Abstract
Lactococcal dairy starter strains are under constant threat from phages in dairy fermentation facilities, especially by members of the so-called 936, P335, and c2 species. Among these three phage groups, members of the P335 species are the most genetically diverse. Here, we present the complete genome sequences of two P335-type phages, Q33 and BM13, isolated in North America and representing a novel lineage within this phage group. The Q33 and BM13 genomes exhibit homology, not only to P335-type, but also to elements of the 936-type phage sequences. The two phage genomes also have close relatedness to phages infecting Enterococcus and Clostridium, a heretofore unknown feature among lactococcal P335 phages. The Q33 and BM13 genomes are organized in functionally related clusters with genes encoding functions such as DNA replication and packaging, morphogenesis, and host cell lysis. Electron micrographic analysis of the two phages highlights the presence of a baseplate more reminiscent of the baseplate of 936 phages than that of the majority of members of the P335 group, with the exception of r1t and LC3.
INTRODUCTION
Lactococcal phages have been the focus of intensive research for many years due to their significant negative impact on the dairy industry. They are currently divided into 10 genetically distinct groups based on morphological and genomic analyses (1). The P335 phage group is among the most often encountered in dairy fermentations globally, along with members of the virulent 936 and c2 species (2). Many reports of the isolation and characterization of members of the P335 group have emerged, particularly since the 1990s. To date, the complete genome sequences of at least eight P335 phages are available (not including predicted prophages that are part of sequenced Lactococcus lactis genomes), namely, the reference phage P335, as well as Tuc2009, TP901-1, BK5-T, r1t, LC3, ul36, and 4268 (3–10). These phages were given this group assignment based on Southern hybridization and morphological analyses with subsequent confirmation by comparative sequence analysis (1). One of the characteristics of these phages is their genome plasticity (11). Their genome sequences are rather divergent and may be grouped loosely into the P335/ul36/Tuc2009/TP901-1, BK5-T/4268, and r1t/LC3 subgroups based on sequence homology or the presence of homologous functional regions (1, 6). While these phages are grouped together based on sequence homology, there is no single genetic feature common to all members of the lactococcal phage group, making it difficult to define new members (1). The presence of a predicted dUTPase-encoding gene on the genome has been used as a marker for members of the group (5); however, the genome of a recent P335-type isolate, phage 4268, does not contain a homologue of this gene, thus further substantiating the polythetic nature of the phage group (9).
The virulent phage Q33 was originally isolated from a buttermilk sample in 1994 in North America (named Q36 in the original description but Q33 in all subsequent publications), and its genome was estimated to be approximately 30 kb through restriction endonuclease profiling (12). In the original description of Q33, a weak serological reaction with antibodies against capsid proteins of ul36 and weak homology to P335 in hybridization studies pointed to potential genomic diversity of the phage. The phage has been used as a representative of the P335-type group in several studies of phage resistance mechanisms (13–16) and is known to be sensitive to the effects of the restriction/modification system LlaDII (15) and engineered phage-encoded resistance (Per) systems (17). In contrast, Q33 is unaffected by the DNA injection-blocking system, Sie2009, encoded by the temperate P335 phage Tuc2009 (16). Through analysis of engineered Per systems, regions of the Q33 genome encoding the replisome organizer (accession no. AF264634.1) and the replication module (accesssion no. AF264635.1) have been sequenced, while the sak (sensitivity to AbiK) gene and another predicted open reading frame (ORF 152) (accession no. AY365424.1) of Q33 were identified and sequenced as part of a study on lactococcal phage sensitivity to AbiK (17, 18).
The above-mentioned studies appear to highlight Q33 as a typical member of the P335-type species. However, a recent study on the structure of the injection devices of lactococcal phages using TP901-1 as a model suggested a bipartite grouping of the P335-type phages based on their requirement for calcium for efficient plaque formation (19). In that study, Q33 was observed to behave like phages belonging to the 936-type species in being strictly dependent on calcium for plaque formation. This finding is in stark contrast to the majority of tested P335-type phages, which do not require calcium for plaque formation. Moreover, phage Q33 has a larger host range than other P335-like phages. These observations prompted the current genome sequence study in order to further investigate its strict requirement for calcium for plaque formation.
The virulent phage BM13 was isolated in March 2011 from cheddar whey in Canada as part of a routine phage detection program by a cheese plant. PCR assay classified the phage in the P335 group (20). Characterization of the phage BM13 host range on several L. lactis strains used at the same cheese plant revealed a broad host range, an unusual characteristic among P335 phages. Phages with the same restriction profiles as BM13 were also isolated within the same cheese factory in August 2011, September 2011, October 2011, December 2011, and February 2012, suggesting that the phage (or close derivatives) can persist for some time in the dairy environment. The titers of phage BM13 varied from 104 PFU/ml to 108 PFU/ml in these samples. However, the phage has not been reisolated since. During the same period, several other, distinct lactococcal phages were isolated from additional cheddar wheys from the cheese factory, but they all belong to the 936 group.
Sequencing of the complete genomes of BM13 and Q33 was undertaken, and a comparison of their sequences relative to each other and other members of the polythetic P335 phage species was performed.
MATERIALS AND METHODS
Bacteria, phages, and growth conditions.
Phage Q33 was propagated on strain L. lactis SMQ-86, while BM13 was propagated on L. lactis SMQ-1212 at 30°C in M17 broth (Oxoid) supplemented with 0.5% glucose (Q33) or 0.5% lactose (BM13) without agitation. Plaque assays were performed using the double-agar method previously described (21). This method was also applied for the host range analysis performed against a bank of 100 L. lactis strains (only the sensitive strains are listed in Table 1).
