Abstract
Reduced to near extinction in the late 1800s, a number of wood bison populations (Bison bison athabascae) have been re-established through reintroduction initiatives. Although an invaluable tool for conservation, translocation of animals can spread infectious agents to new areas or expose animals to pathogens in their new environment. Mycobacterium avium subsp. paratuberculosis, a bacterium that causes chronic enteritis in ruminants, is among the pathogens of potential concern for wood bison management and conservation. In order to inform translocation decisions, our objectives were to determine the M. avium subsp. paratuberculosis infection status of wood bison herds in Canada and to culture and genetically characterize the infective strain(s). We tested fecal samples from bison (n = 267) in nine herds using direct PCR for three M. avium subsp. paratuberculosis-specific genetic targets with different copy numbers within the M. avium subsp. paratuberculosis genome. Restriction enzyme analysis (REA) and sequencing of IS1311 were performed on seven samples from five different herds. We also evaluated a panel of different culture conditions for their ability to support M. avium subsp. paratuberculosis growth from feces and tissues of direct-PCR-positive animals. Eighty-one fecal samples (30%) tested positive using direct IS900 PCR, with positive samples from all nine herds; of these, 75% and 21% were also positive using ISMAP02 and F57, respectively. None of the culture conditions supported the growth of M. avium subsp. paratuberculosis from PCR-positive samples. IS1311 REA and sequencing indicate that at least two different M. avium subsp. paratuberculosis strain types exist in Canadian wood bison. The presence of different M. avium subsp. paratuberculosis strains among wood bison herds should be considered in the planning of translocations.
INTRODUCTION
Historically ranging across most of northwestern North America, wood bison (Bison bison athabascae) were nearly brought to extinction in the late 1800s due to overhunting, changes in the distribution of habitat, and severe winters (1). This subspecies of North American bison is currently listed as threatened by the Committee on the Status of Endangered Wildlife in Canada (2), with approximately 11,000 free-ranging and semicaptive wood bison distributed among 11 conservation herds in North America (3). The establishment of viable, discrete, disease-free populations of wood bison across their historic range has been identified by the Canadian National Wood Bison Recovery Team as one of the most important goals for the recovery of this subspecies (2). Translocation has been an essential management tool for achieving this goal; however, a significant inherent risk of translocating animals is the potential inadvertent introduction of infectious agents to new areas or populations or exposing newly introduced animals to pathogens already present in that environment (4). Nine diseases of particular concern for bison conservation have been identified by the American Bison Specialist Group (ABSG) (3). In addition to their potential impact on bison health, these diseases are relevant to the management of this species because of the restrictions they may impose on animal movements for herd (re)establishment or for improving genetic diversity, and they therefore represent one of the greatest challenges for wood bison conservation in Canada (2).
Among the diseases of concern listed by the ABSG is Johne's disease (JD), caused by Mycobacterium avium subspecies paratuberculosis. It is believed that all ruminants are susceptible to developing JD, which is characterized by chronic granulomatous enteritis leading to weight loss, sometimes accompanied by diarrhea (5). M. avium subsp. paratuberculosis in wood bison was first reported by Sibley and colleagues (6), who detected M. avium subsp. paratuberculosis DNA in fecal samples from clinically normal wood bison using direct nested PCR targeting gene 251. These authors reported PCR-positive fecal samples derived from multiple bison subpopulations within Wood Buffalo National Park, which spans the northeastern area of Alberta and southern Northwest Territories, and from the Slave River Lowlands area to the northeast. Attempts to culture M. avium subsp. paratuberculosis from these samples using Bactec medium were unsuccessful.
Culture is valuable not only as a diagnostic tool but also as a source of large amounts of high-quality DNA for downstream molecular characterization of isolates. Several variables affect the sensitivity of culture or the success of culturing different strains of M. avium subsp. paratuberculosis, and even subtle changes in culture conditions can dramatically impact results (7). For example, the choice of method used for concentrating M. avium subsp. paratuberculosis from feces has been shown to influence sensitivity (8). Additionally, vancomycin, an antibiotic used to suppress the growth of Gram-positive bacteria in fecal and tissue samples, is inhibitory for particular strain types or isolates (9, 10), and other additives such as sodium pyruvate can enhance or inhibit certain strains (11, 12). Furthermore, although some culture media are capable of supporting the growth of a wide variety of M. avium subsp. paratuberculosis strains (13), the medium can selectively support the growth of particular strain types (11). These different variables have never been tested in combination for their ability to culture particularly fastidious strains.
The goals of our study stem from a workshop held in 2005 by the National Wood Bison Recovery Team which established research priorities for M. avium subsp. paratuberculosis infection in wood bison (14). Our specific objectives were to determine the infection status of the major wood bison conservation herds in Canada, and to culture and genetically characterize the infective strain(s). The aim was to facilitate informed management decisions such as the selection of source herds for future translocation.
MATERIALS AND METHODS
Sample collection.
Samples were collected between 2007 and 2012 by us and by wildlife management partners from the governments of the Northwest Territories, Yukon, Alberta, and British Columbia and Parks Canada (Table 1). Many of the herds were sampled specifically for M. avium subsp. paratuberculosis testing; in these cases, fresh fecal samples were collected from the ground after a herd had just moved away from an area, using a new glove for each sample. In some cases, samples were collected from killed animals during recreational hunts (Hay Zama herd) or from animals culled for herd health monitoring (Mackenzie, Etthithun, and Nordquist herds). In these cases, feces were taken from the rectum, and ileocecal lymph nodes (ICLN) and part of the terminal ileum were collected, avoiding cross contamination between specimens. A new sterile scalpel was used for each animal, and intestinal segments were tied off with zip-ties prior to their removal to minimize fecal contamination. Samples were stored at −20°C until shipped in coolers to the University of Calgary, where they were stored at −80°C until processing. The total length of time in storage varied between 7 months and 3 years.
Table 1.
