).
The microarray analyses were carried out in conjunction with the Center for Medical Genomics at the IUSM. We thank Jeanette McClintick at the Center for advice on the microarray analyses. We also thank Christian Konrad and other members of the Sullivan and Wek laboratories for helpful discussions.
Footnotes
Published ahead of print 10 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00021-13.
REFERENCES
- 1. Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086 [DOI] [PubMed] [Google Scholar]
- 2. Wek RC, Cavener DR. 2007. Translational control and the unfolded protein response. Antioxid. Redox Signal. 9:2357–2371 [DOI] [PubMed] [Google Scholar]
- 3. Yoshida H, Haze K, Yanagi H, Yura T, Mori K. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273:33741–33749 [DOI] [PubMed] [Google Scholar]
- 4. Sullivan WJ, Jr, Jeffers V. 2012. Mechanisms of Toxoplasma gondii persistence and latency. FEMS Microbiol. Rev. 36:717–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang M, Joyce BR, Sullivan WJ, Jr, Nussenzweig V. 2012. Translational control in Plasmodium and Toxoplasma parasites. Eukaryot. Cell 12:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luk FC, Johnson TM, Beckers CJ. 2008. N-linked glycosylation of proteins in the protozoan parasite Toxoplasma gondii. Mol. Biochem. Parasitol. 157:169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Black MW, Arrizabalaga G, Boothroyd JC. 2000. Ionophore-resistant mutants of Toxoplasma gondii reveal host cell permeabilization as an early event in egress. Mol. Cell. Biol. 20:9399–9408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naguleswaran A, Alaeddine F, Guionaud C, Vonlaufen N, Sonda S, Jenoe P, Mevissen M, Hemphill A. 2005. Neospora caninum protein disulfide isomerase is involved in tachyzoite-host cell interaction. Int. J. Parasitol. 35:1459–1472 [DOI] [PubMed] [Google Scholar]
- 9. Harbut MB, Patel BA, Yeung BK, McNamara CW, Bright AT, Ballard J, Supek F, Golde TE, Winzeler EA, Diagana TT, Greenbaum DC. 2012. Targeting the ERAD pathway via inhibition of signal peptide peptidase for antiparasitic therapeutic design. Proc. Natl. Acad. Sci. U. S. A. 109:21486–21491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Narasimhan J, Joyce BR, Naguleswaran A, Smith AT, Livingston MR, Dixon SE, Coppens I, Wek RC, Sullivan WJ., Jr 2008. Translation regulation by eukaryotic initiation factor-2 kinases in the development of latent cysts in Toxoplasma gondii. J. Biol. Chem. 283:16591–16601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chow C, Cloutier S, Dumas C, Chou MN, Papadopoulou B. 2011. Promastigote to amastigote differentiation of Leishmania is markedly delayed in the absence of PERK eIF2alpha kinase-dependent eIF2alpha phosphorylation. Cell. Microbiol. 13:1059–1077 [DOI] [PubMed] [Google Scholar]
- 12. Zhang M, Fennell C, Ranford-Cartwright L, Sakthivel R, Gueirard P, Meister S, Caspi A, Doerig C, Nussenzweig RS, Tuteja R, Sullivan WJ, Jr, Roos DS, Fontoura BM, Menard R, Winzeler EA, Nussenzweig V. 2010. The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J. Exp. Med. 207:1465–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roos DS, Donald RG, Morrissette NS, Moulton AL. 1994. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27–63 [DOI] [PubMed] [Google Scholar]
- 14. Duksin D, Mahoney WC. 1982. Relationship of the structure and biological activity of the natural homologues of tunicamycin. J. Biol. Chem. 257:3105–3109 [PubMed] [Google Scholar]
- 15. Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. 2008. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2:448–460 [DOI] [PubMed] [Google Scholar]
- 16. Bahl A, Davis PH, Behnke M, Dzierszinski F, Jagalur M, Chen F, Shanmugam D, White MW, Kulp D, Roos DS. 2010. A novel multifunctional oligonucleotide microarray for Toxoplasma gondii. BMC Genomics 11:603. 10.1186/1471-2164-11-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 19. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 20. Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, Pinney DF, Roos DS, Stoeckert CJ, Jr, Wang H, Brunk BP. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36:D553–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huynh MH, Carruthers VB. 2009. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8:530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donald RG, Carter D, Ullman B, Roos DS. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019 [DOI] [PubMed] [Google Scholar]
- 23. Mori K. 2009. Signalling pathways in the unfolded protein response: development from yeast to mammals. J. Biochem. 146:743–750 [DOI] [PubMed] [Google Scholar]
- 24. Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429:841–847 [DOI] [PubMed] [Google Scholar]
- 25. Lilley BN, Ploegh HL. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429:834–840 [DOI] [PubMed] [Google Scholar]
- 26. Zhang JX, Braakman I, Matlack KE, Helenius A. 1997. Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C-terminal truncations. Mol. Biol. Cell 8:1943–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. 2004. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15:2361–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dixon SE, Stilger KL, Elias EV, Naguleswaran A, Sullivan WJ., Jr 2010. A decade of epigenetic research in Toxoplasma gondii. Mol. Biochem. Parasitol. 173:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Painter HJ, Campbell TL, Llinas M. 2011. The apicomplexan AP2 family: integral factors regulating Plasmodium development. Mol. Biochem. Parasitol. 176:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balaji S, Babu MM, Iyer LM, Aravind L. 2005. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33:3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hollien J, Weissman JS. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313:104–107 [DOI] [PubMed] [Google Scholar]