Table 1.
Host range analysis of Q33 and BM13 against lactococcal strains
| Lactococcal straina | Reference/source | Infectionb |
|
|---|---|---|---|
| Q33 | BM13 | ||
| 223 (c) | UCC Strain Collection | + | − |
| UC77 (c) | UCC Strain Collection | + | − |
| WM1 (l) | UCC Strain Collection | + | − |
| 229 (c) | UCC Strain Collection | + | + |
| 275 | UCC Strain Collection | − | + |
| FD13 (c) | 14 | + | − |
| Bu2 (d) | 47 | + | − |
| ML8 (l) | 1 | + | − |
| IL1403 (l) | 48 | + | − |
| SMQ86 (c) | 11 | + | + |
| R1 (c) | 10 | − | + |
| SMQ347 (c) | Industrial strain used in Canada | − | + |
| SMQ436 (c) | Industrial strain used in Canada | − | + |
| SMQ447 (c) | Industrial strain used in Canada | − | + |
| SMQ748 (c) | Industrial strain used in Canada | − | + |
| SMQ831 (c) | Industrial strain used in Canada | − | + |
| SMQ998 (c) | Industrial strain used in Canada | − | + |
| SMQ1003 (c) | Industrial strain used in Canada | − | + |
| SMQ1009 (c) | Industrial strain used in Canada | − | + |
| SMQ1016 (c) | Industrial strain used in Canada | − | + |
| SMQ1212 | Industrial strain used in Canada | − | + |
| SMQ1214 (l) | Industrial strain used in Canada | − | + |
| SMQ1215 (l) | Industrial strain used in Canada | − | + |
| SMQ1216 (l) | Industrial strain used in Canada | − | + |
| SMQ1217 (l) | Industrial strain used in Canada | − | + |
| SMQ1218 (d) | Industrial strain used in Canada | + | − |
| SMQ1219 (c) | Industrial strain used in Canada | − | + |
| SMQ1220 (d) | Industrial strain used in Canada | − | + |
(l) indicates a strain of L. lactis subsp. lactis, (d) indicates a strain of L. lactis subsp. lactis biovar diacetylactis, and (c) indicates a strain of L. lactis subsp. cremoris.
+, positive; −, negative.
DNA preparation.
DNA for sequencing of the phage Q33 genome was extracted from 50 ml of fresh phage lysate (>108 PFU · ml−1) by treatment with 1 μg · ml−1 DNase and RNase at 37°C for 30 min. After centrifugation at 12,000 rpm for 15 min, the lysate was transferred to a new tube, and 10% polyethylene glycol (PEG) 8000 (Sigma-Aldrich, St. Louis, MO, USA) and 0.5 M sodium chloride were added, after which the suspension was incubated at 4°C overnight. Subsequently, the suspension was centrifuged at 17,700 × g for 15 min and the supernatant was removed. The PEG/salt-induced precipitate was resuspended in 5 ml of Tris-EDTA (TE) buffer (pH 9.0) and treated with 120 μl of 20 mg · ml−1 proteinase K for 20 min at 56°C, followed by treatment with SDS at a final concentration of 2% at 65°C for 20 min. Potassium acetate was added to a final concentration of 1 M, followed by incubation on ice for 20 min before centrifugation at 13,200 × g for 10 min. The supernatant was then treated with phenol-chloroform (25:24:1 phenol-chloroform-isoamyl alcohol; Sigma-Aldrich, St. Louis, MO, USA) at least twice, and the aqueous phase was precipitated with 2.5 volumes of ice-cold 96% ethanol and 0.1 volume of sodium acetate (pH 4.8). Subsequent to centrifugation, the pellet was washed in 70% ethanol and resuspended in 100 μl of TE buffer (pH 8.0). DNA of phage BM13 was extracted using the Qiagen Maxi-Prep kit with modifications as previously described (22).
Genome sequencing, assembly, and annotation.
Five micrograms of DNA was extracted and verified by nanodrop quantification. Confirmatory molecular identification (ID) tests were also conducted on the DNA extract prior to shipment to the contract sequencing facility (Macrogen Inc., Seoul, South Korea, for Q33). A 120-fold sequencing coverage was obtained using pyrosequencing technology on a 454 FLX instrument. Sequencing of phage BM13 was performed using a similar instrument and executed at the Plateforme d'Analyses Génomiques of the Institut de Biologie Intégrative et des Systèmes (Université Laval, Québec, Canada). A 27-fold sequencing coverage was obtained for BM13. The individual sequence files generated by the 454 FLX instrument were assembled with GSassembler (454 Lifesciences, Branford, CT, USA) to generate a consensus sequence. Quality improvement of the genome sequence involved sequencing of 15 (Q33) and 8 (BM13) PCR products across the entire genomes to ensure correct assembly, double stranding, and the resolution of any remaining base conflicts occurring within homopolymer tracts. Protein-encoding ORFs were predicted using Zcurve_V, followed by manual assessment, curation, and correction of the predicted open reading frames (23). ORFs were also determined/verified using the ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) and GeneMark (http://exon.gatech.edu/) software packages and manually by looking for ATG, TTG, and GTG as potential start codons downstream of a consensus ribosome binding sequence (AGAAAGGAGGTG) (3). A functional annotation of ORFs was performed on the basis of BLASTP (1) analysis against the nonredundant (nr) protein database provided by the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The genome was scanned for the presence of potential tRNA genes using tRNA scan SE (24).
Electron microscopy.