Results of direct PCR performed on fecal samples collected from wood bison herds in northern Canada, using three separate genetic targets for M. avium subsp. paratuberculosis
| Wood bison herd | Sample size (estimated herd size) | No. (%) positive fora: |
|||
|---|---|---|---|---|---|
| IS900 | TMRb | F57 | TMRb + F57 | ||
| Elk Island National Park | 31 (361) | 10 (32) | 4 (40) | 2 (20) | 2 (20) |
| Wood Buffalo National Parkc | 23 (4,958) | 11 (48) | 6 (55) | 1 (9) | 2 (18) |
| Aishihikc | 31 (1,230) | 8 (26) | 3 (38) | 0 | 3 (38) |
| Nordquist | 10 (155) | 3 (30) | 3 (100) | 0 | 0 |
| Etthithun | 14 (226) | 7 (50) | 5 (71) | 0 | 0 |
| Nahannic | 48 (400) | 16 (33) | 10 (63) | 0 | 3 (19) |
| Mackenziec | 51 (1,438) | 8 (16) | 4 (50) | 0 | 2 (25) |
| Grande Detourc | 45 (1,790)d | 12 (27) | 9 (75) | 1 (8) | 1 (8) |
| Hay Zama | 14 (524) | 5 (36) | 3 (60) | 0 | 0 |
| Total | 267 | 80 (30) | 47 (59) | 4 (5) | 13 (16) |
Only IS900-positive samples were tested for the other two targets.
TaqMan MAP reagents (Applied Biosystems), a commercially available PCR kit that targets ISMAP02.
IS1311 restriction enzyme analysis performed on samples from this herd.
Estimate is for Slave River Lowlands, which includes Grande Detour and Hook Lake populations.
DNA extraction from feces.
All samples were processed in our laboratory, which has received certification for this test through the USDA National Veterinary Services Laboratories (NVSL) Johne's disease direct PCR proficiency panel. DNA was extracted from all fecal samples using a MagMAX total nucleic acid isolation kit (Applied Biosystems, Carlsbad, CA) following the manufacturer's instructions, with some modifications. Samples were bead beaten twice for 5 min on a mini-Beadbeater (Biospec, Bartlesville, OK) and cooled on ice between beatings. Bead capture was performed using a 96-well magnetic-ring stand (Applied Biosystems). To increase the chances of isolating DNA from small sample volumes (0.3 g, as recommended in the MagMAX protocol), all samples were extracted twice on separate days. No-template controls, in which test reagents were taken through the entire process without the addition of a sample, were included in each extraction.
Quantitative PCR.
All DNA extracted from feces was analyzed using a duplex PCR developed by Slana et al. targeting IS900 and an internal amplification control (IAC) (15). Samples positive for IS900 on either extraction were confirmed using a second duplex PCR targeting the F57 element, also containing an IAC. Primer, probe, and IAC sequences for both assays were identical to those previously described (15), and TaqMan probes were supplied by Biosearch Technologies, Inc. (Novato, CA, USA). Each PCR mixture contained 10 μl of TaqMan Fast Advanced Master Mix (Applied Biosystems), 10 pmol of both forward and reverse primers (either for IS900 or F57), 1 pmol of probes for both the IS900 or F57 target and the IAC, approximately 900 copies of the internal control plasmid (target specific), and 2 μl of template. Cycling conditions were as follows: 50°C for 2 min (to activate UNG included in the commercial Master Mix), denaturation at 95°C for 20 s, followed by 40 cycles of 95°C for 3 s and 61°C for 30 s.
All extracted DNA that was positive using IS900 was also tested using a commercially available PCR kit (TaqMan MAP reagents [TMR]; Applied Biosystems) that includes primers and a probe designed to detect ISMAP02 (Life Technologies, personal communication). Each 25-μl reaction mixture contained 12.5 μl of the supplied master mix, 1 μl of the primer/probe mixture, and 8 μl of extracted DNA as a template. The cycling conditions were as follows: 95°C for 10 min and 40 cycles alternating between 95°C (15 s) and 60°C (1 min). All PCRs included positive and negative controls. qPCR runs were performed using a CFX96 thermocycler (Bio-Rad, Mississauga, Ontario, Canada). CT values lower than 37 cycles were considered positive. We considered a sample to contain M. avium subsp. paratuberculosis DNA if it was positive for IS900 and at least either ISMAP02 or F57.
IS1311 restriction enzyme analysis.
For strain-typing purposes, restriction enzyme analysis (REA) of insertion sequence IS1311 was performed on a subset of samples (n = 7) that were positive using direct PCR for at least two genetic targets (Table 1). A nested PCR was performed as previously described (6), using a Veriti thermocycler (Applied Biosystems). Nested products were digested using HinfI (New England BioLabs, Whitby, ON, Canada), run on a 3% agarose gel including SYBR Safe DNA gel stain (Life Technologies), and viewed using a Chemi Doc MP imaging system (Bio-Rad). Interpretation of the banding patterns was based on the study of Whittington et al. (16), recognizing the modifications to the band size due to the nested PCR (6). Nested PCR products were sequenced by the University Core DNA Services, University of Calgary, Alberta, Canada. Electropherograms were visualized using Finch TV version 1.4.0 and scanned for any double peaks. Sequences were aligned to the 11 sequences published in GenBank (as of March 2013) that cover this area of IS1311 using Geneious Pro version 5.5.5 (Biomatters, Auckland, New Zealand). Alignments were visually inspected, and the electropherograms of the sequences were carefully examined at any sites of discrepancy.
Culture panel.
We investigated the possibility that PCR-positive wood bison samples could be successfully cultured for M. avium subsp. paratuberculosis by making modifications to routine procedures for culturing M. avium subsp. paratuberculosis from cattle and sheep samples. A panel of culture conditions was created that included variations on M. avium subsp. paratuberculosis concentration methods, decontamination protocols, culture media, and additives.
(i) Fecal panel. (a) Samples used.
