Abstract
Consumers trust commercial food production to be safe, and it is important to strive to improve food safety at every level. Several outbreaks of food-borne disease have been caused by Salmonella strains associated with dried food. Currently we do not know the mechanisms used by Salmonella enterica serovar Typhimurium to survive in desiccated environments. The aim of this study was to discover the responses of S. Typhimurium ST4/74 at the transcriptional level to desiccation on a stainless steel surface and to subsequent rehydration. Bacterial cells were dried onto the same steel surfaces used during the production of dry foods, and RNA was recovered for transcriptomic analysis. Subsequently, dried cells were rehydrated and were again used for transcriptomic analysis. A total of 266 genes were differentially expressed under desiccation stress compared with a static broth culture. The osmoprotectant transporters proP, proU, and osmU (STM1491 to STM1494) were highly upregulated by drying. Deletion of any one of these transport systems resulted in a reduction in the long-term viability of S. Typhimurium on a stainless steel food contact surface. The proP gene was critical for survival; proP deletion mutants could not survive desiccation for long periods and were undetectable after 4 weeks. Following rehydration, 138 genes were differentially expressed, with upregulation observed for genes such as proP, proU, and the phosphate transport genes (pstACS). In time, this knowledge should prove valuable for understanding the underlying mechanisms involved in pathogen survival and should lead to improved methods for control to ensure the safety of intermediate- and low-moisture foods.
INTRODUCTION
Salmonella has the ability to survive under low-moisture conditions for extended times and is one of the main biological hazards for dry-food manufacturers (1). Salmonella has been shown to survive for 8 months in halva, for 10 weeks when desiccated on paper discs, and for 12 months in peanut-flavored candy, while storage at 4°C under desiccation on plastic has resulted in 100 weeks of survival (2–5). This bacterium has previously been linked with many food-borne outbreaks associated with low-moisture food products, such as toasted cereal, infant formula, chocolate, and peanut butter (6–12).
Although some studies have found that exposure to low water activity (aw) can increase the tolerance of Salmonella to other, unrelated stresses, including heat and biocidal compounds (13), comparatively little is known about the bacterial mechanisms that ensure its survival under desiccated conditions (14). It is generally thought that organisms enter a dormant state with reduced metabolic activity that maintains viability for several months to years. When moisture is reintroduced, the risk of cross-contamination increases, posing a threat to food safety.
The source of contamination for low-moisture foods is often linked to production environments and manufacturing equipment that allow bacteria to persist for extended periods in a dried state (6, 7, 15). However, the mechanisms employed by cells to survive in such harsh environments, where little to no water is available, have not yet been thoroughly characterized. Studies have reported that the formation of filaments, the production of curli fimbriae, and cellulose can aid long-term persistence under low-moisture conditions (16, 17).
Recently, three studies examining the transcriptome of Salmonella under two different conditions have been published. First, Deng et al. assessed the transcriptomic response of Salmonella enterica serovar Enteritidis to peanut oil, a low-moisture liquid environment (18). The bacterial cells were found to exist in a metabolically dormant state, with <5% of the genome being transcribed (18). Second, Gruzdev et al. studied the transcriptome of an auxotrophic mutant, Salmonella enterica serovar Typhimurium SL1344, which had been desiccated on a plastic surface for 22 h (19). The most highly transcribed genes in that study were those involved in ribosomal structure and potassium ion transport (19). Third, Li et al. examined the transcriptomic responses of two Salmonella isolates to desiccation at a very low relative humidity (RH) (11%) (20). In that case, fatty acid metabolism was significantly upregulated after 2 h of desiccation on paper discs (20). From these studies, it is clear that a diverse range of responses can be induced in response to desiccation, depending on the precise experimental conditions involved.
The aim of this study was to examine the transcriptomic response of Salmonella Typhimurium ST4/74 desiccated on a stainless steel surface. A limited set of conditions was chosen for desiccation, and we acknowledge that this study represents only one potential condition during typical manufacture of low-moisture food products. These results provide an improved understanding of this bacterium's response upon transition into a dried state on an industrially relevant contact surface. Several S. Typhimurium genes required for survival of desiccation under these conditions were identified; among these, ProP is of critical importance.
MATERIALS AND METHODS
Bacterial strains and construction of static growth curves.
Salmonella Typhimurium ST4/74 was used in this study. Strain ST4/74 was originally isolated from the bowel of a calf with salmonellosis (21), and it is the prototrophic parent of the S. Typhimurium strain SL1344 (22). Strain SL1344 is a histidine auxotroph and has been widely used for the study of Salmonella virulence and gene regulation (23). There are only 8 single nucleotide polymorphisms that differentiate strains ST4/74 and SL1344 (24). The growth medium was Luria-Bertani (LB) medium (pH 7.0) that had been filter sterilized (0.22-μm polyethersulfone membrane) using Stericup filter units (FDR-125-010Q; Millipore) into 1-liter Millipore disposable flasks. To avoid any batch-to-batch variations, the same batch of powdered medium constituents was used throughout this study.