Purification of phages Q33 and BM13 by CsCl gradient was performed as previously described (25). Adsorption of CsCl-purified phages to freshly prepared carbon film floated from a freshly coated mica sheet and negative staining with 2% (wt/vol) uranyl acetate were done as described previously (26). The film was picked up with a 400-mesh copper grid (Agar Scientific, Essex, United Kingdom), and specimens were examined with a Tecnai 10 transmission electron microscope (TEM) (FEI, Eindhoven, The Netherlands) operated at an acceleration voltage of 80 kV. Images of other P335 species phage members (BK5-T, Tuc2009, r1t, and phiLC3 [1] and P335 [6]) were taken from the TEM records at Max Rubner-Institut, Kiel, Germany (MRI) to compare the morphology of Q33 and BM13 to that of previously characterized members of the phage species.
Identification of structural proteins.
Purified particles of BM13 (∼1011 PFU/ml) were concentrated with a NanoSep 300K column (Pall Corporation) and treated as previously described (27, 28). Phage proteins were separated by SDS-PAGE on a 15% precast polyacrylamide gel (Bio-Rad). Coomassie-stained protein bands of interest were excised from the gel and identified by liquid chromatography-tandem mass spectrometry (LC–MS-MS) at the Centre Protéomique de l'Est du Québec (Université Laval, Quebec, Canada). These results were analyzed using the Scaffold Proteome software. Purified phages were also directly analyzed by LC–MS-MS.
Proteomic tree construction.
The lactococcal phage genome sequences were downloaded and arranged as previously described (28). The resulting concatemeric protein sequences were aligned using the Mega 5.05 software and a BLOSUM matrix (29). A phylogenetic tree was constructed by the neighbor-joining method according to the number-of-differences model (30). The parameters used to test the phylogeny were bootstraps from 2,000 replicates, and the tree was transformed with iTOL (http://itol.embl.de/) and Adobe Illustrator CS6.
GenBank accession numbers.
The GenBank accession numbers for the phages sequenced in this study are as follows: JX564242 (Q33) and JX567312 (BM13).
RESULTS AND DISCUSSION
Host ranges of phages Q33 and BM13.
One hundred lactococcal strains were assessed for their susceptibilities to Q33 and BM13 using the spot assay method (31) and verified by the double-agar plaque assay (21). Of the industrial strains, Q33 infects 10 strains while BM13 infects 20, highlighting the rather broad host ranges of these phages. Therefore, the phages were able to infect at least 10% of the strains assessed. The phages' host ranges are not restricted to L. lactis subsp. lactis or subsp. cremoris, as strains of both subspecies were shown to be infected by the phages (Table 1).
Morphological analysis of Q33 and BM13.
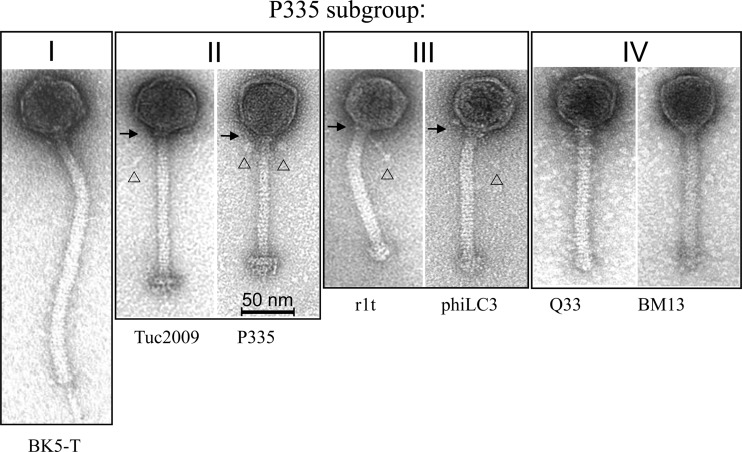
Phage Q33 possesses a long, noncontractile tail of approximately 148 ± 4 nm (n = 15). A distinct baseplate is not observed, but rather, a seemingly “stubby” tail end, which is reminiscent of that observed in the lactococcal 936-type phages and the P335-like phages, phiLC3 and r1t (Fig. 1) (32, 33). Its isometric capsid measures approximately 56 ± 2 nm (n = 15), which is similar to those of other members of the P335 phage species (Fig. 1). Phage BM13 has a similar morphology, with a capsid diameter of 56 ± 3 nm and a tail length of 125 ± 3 nm (n = 17) (Fig. 1).
Fig 1.
Transmission electron micrographs of lactococcal phages representing P335 subgroups I to IV. The phage particles are shown at the same magnification (cf. 50-nm size bar). The details of neck passage structures of subgroup II and III phages are indicated as follows: 32 arrows, collar; arrowheads, whiskers of different lengths terminating in globular structures of different diameters.
Genome sequence analysis.