The fecal culture panel consisted of the following. (i) Feces from two wood bison (A and B) from the Mackenzie bison herd that tested positive by direct fecal PCR using all three genetic targets on two separate extractions and from which tissues were also available for testing were used. (ii) Feces from one plains bison (C) that tested positive by direct PCR on two separate extractions were used. This sample was included as a positive control since it had already been cultured successfully using the TREK ESP system and had a CT value on direct PCR (CT = 30) similar to that of the wood bison samples included in the panel (CT = 32). (iii) Positive- and negative-control samples, which consisted of pooled bovine feces from the NVSL 2011 direct PCR proficiency panel (17) were also included. Each of the three bison samples and the positive and negative controls (total of 5) were thoroughly homogenized and then separated into nine aliquots of 2 g each, which were subjected to different protocol variations (Fig. 1).
Fig 1.
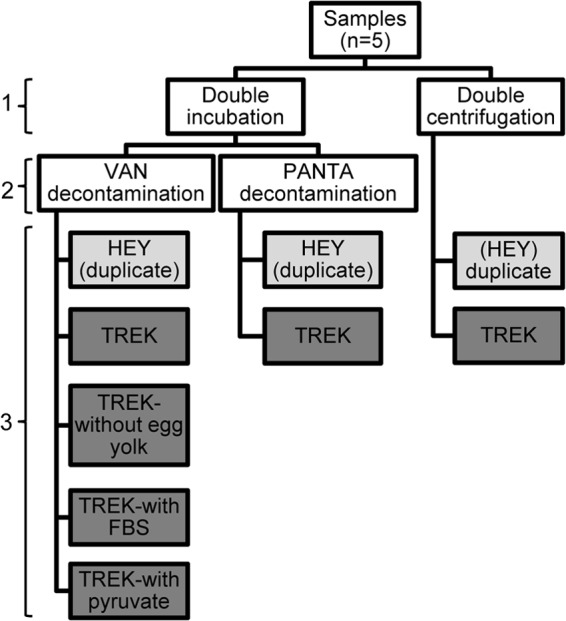
Culture panel tested for culturing Mycobacterium avium subsp. paratuberculosis from wood bison feces. Variations included the M. avium subsp. paratuberculosis concentration method (1), the decontamination method (2), and the type of medium and supplements used (3). The nine final culture variations for each of the five samples are shown in light gray (HEY) and dark gray (TREK). VAN, vancomycin, amphotericin B, and nalidixic acid; HEY, Herrold's egg yolk; FBS, fetal bovine serum.
(b) M. avium subsp. paratuberculosis concentration and decontamination methods.
The double-incubation (DI) method described by Whittington et al. was used as the standard protocol for concentrating M. avium subsp. paratuberculosis from feces (18), modified by incubating the samples for only 24 h in the antibiotic brew. Seven aliquots of each sample were processed with this method (Fig. 1). For five of these aliquots, the antibiotic brew consisted of vancomycin, amphotericin B, and nalidixic acid (VAN) at the concentrations recommended by the authors. For the other two aliquots, this antibiotic mix was replaced with PANTA Plus (Becton Dickinson, Sparks, MD), which was prepared by adding one reconstituted vial of PANTA Plus to 15 ml of half-strength brain heart infusion (BHI). Two additional aliquots of each sample were processed using the double centrifugation (NADC) method with VAN decontamination (8), except that half-strength BHI was used for the 0.9% hexadecylpyridinium chloride (HPC)-BHI decontamination step.
(c) Sample inoculation.
Six fecal aliquots from each of the five samples were inoculated into TREK para-JEM bottles, adding 1 ml of sample per bottle (Fig. 1). Standard supplements were added to the para-JEM bottles prior to sample inoculation as recommended by the manufacturer, with the following exceptions (Fig. 1). For one aliquot with DI-VAN decontamination, no egg yolk supplement was added; for two other DI-VAN aliquots, 500 μl of the culture medium in the TREK bottle (∼10% volume) was removed and replaced with an equal volume of either fetal bovine serum (FBS) or 100 mM minimum essential medium with sodium pyruvate (Life Technologies, Burlington, ON, Canada) prior to the addition of supplements. For the aliquot that had been decontaminated with PANTA, the standard para-JEM AS supplement (VAN) was replaced with an equal volume of reconstituted PANTA Plus. A negative culture control (adding only the culture supplements to the TREK bottle) was processed along with the samples. Three aliquots from each of the samples (Fig. 1) were inoculated in duplicate onto commercially prepared Herrold's egg yolk (HEY) slants (Becton Dickinson) containing mycobactin J and VAN, using 50 μl per slant.
(d) Incubation and DNA extraction.
Bottles were incubated in the TREK ESP II culture system for 60 days, after which DNA was extracted from all bottles following a standard protocol (19). Following this first extraction, bottles were placed in an incubator at 37°C for a further 4 months and samples re-extracted. HEY slants were kept in a 37°C incubator and observed every 2 weeks for growth or contamination for a total of 6 months, after which DNA was extracted from visible colonies. Briefly, 2 to 3 colonies were removed from the slant and added to a 2-ml conical bead-beating tube containing 20 μl of Dulbecco's phosphate-buffered saline (PBS; Life Technologies), 180 μl of ATL tissue lysis buffer (Qiagen, Mississauga, ON, Canada), and 0.4 g of 0.1-mm zirconia/silica beads (Biospec). This mixture was bead beaten for 2 min, and then the spin column protocol from the DNeasy blood and tissue kit (Qiagen) was followed beginning at step 2. DNA extracted from para-JEM broth or from colonies was subjected to the same qPCR protocols as DNA extracted from feces.
(ii) Tissue panel. (a) Samples used.
Tissues were available from the same two wood bison (A and B) and plains bison (C) that were used in the fecal panel. Additionally, tissues were available from a captive wood bison from a wildlife preserve (D) that was positive by fecal culture using the standard TREK ESP protocol; tissues from this animal were included as a second positive control. Negative-control tissues were not available; however, a negative TREK culture control with supplements was included. To allow sufficient tissue aliquots, ileum and ICLN from each individual animal were pooled. The tissues were placed in a sterile petri dish, and the fat was trimmed away using a sterile disposable scalpel and flame-sterilized forceps. After opening the ileum to expose the epithelium, the tissues were rinsed three times with sterile 1× PBS and diced together.