- 32. Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186:323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quenault T, Lithgow T, Traven A. 2011. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21:104–112 [DOI] [PubMed] [Google Scholar]
- 34. Muller K, Matuschewski K, Silvie O. 2011. The Puf-family RNA-binding protein Puf2 controls sporozoite conversion to liver stages in the malaria parasite. PLoS One 6:e19860. 10.1371/journal.pone.0019860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gomes-Santos CS, Braks J, Prudencio M, Carret C, Gomes AR, Pain A, Feltwell T, Khan S, Waters A, Janse C, Mair GR, Mota MM. 2011. Transition of Plasmodium sporozoites into liver stage-like forms is regulated by the RNA binding protein Pumilio. PLoS Pathog. 7:e1002046. 10.1371/journal.ppat.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326–332 [DOI] [PubMed] [Google Scholar]
- 37. Trusina A, Papa FR, Tang C. 2008. Rationalizing translation attenuation in the network architecture of the unfolded protein response. Proc. Natl. Acad. Sci. U. S. A. 105:20280–20285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 100:3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 101:11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsuura H, Ishibashi Y, Shinmyo A, Kanaya S, Kato K. 2010. Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol. 51:448–462 [DOI] [PubMed] [Google Scholar]
- 41. Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. 2006. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev. Dyn. 235:2449–2459 [DOI] [PubMed] [Google Scholar]
- 42. Euskirchen G, Auerbach RK, Snyder M. 2012. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 287:30897–30905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seeber F. 1997. Consensus sequence of translational initiation sites from Toxoplasma gondii genes. Parasitol. Res. 83:309–311 [DOI] [PubMed] [Google Scholar]
- 44. Yamagishi J, Watanabe J, Goo YK, Masatani T, Suzuki Y, Xuan X. 2012. Characterization of Toxoplasma gondii 5′ UTR with encyclopedic TSS information. J. Parasitol. 98:445–447 [DOI] [PubMed] [Google Scholar]
- 45. Hinnebusch AG. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407–450 [DOI] [PubMed] [Google Scholar]
- 46. Baird TD, Wek RC. 2012. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 3:307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reineke LC, Merrick WC. 2009. Characterization of the functional role of nucleotides within the URE2 IRES element and the requirements for eIF2A-mediated repression. RNA 15:2264–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reineke LC, Komar AA, Caprara MG, Merrick WC. 2008. A small stem loop element directs internal initiation of the URE2 internal ribosome entry site in Saccharomyces cerevisiae. J. Biol. Chem. 283:19011–19025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 285:26779–26787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thakor N, Holcik M. 2012. IRES-mediated translation of cellular messenger RNA operates in eIF2alpha-independent manner during stress. Nucleic Acids Res. 40:541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim JH, Park SM, Park JH, Keum SJ, Jang SK. 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 30:2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lytton J, Westlin M, Hanley MR. 1991. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 266:17067–17071 [PubMed] [Google Scholar]
- 53. Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR, III, Delahunty CM, Hahn P, Lengeling A, Mann M, Proudfoot NJ, Schofield CJ, Bottger A. 2009. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325:90–93 [DOI] [PubMed] [Google Scholar]
- 54. Cui P, Qin B, Liu N, Pan G, Pei D. 2004. Nuclear localization of the phosphatidylserine receptor protein via multiple nuclear localization signals. Exp. Cell Res. 293:154–163 [DOI] [PubMed] [Google Scholar]
- 55. Chang B, Chen Y, Zhao Y, Bruick RK. 2007. JMJD6 is a histone arginine demethylase. Science 318:444–447 [DOI] [PubMed] [Google Scholar]
- 56. Unoki M, Masuda A, Dohmae N, Arita K, Yoshimatsu M, Iwai Y, Fukui Y, Ueda K, Hamamoto R, Shirakawa M, Sasaki H, Nakamura Y. 2013. Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem. 288:6053–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boeckel JN, Guarani V, Koyanagi M, Roexe T, Lengeling A, Schermuly RT, Gellert P, Braun T, Zeiher A, Dimmeler S. 2011. Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc. Natl. Acad. Sci. U. S. A. 108:3276–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hong X, Zang J, White J, Wang C, Pan CH, Zhao R, Murphy RC, Dai S, Henson P, Kappler JW, Hagman J, Zhang G. 2010. Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 107:14568–14572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gosline SJ, Nascimento M, McCall LI, Zilberstein D, Thomas DY, Matlashewski G, Hallett M. 2011. Intracellular eukaryotic parasites have a distinct unfolded protein response. PLoS One 6:e19118. 10.1371/journal.pone.0019118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, Aragon T, Li H, Walter P. 2012. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLife 1:e00048. 10.7554/eLife.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bernales S, Papa FR, Walter P. 2006. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22:487–508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.