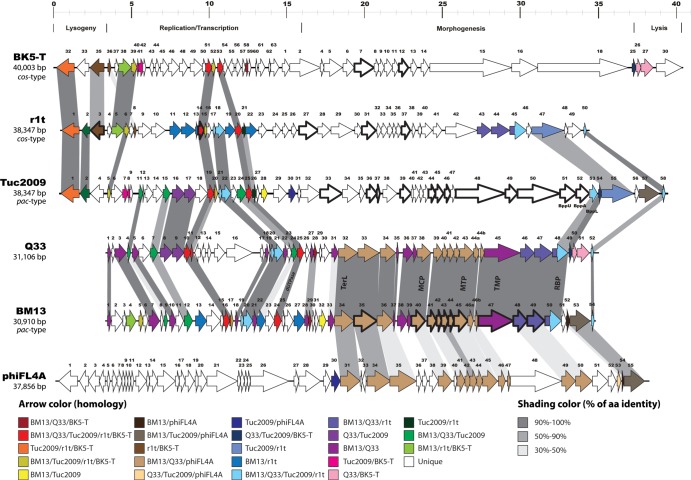
The genome of Q33 was elucidated to be 31,106 bp in length, with a GC content of 36.22% and containing 53 predicted protein-encoding ORFs, while the BM13 genome is 30,910 bp long with a GC content of 35.98% and with 55 predicted ORFs. Neither phage is predicted to possess tRNA-encoding regions as determined using the tRNA-scanning program tRNA scan E (24). Q33 is presumed to use the cos method of genome packaging, since it possesses a cos site and surrounding sequences identical to those of r1t and LC3 at positions 5873 to 5886 on the Q33 genome. BM13 does not contain an identical cos site, although the surrounding regions are similar to those of Q33. A list of the predicted genes and their functional annotation based on database searches is presented in Table S1 in the supplemental material. A total of 39.6% of Q33 ORFs (21 of 53 ORFs) and 40% of BM13 ORFs (22 of 55 ORFs) have been assigned a putative function based on the latest available annotation data and Pfam searches, and the genomes are predicted to have approximately 85% coding regions within their sequences. Three functional modules can be identified based on observed similarities to phage sequences in the database: (i) genes involved in replication functions, (ii) structural/morphogenesis genes, and (iii) remnants of a lysogeny module. These functional modules are organized in a rightward direction with the exception of two open reading frames of Q33, which are present in a leftward orientation (Fig. 2). Both genomes appear rather mosaic in terms of their content, containing genes with homology to (pro)phages of L. lactis, Streptococcus suis, Bifidobacterium catenulatum, and Enterococcus faecalis, among others (see Table S1 in the supplemental material). Each of the functional modules is discussed below.
Fig 2.
Genomic organizations of three P335-type phages and an E. faecalis phage. ORFs are represented by arrows, and arrows of the same color have at least 30% identity at the amino acid level. ORFs identified by LC–MS-MS are indicated by a thick outline. TerL, large terminase subunit; MCP, major capsid protein; MTP, major tail protein; TMP, tail tape measure protein; RBP, receptor binding protein; Bpp, base plate protein; aa, amino acid.
Lysogeny module.
Remnants of a lysogeny module appear to be present in the genomes of these two virulent lactococcal phages (ORFs 1 to 3 of Q33 and ORFs 1, 2, and 5 of BM13). Genes predicted to encode a Cro-like DNA binding protein and an antirepressor-like function are identifiable based on homology with the reference phage P335 and some prophages found in the genomes of laboratory strains L. lactis MG1363 and IL1403 (Fig. 2; see Table S1 in the supplemental material). Within the replication module of phage Q33, genes for a seemingly misplaced and truncated integrase (ORF 12) and repressor (ORF 14) were also identified and may have been acquired through homologous recombination with a prophage (11) or may represent the result of a DNA rearrangement event from its ancestral genome. The presence of a presumably incomplete and thus inactive lysogeny module (which in the case of Q33 is apparently dispersed through the genome) validates the virulent nature of the phages, while it also suggests that Q33 and BM13 may have had lysogenic predecessors.
Replication module.
Within the replication module, several features that are typical of this functional genomic segment in P335-like phages were identified, which included genes that are predicted to encode a putative topoisomerase, a translation initiation factor, RusA resolvase, a dUTPase, and DNA binding proteins (see Table S1 in the supplemental material). The majority of these open reading frames clearly display conserved synteny and highly significant homology to corresponding regions in lactococcal prophage genomes, such as those of bIL285 and bIL286, as well as more characterized P335-type members, including phages ul36, r1t, Tuc2009, and phi31. The dUTPase-encoding gene was originally thought to represent a genetic signature of all P335-type phages (6), and accordingly, it was also found in phages Q33 and BM13. However, as indicated previously, analysis of phage 4268 demonstrated that this P335-type phage did not contain a dUTPase-encoding gene (9). It is unclear at this time why the enzyme involved in nucleotide metabolism is absent from 4268.
The organization of the Q33 and BM13 replication modules is typical of those of other P335-type phages (with the exception of the integrase- and repressor-encoding genes in Q33). A number of genes whose encoded products possess greater than 80% identity at the amino acid level to the temperate P335 phages Tuc2009 and r1t are observed within the replication module, signifying its relatedness to these P335 phages (Fig. 2), although the phages do not infect the hosts for r1t or Tuc2009. Within the replication region of Q33 there appears to be an insertion of six open reading frames (ORFs 12 to 17) relative to Tuc2009 and BM13. The origins of this insertion appear to be prophages of Lactococcus and Streptococcus, and therefore, it may have been acquired through homologous-recombination events (Fig. 2). However, the relatedness of Tuc2009/r1t and Q33/BM13 appears to be largely confined to the replication module, with the exception of two genes in the morphogenesis module (see below).
Morphogenesis module.
The most striking genetic distinction between phages Q33/BM13 and the other members of the lactococcal P335 group is found within the morphogenesis module. This module harbors identifiable features, including genes that are predicted to encode the tail tape measure protein, baseplate complex, and capsid and tail proteins, displaying homology to presumed prophage sequences found in the genomes of various bacteria from the phyla Firmicutes and Actinobacteria that are typical colonizers of the human gastrointestinal tract, including E. faecalis and B. catenulatum (see Table S1 in the supplemental material). This is reminiscent of the case of lactococcal phage 1706 (1706 species), which encodes proteins displaying similarity to proteins of Ruminococcus torques and Clostridium leptum, both gut colonizers (34). It has been suggested that phage 1706 may have infected another host before acquiring the appropriate machinery to infect lactococcal strains, a suggestion that may also be pertinent to Q33 and BM13. Similarly, P087 (P087 species) possesses a morphogenesis module with similarity to an enterococcal prophage (35).