(b) Sample preparation.
Two-gram aliquots of the homogenized ileum/ICLN tissue were added to 25 ml of 0.75% HPC in a stomacher bag. The sample was processed in a Stomacher 80 Biomaster (Seward, Port Saint Lucie, FL) for 4 min on the high setting, before the entire contents of the bag were transferred to a 50-ml centrifuge tube and left to settle for 30 min. A 10-ml portion of the supernatant was transferred to a new 50-ml tube containing 25 ml 0.75% HPC, incubated for 3 h at room temperature, and centrifuged at 900 × g for 30 min. The supernatant was discarded, and the pellet was resuspended in 1 ml antibiotic brew by pipetting; VAN was used at standard concentrations, except for one aliquot that was processed with PANTA (prepared in the same way as the fecal panel).
One tissue aliquot (1 ml) was added to a single TREK para-JEM bottle, following the same protocols as for the fecal panel. Two aliquots of each sample were sufficient to inoculate 20 slants (four each of HEY, Middlebrook 7H11 with and without pyruvate, and Lowenstein-Jensen [LJ] medium with and without pyruvate), with 100 μl inoculated onto each slant. The culture variations are shown in Fig. 2. There was insufficient tissue to perform all variations on each sample; omissions are shown in Table 2. Observations and DNA extraction were performed as described for the fecal panel.
Fig 2.
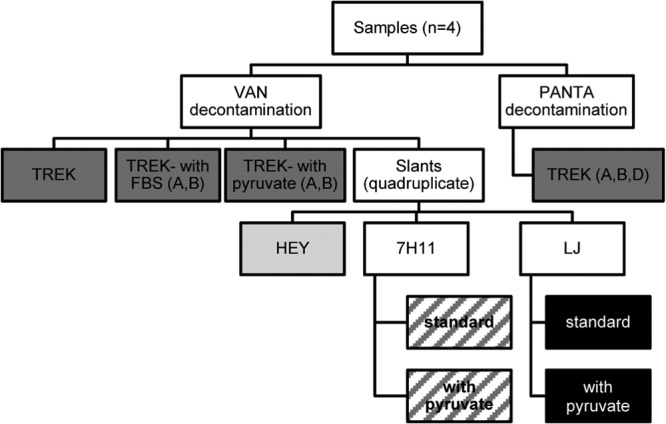
Culture panel tested for culturing Mycobacterium avium subsp. paratuberculosis from wood bison tissues. Variations included the decontamination method and the type of medium and supplements used. Final culture variations are shown in light gray (HEY), dark gray (TREK), hatching (7H11) and black (LJ). Letters refer to bison samples included in each variation; all four are included unless otherwise indicated. VAN, vancomycin, amphotericin B, and nalidixic acid; HEY, Herrold's egg yolk; FBS, fetal bovine serum; 7H11, Middlebrook 7H11; LJ, Lowenstein-Jensen.
Table 2.
Results of fecal and tissue panels for culturing Mycobacterium avium subsp. paratuberculosisa
| Sample type and test | Result for: |
|||||
|---|---|---|---|---|---|---|
| Bison A | Bison B | Bison C | Bison D | USDA positive control | USDA negative control | |
| Fecal | ||||||
| TREK | − | − | + | + | + | − |
| TREK-PD | − | − | + | NI | + | − |
| TREK-DC | − | − | + | NI | + | − |
| TREK-NE | − | − | − | NI | − | − |
| TREK-FBS | − | − | + | NI | + | − |
| TREK-pyr | − | − | + | NI | + | − |
| HEY | − | − | + | NI | + | − |
| HEY-PD | − | − | + | NI | + | − |
| HEY-DC | − | − | + | NI | + | − |
| Tissue | ||||||
| TREK | − | − | + | + | NI | NI |
| TREK-PD | − | − | NI | + | NI | NI |
| TREK-FBS | − | − | NI | NI | NI | NI |
| TREK-pyr | − | − | NI | NI | NI | NI |
| HEY | − | − | + | − | NI | NI |
| LJ | − | − | + | − | NI | NI |
| LJ-pyr | − | − | + | − | NI | NI |
| 7H11 | − | − | − | − | NI | NI |
| 7H11-pyr | − | − | − | − | NI | NI |
A culture variation was recorded as positive if at least one of the replicate slants had growth. Bison A and B are wood bison from the Mackenzie bison herd for which samples were positive on direct fecal PCR and from which tissues were also available. Bison C is a commercial plains bison for which samples were positive on direct fecal PCR and which is known to be culture positive according to TREK ESP. Bison D is a wood bison from a wildlife preserve that was known to be fecal culture positive according to TREK ESP. Positive and negative control feces are from the NVSL 2011 direct PCR proficiency panel. NI, not included; PD, PANTA decontamination; DC, double centrifugation; NE, no egg yolk; FBS, fetal bovine serum; pyr, pyruvate; HEY, Herrold's egg yolk; LJ, Lowenstein-Jensen; 7H11, Middlebrook 7H11.
(c) Medium preparation.
Middlebrook 7H11 medium was prepared using Difco 7H11 agar (Becton Dickinson) according to the manufacturer's instructions. Once the autoclaved medium had cooled to 55°C, OADC (oleic acid-albumin-dextrose-catalase) enrichment medium (Becton Dickinson), PANTA Plus, and mycobactin J (Allied Monitor, Fayette, MO) were added. Lowenstein-Jensen (LJ) medium was prepared using Difco Lowenstein medium base (Becton Dickinson) according to the manufacturer's instructions. A whole-egg supplement was prepared by soaking 14 antibiotic-free eggs in 70% ethanol, briefly flaming, and then breaking them into an autoclaved beaker. The eggs were mixed gently before being poured into another beaker through two layers of sterile gauze to remove any clumps. The total volume (approximately 660 ml) was added to the cooled medium (55°C) and mixed well before the addition of Mycobactin J. The medium was set in the culture tubes by steaming at 85°C for approximately 35 to 40 min or until visibly solidified (13). For both types of medium, a subset of slants with pyruvate was made by adding 100 ml 100 mM sodium pyruvate to 250 ml of the liquid medium.