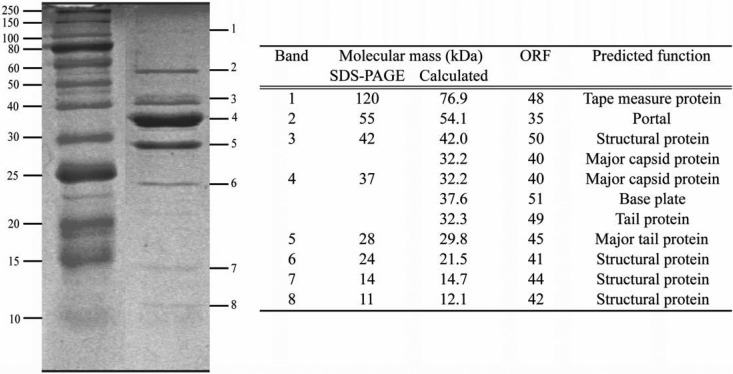
The Q33 and BM13 phage genomes encode a single identifiable terminase subunit (the large subunit), although the gene is preceded by an ORF of unknown function, and it may be possible that the latter ORF encodes a small-subunit terminase with no obvious similarity to known small-subunit terminases. Downstream (Fig. 2) of the ORFs putatively involved in DNA packaging, several genes are predicted to specify the portal protein, a minor and major capsid protein, a tail protein, and four other genes encoding proteins that show similarity to proteins specified by E. faecalis phage FL4A. Furthermore, the putative tail tape measure proteins encoded by Q33 and BM13 exhibit the most significant similarity to that of a presumed prophage of B. catenulatum and Bifidobacterium bifidum, respectively (see Table S1 in the supplemental material). Furthermore, the tail of BM13 is slightly shorter than that of Q33, which is substantiated by the shorter gene encoding the tail tape measure protein of BM13 (see Table S1 in the supplemental material). Nine structural proteins of BM13 were identified by LC–MS-MS analysis, namely, those encoded by ORFs 35, 40, 41, 42, 44, 45, 48, 49 and 50, among which are the predicted tail tape measure, portal, major tail, major capsid, and receptor binding proteins (RBPs) (Fig. 3). BM13 was chosen as a representative of both BM13 and Q33, given the similarity of their structural proteins (Fig. 2). The lysis cassettes of these phages are disparate, with no significant homology between the holins or lysins of BM13 and Q33 (see Table S1 in the supplemental material). The holin of Q33 has homology to that of the P335 phage BK-5T, and its lysin bears similarity to that of phi31, while those of BM13 bear most similarity to the holin and lysine of ul36 (see Table S1 in the supplemental material).
Fig 3.
Structural-protein profile of phage BM13. Bands excised for LC–MS-MS analysis are marked on the right, with the corresponding protein ORF assignment associated with the relevant bands presented in the accompanying table.
The RBPs of Q33 and BM13 are predicted to be encoded by ORF 48 and ORF 51, respectively. The RBP is known to be the primary interaction indicator for phages, and while the presumed RBPs of Q33 and BM13 are well conserved, there are amino acid variations in the respective carboxy termini that may be responsible for the host range variation of the two phages, a notion also suggested for other phages (36–38). The deduced amino terminus of the protein product of each of the ORFs bears similarity to the predicted baseplate of the temperate E. faecalis phage FL4A and, though with a lower level of similarity, to lactococcal P335 phage baseplate proteins. The carboxy terminus is highly similar to the lower baseplate protein, BppL, of the temperate P335 phage Tuc2009 (Fig. 2), and also to the putative RBPs encoded by the 936-type phages ASCC531, ASCC358, and ASCC497 (data not shown). The lineage of the RBP may explain the behavior of the phage in terms of its strict requirement for calcium to produce plaques and to replicate efficiently. Q33 and BM13 behave akin to Tuc2009 and the 936-type phages in their requirement for calcium for plaque formation, and therefore, it is conceivable that the RBP, as the first point of contact with the host, would be involved in such a phenotype (19). In the case of the 936-type phages, calcium is required for the conformational switch in the baseplate to the downward-pointing or “infection-ready” conformation in order to interact with its receptor (39). In contrast, the P335 phage TP901-1 does not require calcium for plaque formation, and its baseplate is maintained in an infection-ready position (19). It is more likely that some P335 phages, including BM13, Q33, and Tuc2009, require calcium to facilitate initiation of the infection process. While a large conformational change of the baseplate, which is required for infection by 936-type phages, has not been demonstrated for P335-type phages, the calcium requirement of certain P335-type phages may be based on other infection events, such as initial receptor binding or DNA injection.
Baseplate components: insights into phage-host interactions.