Additional samples cultured.
In addition to the samples included in the culture panels, 24 PCR-positive fecal samples with the lowest CT values on qPCR from seven of the different bison herds were also cultured following the standard TREK ESP protocol and incubated for 8 weeks prior to DNA extraction.
Nucleotide sequence accession numbers.
Sequences are available from GenBank under accession numbers KC990352 and KC990353.
RESULTS
Direct fecal PCR.
Of the 267 fecal samples processed using direct extraction, 80 (30%) were positive by IS900 PCR on at least one extraction. Nineteen of these samples were positive on two separate extractions. Of the IS900-positive samples, 60 (75%) were also positive using the TaqMan MAP reagents (TMR) targeting ISMAP02. Seventeen of the IS900-positive samples (21%) were positive with F57 PCR, only four of which were negative for ISMAP02. Therefore, a total of 64 of the IS900-positive samples (80%) were also positive for at least either ISMAP02 or F57. Results, grouped by herd, are shown in Table 1.
IS1311 REA and sequencing.
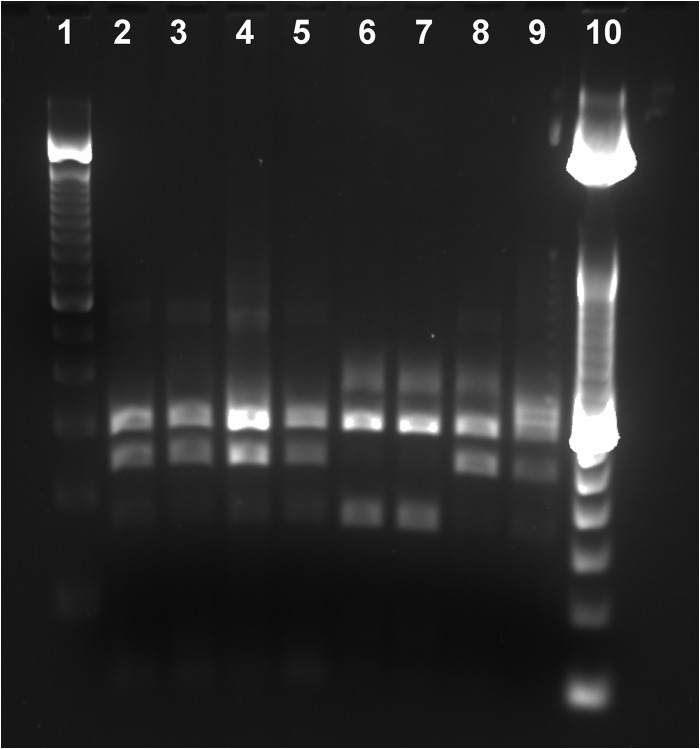
Samples from the Aishihik (n = 3), Grande Detour (n = 1), and Nahanni (n = 1) herds produced a banding pattern consistent with a C-type strain (Fig. 3). This was confirmed using sequencing, with a clear double peak on the electropherogram at base pair position 160 of the nested PCR product (Fig. 4A). Samples from the Mackenzie (n = 1) and Wood Buffalo National Park (n = 1) herds gave a banding pattern consistent with a B-type (bison-type) strain, although a very faint band was observed at the position that would be expected in C-type strains (Fig. 3). Sequencing results were consistent with the two samples being classified as B-type (Fig. 4B). Aside from the variability at this position, the sequences obtained from all seven samples were identical to the GenBank reference sequences, except that a double peak (A/T) was observed in the electropherogram at base position 458 of the nested product in all seven wood bison samples. This double peak was not present in the sequenced amplicon from the positive cattle control sample (A only).
Fig 3.
Gel image of IS1311 restriction enzyme analysis using HinfI on nested PCR products from DNA extracted directly from wood bison feces. Lane 1, 100-bp ladder; lane 2, Aishihik bison 1; lane 3, Aishihik bison 2; lane 4, Aishihik bison 3; lane 5, Grande Detour bison; lane 6, Mackenzie bison; lane 7, Wood Buffalo National Park bison; lane 8, Nahanni bison; lane 9, positive control, C-type cattle strain; lane 10, 50-bp ladder.
Fig 4.
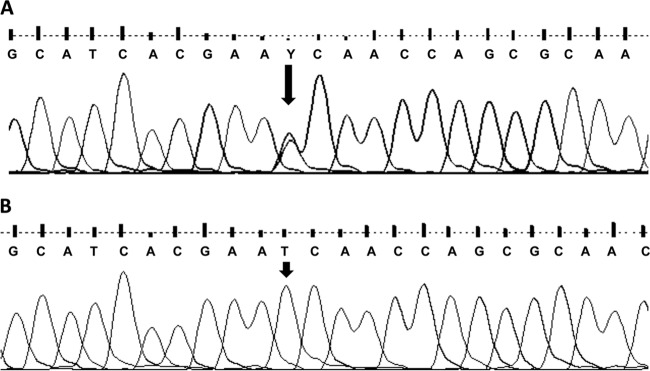
Typical electropherogram obtained from samples with a C-type banding pattern on IS1311 restriction enzyme analysis (A) and with a B-type banding pattern (B).
Culture panels.
Results of the fecal and tissue culture panels are summarized in Table 2. M. avium subsp. paratuberculosis was not cultured from either of the two wood bison fecal samples (A and B) on any of the culture variations. Both the plains bison feces (C) and the positive-control feces were PCR positive on five of the six TREK variations; neither was positive when egg yolk was omitted. All of the HEY slants inoculated with the positive-control sample had growth after 5 weeks. For bison C, the duplicate slants of HEY with and without PANTA decontamination had growth after 7 weeks. One of the slants with double centrifugation was positive after 5 weeks, while the other was only positive after 27 weeks. No M. avium subsp. paratuberculosis growth was observed on any of the media for the negative-control sample.