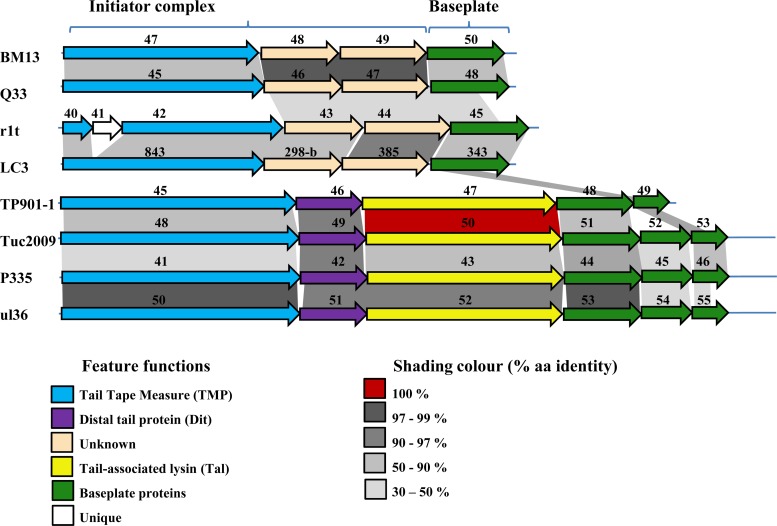
Recent structural analysis of the baseplates of the P335-like phages TP901-1 and Tuc2009 has revealed interactive data on the distal tail protein (Dit), the tail-associated lysin (Tal), and the baseplate components (19, 40). Phages Q33 and BM13, like the P335-like phages r1t and LC3, do not encode recognizable (based on sequence similarity) Dit or Tal proteins but nevertheless possess an overall genomic organization found in all P335 phages studied to date (Fig. 4). Furthermore, domain searches failed to reveal lytic domains in the presumed tail structure-encoding genes of Q33 or BM13, which is reminiscent of the genes in the same relative positions in the P335-type phage genomes of r1t and LC3 (41). Nonetheless, the relatedness of the lower baseplate protein (BppL) of Tuc2009 to the carboxy termini of Q33 ORF 48 and BM13 ORF 51 is reminiscent of the r1t-like phages, such as LC3, which possesses identity to BppL of TP901-1 at its carboxy terminus (Fig. 4) (41). Notable differences in the morphologies of the baseplates among the P335 phages are observed. For example, TP901-1 and Tuc2009 possess a double-disc baseplate structure (42), while r1t and LC3 do not possess such a complex baseplate structure and appear more like members of the 936 phage species, with a stubby end (32, 33).
Fig 4.
Genomic organizations of the initiator complex and baseplate regions of BM13 and Q33 in comparison to six previously sequenced lactococcal P335 phages. The schematic demonstrates the comparable architecture of the Q33/BM13 initiator complex and baseplate regions with those of the r1t/LC3 subgroup of the P335 phages, with the Tuc2009/TP901-1 subgroup displaying a distinct genetic organization in the region.
These data therefore suggest that r1t-like phages (Q33, BM13, r1t, and LC3) possess an alternative P335 baseplate type, which possibly represents an as-yet-uncharacterized receptor recognition platform and injection pathway. Furthermore, this alternative baseplate appears to be an architectural feature in many phages infecting various genera of the phyla Firmicutes and Actinobacteria. The prevalence of this baseplate type in phages of Lactococcus, Enterococcus, Ruminococcus, Clostridium, and Eubacteria, among others (based on sequence similarity) (data not shown), highlights the conservation of such structural features while also highlighting the usefulness of studying individual phages as models for the broader community with similar architectural features. It has previously been suggested that the temperate phages TP901-1 and LC3 follow different injection pathways (43), and our data support this theory if the analogies between Q33/BM13 and LC3 are indeed correct.
Phylogeny of lactococcal phages.
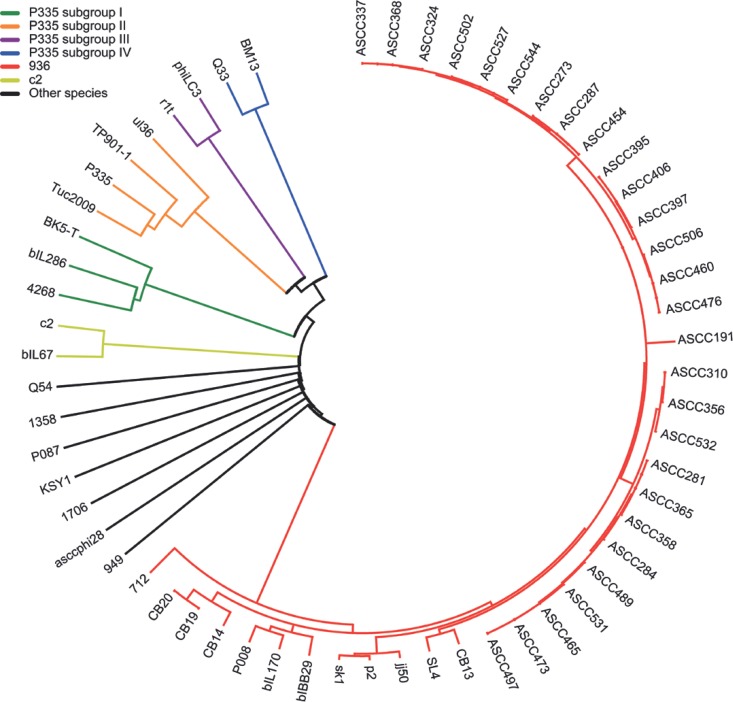
In order to phylogenetically classify BM13 and Q33 phages, a multiple alignment was performed using all 62 lactococcal phage genomes available in the NCBI database (http://www.ncbi.nlm.nih.gov). Mega 5.05 software and a neighbor-joining method were used for the construction of a phylogenetic tree based on predicted proteomic data (30). The phage proteomic tree yields a clear view of the groups and subgroups of lactococcal phages, like the P335 group (Fig. 5) (28, 44). The 936-like phages (displayed in red in Fig. 5) are grouped together, as expected, by the high sequence identity among them, as well as the few phages that belong to the c2-like group (indicated in yellow in Fig. 5) for which the genome has been sequenced. Samson and Moineau previously proposed three different subgroups within the P335 group, based in part on the level of amino acid identity in the structural module (packaging, capsid morphogenesis, and tail morphogenesis modules) (28). BK5-T, 4268, and bIL286 form a subgroup (Fig. 5, green); another subgroup contains P335, Tuc2009, TP901-1, and ul36 (Fig. 5, orange), while a third subgroup contains r1t and phiLC3 (Fig. 5, purple). Each subgroup is indeed well separated based on sequence similarity (Fig. 1, 4, and 5) (28). Identity within each subgroup member morphogenesis module is more than 60%. Of note, the seven other genetically distinct lactococcal phage groups (1706, asccphi28/P034, 1358, Q54, P087, KSY1, and 949) were also well separated (Fig. 5, black).
Fig 5.
Comparison of the complete proteome of lactococcal phages aligned using Mega 5.05 software and the neighbor-joining method based on the number of differences in amino acids. The parameters used to test the phylogeny were bootstraps from 2,000 replicates.
As expected, BM13 and Q33 were grouped within the large polythetic P335 group. However, the structural modules of BM13 and Q33 are not homologous to those of the other P335-like phages, setting them apart (Fig. 5, blue). Furthermore, a comparison between their structural modules (ORF 34 to ORF 50 of BM13 and ORF 32 to ORF 48 of Q33) shows 94.3% and 96.8% identity in amino acids and in nucleic acids, respectively. Therefore, a fourth P335 subgroup is proposed, with the sole current members being phages BM13 and Q33 (Fig. 1 and 5). The high level of identity between BM13 and Q33 structural modules suggests that the two phages have coexisted at some point and/or have acquired the same module by recombination with a similar phage or a prophage (1, 45, 46).
Conclusions.
Phages Q33 and BM13 were isolated in North America almost 20 years apart, yet their relatedness emphasizes the persistence of successful virulent phages in dairy settings. Their broad host range likely facilitated such persistence. The presence of genes with encoded products that have higher levels of similarity to prophages of gut commensals than other lactococcal phages suggests a corresponding ancestry of these phages and the role of their genomic plasticity in becoming effective virulent phages targeting dairy lactococcal strains. Q33 and BM13 are proposed to represent members of a novel, broad-host-range subgroup of the P335-type group. Our work highlights the relevance of sequencing the genomes of representatives of this lactococcal phage group in order to adapt our molecular phage detection tools and to develop antiphage strategies. Finally, genomic analysis has highlighted the variation of the P335 phages in terms of their initiator complex and baseplate components, which may present a phage-host interaction model distinct from the well-characterized TP901-1 and Tuc2009 model system.
Supplementary Material
ACKNOWLEDGMENTS
D. van Sinderen is the recipient of a Science Foundation Ireland (SFI) Principal Investigator award (08/IN.1/B1909). B. Martel is the recipient of a graduate student scholarship from the Commission Canadienne du Lait and Novalait Inc. S.M. is supported by a strategic grant from the Natural Sciences and Engineering Council (NSERC) of Canada. S.M. holds a Tier 1 Canada Research Chair in Bacteriophages.
Footnotes
Published ahead of print 10 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00832-13.
REFERENCES
- 1.Deveau H, Labrie SJ, Chopin MC, Moineau S. 2006. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72:4338–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahony J, Ainsworth S, Stockdale S, van Sinderen D. 2012. Phages of lactic acid bacteria: the role of genetics in understanding phage-host interactions and their co-evolutionary processes. Virology 434:143–150 [DOI] [PubMed] [Google Scholar]
- 3.Blatny JM, Godager L, Lunde M, Nes IF. 2004. Complete genome sequence of the Lactococcus lactis temperate phage phiLC3: comparative analysis of phiLC3 and its relatives in lactococci and streptococci. Virology 318:231–244 [DOI] [PubMed] [Google Scholar]
- 4.Brondsted L, Ostergaard S, Pedersen M, Hammer K, Vogensen FK. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93–109 [DOI] [PubMed] [Google Scholar]
- 5.Labrie S, Moineau S. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308–320 [DOI] [PubMed] [Google Scholar]
- 6.Labrie SJ, Josephsen J, Neve H, Vogensen FK, Moineau S. 2008. Morphology, genome sequence, and structural proteome of type phage P335 from Lactococcus lactis. Appl. Environ. Microbiol. 74:4636–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahanivong C, Boyce JD, Davidson BE, Hillier AJ. 2001. Sequence analysis and molecular characterization of the Lactococcus lactis temperate bacteriophage BK5-T. Appl. Environ. Microbiol. 67:3564–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seegers JF, McGrath S, O'Connell-Motherway M, Arendt EK, van de Guchte M, Creaven M, Fitzgerald GF, van Sinderen D. 2004. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 329:40–52 [DOI] [PubMed] [Google Scholar]
- 9.Trotter M, McAuliffe O, Callanan M, Edwards R, Fitzgerald GF, Coffey A, Ross RP. 2006. Genome analysis of the obligately lytic bacteriophage 4268 of Lactococcus lactis provides insight into its adaptable nature. Gene 366:189–199 [DOI] [PubMed] [Google Scholar]
- 10.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters MH, Venema G, Nauta A. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343–1355 [DOI] [PubMed] [Google Scholar]
- 11.Labrie SJ, Moineau S. 2007. Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J. Bacteriol. 189:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moineau S, Fortier J, Ackermann HW, Pandian S. 1992. Characterization of lactococcal bacteriophages from Quebec cheese plants. Can. J. Microbiol. 38:875–882 [Google Scholar]
- 13.Emond E, Holler BJ, Boucher I, Vandenbergh PA, Vedamuthu ER, Kondo JK, Moineau S. 1997. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl. Environ. Microbiol. 63:1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madsen A, Josephsen J. 1998. Characterization of LlaCI, a new restriction-modification system from Lactococcus lactis subsp. cremoris W15. Biol. Chem. 379:443–449 [DOI] [PubMed] [Google Scholar]