On the tissue culture panel, none of the culture variations supported the growth of M. avium subsp. paratuberculosis from the tissues of wood bison A or B. Bison C and D were culture positive using TREK; however, only the sample from bison C had growth on HEY (4/4 slants) and LJ (4/4 without pyruvate; 3/4 with) media. All four HEY slants were positive after 6 weeks. The LJ slants had visible colonies between 10 and 24 weeks. M. avium subsp. paratuberculosis was not grown from any sample on 7H11 medium.
None of the additional fecal samples cultured using the TREK ESP system were positive.
DISCUSSION
We have shown that in all nine wood bison populations tested, fragments of at least two genes unique to M. avium subsp. paratuberculosis were present in fecal samples. This complements the findings of Sibley et al., who found several PCR-positive wood bison feces using an M. avium subsp. paratuberculosis-specific genetic target distinct from those used in our study (6). Like those authors, we were unable to culture M. avium subsp. paratuberculosis from any of the PCR-positive samples tested, despite using a variety of different culture conditions. Although the inability to culture M. avium subsp. paratuberculosis from wood bison samples limited the extent to which the isolates could be genetically characterized, we were able to identify different strains among the herds in northern Canada.
The differences observed in the sensitivity of the three genetic targets were expected. The insertion sequence IS900 is present in 15 to 20 copies in the M. avium subsp. paratuberculosis genome, making it a highly sensitive target (20). However, it has been suggested that IS900 PCR is unsuitable as a stand-alone test for confirming M. avium subsp. paratuberculosis detection in a species or population of unknown infection status (21), since several related mycobacteria possess similar elements with up to 94% homology that can be amplified with different IS900 primer sets (22–24). ISMAP02 (present in six copies) and the single-copy target F57 are believed to be unique to M. avium subsp. paratuberculosis (25), but their lower copy numbers result in reduced sensitivity (26). Taking a conservative approach and considering a sample to be positive for M. avium subsp. paratuberculosis only if at least either the ISMAP02 or F57 assay was also positive, 80% of the IS900 positive samples would be considered “true positives.” At least three positive samples were identified in each herd.
There are a number of possible explanations as to why the M. avium subsp. paratuberculosis PCR-positive fecal samples could not be cultured. First, collecting wildlife samples frequently entails working under challenging field conditions, where optimal storage facilities are not readily available. All of the samples that we tested were stored at −20°C for at least 24 h (and in some cases up to several years) prior to storage in a −80°C freezer. Storage at −20°C can impact the viability of M. avium subsp. paratuberculosis (27), as can repeated freezing and thawing of samples (20), and this would be of particular relevance for samples with low concentrations of the bacterium. Although one study has shown that storing feces at −70°C for 15 weeks does not reduce the viability of M. avium subsp. paratuberculosis any further than storage for 3 weeks (28), to our knowledge, the effect of long-term storage on M. avium subsp. paratuberculosis survival has never been evaluated. The wood bison samples included in the culture panel were stored for 2 and 3 years, respectively, prior to testing, while the positive-control samples were stored for several months.
A second possible reason is that the direct PCR assay we used could be more sensitive than the culture techniques we explored. In recent years, major advances have been made in protocols wherein DNA is directly extracted from fecal samples (20), and some of these techniques may now be more sensitive than fecal culture (29). The CT values obtained during direct quantitative PCR on wood bison feces were similar or lower (indicative of a higher DNA concentration) than some of the CT values we have attained using proficiency panel fecal samples, which we have subsequently cultured successfully using the TREK ESP system (data not shown). Therefore, a low concentration of M. avium subsp. paratuberculosis in these fecal samples does not on its own explain the failure to culture M. avium subsp. paratuberculosis. However, the fact that PCR detects DNA from both live and dead organisms is particularly relevant for samples that have been stored under suboptimal conditions.
A third factor that could contribute to our inability to culture M. avium subsp. paratuberculosis from wood bison samples is that the strains present in these populations are different from those that have been described to date and that the culture conditions we attempted were inadequate. The inability to culture the organism precluded in-depth genetic characterization using full-genome sequencing, which would have enabled valuable comparisons to be made between the M. avium subsp. paratuberculosis strains found in wood bison and other sequenced isolates, as well as a better understanding of the genetic variability of M. avium subsp. paratuberculosis within and among herds. IS1311-REA results indicated that C-type M. avium subsp. paratuberculosis is present in at least three of the wood bison herds, while B-type M. avium subsp. paratuberculosis is present in two separate herds. Previously, Sibley et al. found both C-type and S-type strains in samples from the Wood Buffalo National Park herd (6), which is one of the two populations in which we found B-type M. avium subsp. paratuberculosis, suggesting that even more strains of M. avium subsp. paratuberculosis may be present in these herds.
Although we have demonstrated that M. avium subsp. paratuberculosis DNA is present in feces from wood bison populations across northern Canada, the effect that M. avium subsp. paratuberculosis infection may have on bison health remains unclear. Clinically affected animals might be quickly removed from the population through predation and thus go unobserved. Additionally, it is unlikely that underweight animals would be identified by wildlife managers, given the fact that these herds are monitored only from a distance; population surveys may be conducted, or health testing may be performed on a few individuals, but animals are not regularly observed. The one exception is the herd kept in semicaptivity at Elk Island National Park, where animals are monitored much more closely, and most fatalities are subjected to a full postmortem examination. Only one clinical case of JD has been observed in this herd; this occurred in 2006 (Alberta Agriculture, Food and Rural Development, unpublished data). Johne's disease has been described in commercial plains bison (30, 31), which show lesions and clinical disease similar to what is typical in cattle. However, given that captive plains bison have different habitats, population densities, and genetic backgrounds from those of wood bison and may be infected with different M. avium subsp. paratuberculosis strains, it is uncertain how much comparison can be made between these subspecies. Although it is unlikely that M. avium subsp. paratuberculosis infection causes a high level of mortality in wood bison, chronic diseases can have an important impact on a population by reducing survival or reproduction (32), thereby potentially decreasing the resilience of a population to disturbances and environmental perturbation (33).