- 15.Madsen A, Josephsen J. 1998. Cloning and characterization of the lactococcal plasmid-encoded type II restriction/modification system, LlaDII. Appl. Environ. Microbiol. 64:2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGrath S, Fitzgerald GF, van Sinderen D. 2002. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43:509–520 [DOI] [PubMed] [Google Scholar]
- 17.McGrath S, Fitzgerald GF, van Sinderen D. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67:608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouchard JD, Moineau S. 2004. Lactococcal phage genes involved in sensitivity to AbiK and their relation to single-strand annealing proteins. J. Bacteriol. 186:3649–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veesler D, Spinelli S, Mahony J, Lichiere J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, Cambillau C. 2012. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. U. S. A. 109:8954–8958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labrie S, Moineau S. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85–90 [DOI] [PubMed] [Google Scholar]
- 22.Deveau H, Van Calsteren MR, Moineau S. 2002. Effect of exopolysaccharides on phage-host interactions in Lactococcus lactis. Appl. Environ. Microbiol. 68:4364–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo FB, Zhang CT. 2006. ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinformatics 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Deasy T, Mahony J, Neve H, Heller KJ, van Sinderen D. 2011. Isolation of a virulent Lactobacillus brevis phage and its application in the control of beer spoilage. J. Food Prot. 74:2157–2161 [DOI] [PubMed] [Google Scholar]
- 27.Fortier LC, Bransi A, Moineau S. 2006. Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J. Bacteriol. 188:6101–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson JE, Moineau S. 2010. Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages. Appl. Environ. Microbiol. 76:6843–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dupont K, Vogensen FK, Neve H, Bresciani J, Josephsen J. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70:5818–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lillehaug D, Lindqvist B, Birkeland NK. 1991. Characterization of phiLC3, a Lactococcus lactis subsp. cremoris temperate bacteriophage with cohesive single-stranded DNA ends. Appl. Environ. Microbiol. 57:3206–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rousseau GM, Moineau S. 2009. Evolution of Lactococcus lactis phages within a cheese factory. Appl. Environ. Microbiol. 75:5336–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garneau JE, Tremblay DM, Moineau S. 2008. Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 373:298–309 [DOI] [PubMed] [Google Scholar]
- 35.Villion M, Chopin MC, Deveau H, Ehrlich SD, Moineau S, Chopin A. 2009. P087, a lactococcal phage with a morphogenesis module similar to an Enterococcus faecalis prophage. Virology 388:49–56 [DOI] [PubMed] [Google Scholar]
- 36.Duplessis M, Moineau S. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325–336 [DOI] [PubMed] [Google Scholar]
- 37.Dupont K, Vogensen FK, Josephsen J. 2005. Detection of lactococcal 936-species bacteriophages in whey by magnetic capture hybridization PCR targeting a variable region of receptor-binding protein genes. J. Appl. Microbiol. 98:1001–1009 [DOI] [PubMed] [Google Scholar]
- 38.Tremblay DM, Tegoni M, Spinelli S, Campanacci V, Blangy S, Huyghe C, Desmyter A, Labrie S, Moineau S, Cambillau C. 2006. Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site. J. Bacteriol. 188:2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sciara G, Bebeacua C, Bron P, Tremblay D, Ortiz-Lombardia M, Lichiere J, van Heel M, Campanacci V, Moineau S, Cambillau C. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc. Natl. Acad. Sci. U. S. A. 107:6852–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sciara G, Blangy S, Siponen M, McGrath S, van Sinderen D, Tegoni M, Cambillau C, Campanacci V. 2008. A topological model of the baseplate of lactococcal phage Tuc2009. J. Biol. Chem. 283:2716–2723 [DOI] [PubMed] [Google Scholar]
- 41.McGrath S, Neve H, Seegers JF, Eijlander R, Vegge CS, Brondsted L, Heller KJ, Fitzgerald GF, Vogensen FK, van Sinderen D. 2006. Anatomy of a lactococcal phage tail. J. Bacteriol. 188:3972–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vegge CS, Vogensen FK, McGrath S, Neve H, van Sinderen D, Brondsted L. 2006. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009. J. Bacteriol. 188:55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ostergaard Breum S, Neve H, Heller KJ, Vogensen FK. 2007. Temperate phages TP901-1 and phiLC3, belonging to the P335 species, apparently use different pathways for DNA injection in Lactococcus lactis subsp. cremoris 3107. FEMS Microbiol. Lett. 276:156–164 [DOI] [PubMed] [Google Scholar]
- 44.Rohwer F, Edwards R. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouchard JD, Moineau S. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65–75 [DOI] [PubMed] [Google Scholar]
- 46.Moineau S, Pandian S, Klaenhammer TR. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jahns A, Schafer A, Geis A, Teuber M. 1991. Identification, cloning and sequencing of the replication region of Lactococcus lactis ssp. lactis biovar diacetylactis Bu2 citrate plasmid pSL2. FEMS Microbiol. Lett. 64:253–258 [DOI] [PubMed] [Google Scholar]
- 48.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.