Management decisions, including whether to translocate bison between herds, must be made using the best available data, and a complete risk-benefit assessment is recommended prior to any such animal movements. In the absence of the gold standard diagnosis, which in the case of M. avium subsp. paratuberculosis is bacterial culture, PCR results offer valuable information for evaluating the risks associated with translocation. We have shown that M. avium subsp. paratuberculosis DNA is present in all of the herds tested; however, we have also demonstrated that different strains exist among the wood bison herds in northern Canada. It is recommended that animals be translocated between herds of similar health status (34). Since we have yet to determine the extent to which the strains differ among herds and whether these differences are important in terms of disease dynamics and pathogenesis, fully evaluating the risks remains a challenge. Given the value of translocation for wood bison conservation, the benefits may outweigh the risks of moving animals among herds of M. avium subsp. paratuberculosis PCR-positive status.
ACKNOWLEDGMENTS
This project was possible thanks to the efforts of several collaborators who contributed to the sample collection. Thanks go to Terry Armstrong, Karl Cox, and Nic Larter (Government of the Northwest Territories), Helen Schwantje (Government of British Columbia), Lyle Fullerton, Mark Ball, and Margo Pybus (Government of Alberta), N. Jane Harms, Mary Vanderkop, and Tom Jung (Government of Yukon), Archie Handel (Elk Island National Park), Rhona Kindopp (Wood Buffalo National Park), and members of the community of Fort Providence, NWT. The National Wood Bison Recovery Team supported this work and greatly facilitated our contact within the network of bison managers and researchers. We thank N. Jane Harms, Margo Pybus, and Helen Schwantje for their critical review of the manuscript. Additional thanks go to Jessica Hillier, who provided excellent laboratory assistance.
Funding for this project was provided by the ACA Grants in Biodiversity (supported by the Alberta Conservation Association) and the Arctic Institute of North America's Grant-in-Aid and Northern Scientific Training Program. A trainee stipend was provided by the Natural Science and Engineering Research Council of Canada, Killam Trusts, the Canadian Wildlife Foundation, and the University of Calgary's Faculty of Veterinary Medicine.
Footnotes
Published ahead of print 17 May 2013
REFERENCES
- 1. Reynolds HW, Gates CC, Glaholt RD. 2003. Bison, p 1009–1060 In Feldhamer JA, Thompson BC, Chapman JA. (ed), Wild mammals of North America: biology, management, and conservation, 2nd ed. JHU Press, Baltimore, MD [Google Scholar]
- 2. Gates CC, Stephenson RO, Reynolds HW, van Zyll de Jong CG, Schwantje H, Hoefs M, Nishi J, Cool N, Chisholm J, James A, Koonz B. 2001. National recovery plan for the wood bison (Bison bison athabascae). National Recovery Plan No. 21. Recovery of Nationally Endangered Wildlife (RENEW), Ottawa, Ontario, Canada [Google Scholar]
- 3. Gates CC, Freese CH, Gogan PJP, Kotzman M. 2010. American bison: status survey and conservation guidelines 2010. IUCN, Gland, Switzerland [Google Scholar]
- 4. Kock RA, Woodford MH, Rossiter PB. 2010. Disease risks associated with the translocation of wildlife. Rev. Sci. Tech. 29: 329– 350 [DOI] [PubMed] [Google Scholar]
- 5. Manning EJB. 2011. Paratuberculosis in captive and free-ranging wildlife. Vet. Clin. North Am. Food Anim. Pract. 27: 621– 630 [DOI] [PubMed] [Google Scholar]
- 6. Sibley JA, Woodbury MR, Appleyard GD, Elkin B. 2007. Mycobacterium avium subspecies paratuberculosis in Bison (Bison bison) from northern Canada. J. Wildl. Dis. 43: 775– 779 [DOI] [PubMed] [Google Scholar]
- 7. Eamens GJ, Whittington RJ, Marsh IB, Turner MJ, Saunders V, Kemsley PD, Rayward D. 2000. Comparative sensitivity of various faecal culture methods and ELISA in dairy cattle herds with endemic Johne's disease. Vet. Microbiol. 77: 357– 367 [DOI] [PubMed] [Google Scholar]
- 8. Stabel JR. 1997. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J. Vet. Diagn. Invest. 9: 375– 380 [DOI] [PubMed] [Google Scholar]
- 9. Reddacliff LA, Vadali A, Whittington RJ. 2003. The effect of decontamination protocols on the numbers of sheep strain Mycobacterium avium subsp. paratuberculosis isolated from tissues and faeces. Vet. Microbiol. 95: 271– 282 [DOI] [PubMed] [Google Scholar]
- 10. Thornton CG, MacLellan KM, Stabel JR, Carothers C, Whitlock RH, Passen S. 2002. Application of the C18-carboxypropylbetaine specimen processing method to recovery of Mycobacterium avium subsp. paratuberculosis from ruminant tissue specimens. J. Clin. Microbiol. 40: 1783– 1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Juan L, Alvarez J, Romero B, Bezos J, Castellanos E, Aranaz A, Mateos A, Domínguez L. 2006. Comparison of four different culture media for isolation and growth of type II and type I/III Mycobacterium avium subsp. paratuberculosis strains isolated from cattle and goats. Appl. Environ. Microbiol. 72: 5927– 5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whitlock R, West S, Layton B, Ellingson J, Stabel J. 1999. Paratuberculosis in Bison: A comparison of PCR, culture and histopathology, p 424–438 In Manning EJB, Collins MT. (ed), Proceedings of the 6th International Colloquium on Paratuberculosis, Madison, WI: [Google Scholar]
- 13. Whittington RJ, Marsh IB, Saunders V, Grant IR, Juste R, Sevilla IA, Manning EJB, Whitlock RH. 2011. Culture phenotype of genomically and geographically diverse Mycobacterium avium subsp. paratuberculosis isolated from different hosts. J. Clin. Microbiol. 49: 1822– 1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woodbury M, Garde E, Schwantje H, Nishi J, Elkin B. 2006. Manuscript report no. 170. Workshop on Mycobacterium avium subsp. paratuberculosis in North American bison (Bison bison), Yellowknife, NT, Canada [Google Scholar]
- 15. Slana I, Kralik P, Kralova A, Pavlik I. 2008. On-farm spread of Mycobacterium avium subsp. paratuberculosis in raw milk studied by IS900 and F57 competitive real time quantitative PCR and culture examination. Int. J. Food Microbiol. 128: 250– 257 [DOI] [PubMed] [Google Scholar]
- 16. Whittington RJ, Marsh IB, Whitlock RH. 2001. Typing of IS1311 polymorphisms confirms that bison (Bison bison) with paratuberculosis in Montana are infected with a strain of Mycobacterium avium subsp. paratuberculosis distinct from that occurring in cattle and other domesticated livestock. Mol. Cell. Probes 15: 139– 145 [DOI] [PubMed] [Google Scholar]
- 17. Robbe-Austerman S. 2011. 2011 Johne's Disease Fecal Proficiency Panel: general summary. Animal and Plant Health Inspection Service, U.S. Department of Agriculture. http://www.aphis.usda.gov/animal_health/lab_info_services/downloads/PTReport2011Johnes.pdf
- 18. Whittington RJ, Marsh I, Turner MJ, McAllister S, Choy E, Eamens GJ, Marshall DJ, Ottaway S. 1998. Rapid detection of Mycobacterium paratuberculosis in clinical samples from ruminants and in spiked environmental samples by modified BACTEC 12B radiometric culture and direct confirmation by IS900 PCR. J. Clin. Microbiol. 36: 701– 707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forde T, Kutz S, De Buck J, Warren A, Ruckstuhl K, Pybus M, Orsel K. 2012. Occurrence, diagnosis, and strain typing of Mycobacterium avium subspecies paratuberculosis infection in Rocky Mountain bighorn sheep (Ovis canadensis canadensis) in southwestern Alberta. J. Wildl. Dis. 48: 1– 11 [DOI] [PubMed] [Google Scholar]
- 20. Bölske G, Herthnek D. 2010. Diagnosis of paratuberculosis by PCR, p 267–283 Behr MA, Collins DM. (ed), Paratuberculosis: organism, disease, control. CAB International, Cambridge, MA [Google Scholar]
- 21. Whittington R. 2010. Cultivation of Mycobacterium avium subsp. paratuberculosis, p 244–266 Behr MA, Collins DM. (ed), Paratuberculosis: organism, disease, control. CAB International, Cambridge, MA [Google Scholar]
- 22. Cousins DV, Whittington R, Marsh I, Masters A, Evans RJ, Kluver P. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13: 431– 442 [DOI] [PubMed] [Google Scholar]
- 23. Englund S, Bölske G, Johansson K-E. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209: 267– 271 [DOI] [PubMed] [Google Scholar]
- 24. Turenne CY, Wallace R, Jr, Behr MA. 2007. Mycobacterium avium in the postgenomic era. Clin. Microbiol. Rev. 20: 205– 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castellanos E, De Juan L, Domínguez L, Aranaz A. 2012. Progress in molecular typing of Mycobacterium avium subspecies paratuberculosis. Res. Vet. Sci. 92: 169– 179 [DOI] [PubMed] [Google Scholar]
- 26. Kralik P, Slana I, Kralova A, Babak V, Whitlock RH, Pavlik I. 2011. Development of a predictive model for detection of Mycobacterium avium subsp. paratuberculosis in faeces by quantitative real time PCR. Vet. Microbiol. 149: 133– 138 [DOI] [PubMed] [Google Scholar]
- 27. Khare S, Adams LG, Osterstock J, Roussel A, David L. 2008. Effects of shipping and storage conditions of fecal samples on viability of Mycobacterium paratuberculosis. J. Clin. Microbiol. 46: 1561– 1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richards WD, Thoen CO. 1977. Effect of freezing on the viability of Mycobacterium paratuberculosis in bovine feces. J. Clin. Microbiol. 6: 392– 395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marsh IB, Plain KM, Galea F, Waldron AM, Whittington AM, Whittington RJ. 2012. New and improved direct faecal PCR test for Johne's disease, p 15–16 In Neilsen SS. (ed), Proceedings of the 11th International Colloquium on Paratuberculosis, Sydney, Australia: [Google Scholar]
- 30. Buergelt CD, Layton AW, Ginn PE, Taylor M, King JM, Habecker PL, Mauldin E, Whitlock R, Rossiter C, Collins MT. 2000. The pathology of spontaneous paratuberculosis in the North American bison (Bison bison). Vet. Pathol. 37: 428– 438 [DOI] [PubMed] [Google Scholar]
- 31. Huntley JFJ, Whitlock RH, Bannantine JP, Stabel JR. 2005. Comparison of diagnostic detection methods for Mycobacterium avium subsp. paratuberculosis in North American bison. Vet. Pathol. 42: 42– 51 [DOI] [PubMed] [Google Scholar]
- 32. Joly DO, Messier F. 2005. The effect of bovine tuberculosis and brucellosis on reproduction and survival of wood bison in Wood Buffalo National Park. J. Anim. Ecol. 74: 543– 551 [Google Scholar]
- 33. Jolles AE, Cooper DV, Levin SA. 2005. Hidden effects of chronic tuberculosis in African buffalo. Ecology 86: 2358– 2364 [Google Scholar]
- 34. Gardner IA, Hietala S, Boyce WM. 1996. Validity of using serological tests for diagnosis of diseases in wild animals. Rev. Sci. Tech. 15: 323– 335 [DOI] [PubMed] [Google Scholar]