Bacteria were resuscitated from storage on cryobeads (Technical Service Consultants Ltd., Heywood, Lancashire, England) at −80°C onto LB agar (Difco) and were incubated overnight at 37°C. Experiments were then carried out at 24°C using a static system. This static system was selected for use because it is representative of a storage environment the bacteria are likely to encounter in the food production setting. The method used for the production of a standard inoculum was similar to that described by Rolfe et al. (25). One colony from an LB agar plate was used to inoculate 10 ml of LB medium in a 25-ml disposable universal tube, and this culture was incubated statically at 24°C for 48 h. The culture was serially diluted 1:100, and 200 μl of the diluted culture was then used to inoculate 500 ml of fresh LB medium in a 1-liter Stericup flask (approximately 4 × 103 CFU/ml) and was incubated statically at 24°C. This bacterial culture would then serve as the standardized inoculum for subsequent experiments. Viable counts were used to measure cell numbers periodically. Serial decimal dilutions were carried out in sterile LB medium, and 20 μl of each dilution was plated in triplicate onto LB agar. The plates were incubated for 24 h at 37°C. The CFU/ml was then calculated from dilutions that produced 4 to 50 colonies per 20-μl droplet. It should be noted that because approximately 20 h was required for the cells to reach stationary phase, it was necessary to use two identical cultures that had been staggered by 12 h for each of the three independent replicates.
Viability of ST4/74 on stainless steel.
Stainless steel (grade 304) was cut to dimensions of 5 cm by 0.7 cm by 0.1 cm. These stainless steel coupons were sterilized by soaking overnight in a 1% (wt/vol) solution of TriGene disinfectant. The stainless steel coupons were then placed in 70% ethanol for 3 h to remove any residue and were rinsed with sterile water. Ten coupons were then placed in an aluminum foil package and were heated to 200°C for 1 h. Dry heat has been used for sterilization previously (26, 27). Total viable counts were carried out to ensure the sterility of the stainless steel following this procedure (data not shown). The traditional method of autoclaving was not used to sterilize the stainless steel coupons, because the moist heat caused slight bends in some of the coupons. This process was repeated after each use.
A standard inoculum of Salmonella Typhimurium ST4/74 was prepared as described above and was grown to early stationary phase (ESP). Samples (10 ml) of ESP culture were centrifuged at 3,200 × g for 10 min; the supernatant was removed; and the cell pellet was resuspended in 1 ml fresh LB medium. This cell suspension was spread in a zigzag fashion across the steel coupons with 100 μl of culture (approximately 4 × 108 CFU) on each coupon. The stainless steel coupons were allowed to dry in a sterile laminar flow cabinet at 24°C for 4 h, at ca. 45% relative humidity (RH). At 0, 1, 2, 3, and 4 h, one steel coupon was placed in 5 ml phosphate-buffered saline (PBS) and was vortexed at full speed for 1 min to remove cells from the steel (27). The coupon was removed from the PBS by using sterile tweezers, and the liquid was centrifuged for 5 min at 3,200 × g. The supernatant was removed; the pellet was resuspended in 1 ml PBS; the suspension was serially diluted 1:10; and 100 μl was spread onto LB agar to measure viability. To examine the effect of rehydration, at the end of the 4 h-period, one steel coupon was placed in sterile H2O for 30 min before viable counts. This procedure was carried out in triplicate. The whole experiment was repeated on three separate, independent occasions.
RNA extraction from cells desiccated on stainless steel.
To examine the transcriptome of Salmonella Typhimurium ST4/74 after 4 h of desiccation on stainless steel, an experiment was designed to extract sufficient RNA from dried cells for use in microarray experiments.
LB medium (500 ml) was inoculated as described above and was incubated at 24°C until the culture reached ESP. The stainless steel coupons were inoculated as described above with 100 μl of culture per coupon (a total of 1 ml per pack of 10 coupons). The bacteria were air dried on the steel for 4 h at 24°C in a laminar flow cabinet. After 4 h, the coupons from one pack (10 coupons) were placed in 20 ml RNAlater (Ambion) to stabilize the RNA, incubated for 30 min at 24°C, and vortexed at full speed intermittently until the cells had been removed. The resulting suspension was centrifuged for 15 min at 3,200 × g; the majority of the supernatant was discarded; and the cell pellet was first resuspended in the remaining liquid and then transferred to a 1.5-ml microcentrifuge tube. The suspension was centrifuged for 1 min at 20,800 × g, after which the supernatant was discarded and the pellet resuspended on ice in 1 ml TRIzol (Invitrogen). To examine the effect of rehydration on the cells, after the 4 h of drying, 10 stainless steel coupons were placed in 20 ml sterile H2O for 30 min instead of RNAlater. A 10-ml sample of the original ESP culture was also harvested in a similar manner and was resuspended in 1 ml TRIzol on ice. Total RNA was then extracted as described previously by Kröger et al. and was resuspended in a final volume of 80 μl RNase-free water (24). Samples were treated with DNase I (catalog no. EN0521; Fermentas) and SUPERase·In RNase inhibitor (1 μl; catalog no. AM2694; Ambion). Samples were adjusted to a concentration of >1,300 ng/μl with RNase-free water.
A NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA) was used to quantify RNA concentrations. The Agilent 2100 Bioanalyzer (Agilent, Stockport, United Kingdom) was used to assess RNA quality according to the manufacturer's instructions. Ladders, gels, dye, and chips were taken from the Agilent RNA 6000 Nano kit (catalog no. 5067-1511).
Microarray preparation and transcriptome analysis.
Microarray analysis was carried out using the SALSIFY2 array (Agilent microarray design identifier [AMADID] 037367; Agilent Technologies, Santa Clara, CA). In order to enable an optimum comparison between our samples here and those in future experiments, a common-reference (indirect) approach was used (28). This method avoids the use of dye swap experiments and can allow the comparison of expression profile results obtained under different growth conditions (28, 29). The methods used have been described previously (30). Using this approach, RNA produced under the defined environmental condition being tested is reverse transcribed to cDNA, which is fluorescently labeled with Cy3-dCTP using a random priming reaction. Genomic DNA (gDNA) was purified from Salmonella Typhimurium ST4/74 and was fluorescently labeled with Cy5-dCTP. This gDNA was then used as the standard reference in each experiment. Dye-labeled cDNA and gDNA were mixed, denatured, and hybridized to the array over an 18-h period at 65°C; then they were washed according to the manufacturer's instructions (Agilent). Microarray slides were cleaned with inert gas to remove any debris before scanning with an Agilent microarray scanner (Agilent Technologies, Santa Clara, CA). Scans were carried out at 5-μm resolution with green and red photomultiplier tube (PMT) values set to 100% and an extended-dynamic-range (XDR) value of 0.1. The images generated were saved as multi-image TIFF files. Feature extraction software (Agilent Technologies) was used to extract data, which were then analyzed using GeneSpring, version 7.3 (Agilent Technologies, Santa Clara, CA). Two biological replicates were carried out for each condition. Expression profiles were compared to that of the control early-stationary-phase culture. Differentially expressed genes were identified using a t test (P < 0.05) with a 5-fold-change cutoff. Only genes that were detected by two or more probes above the 5-fold cutoff were considered.
qRT-PCR.
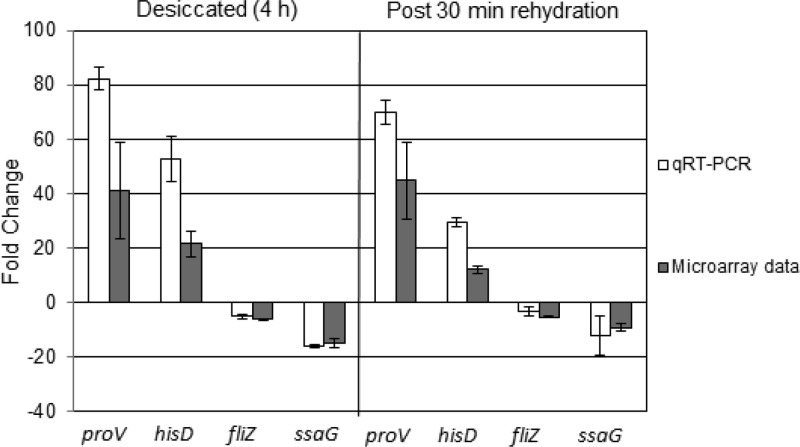
Quantitative real-time PCR (qRT-PCR) was used to validate the results obtained from microarray experiments. The primers used are listed in Table S1 in the supplemental material. From the microarray results, two upregulated (proV, hisD) and two downregulated (fliZ, ssaG) genes were chosen for analysis, and their expression profiles were normalized against that of the nondifferentially expressed housekeeping gene gapA. qRT-PCR was carried out using the Qiagen QuantiTect SYBR green RT-PCR kit in a Mastercycler ep realplex system, with a total-RNA concentration of 50 ng for each sample. Three biological replicates were carried out, and the fold change was calculated using the 2−ΔΔCT method (31).
Construction of deletion mutants and P22 transduction.
The lambda Red method for gene deletions was used for the construction of gene knockouts as described previously (32). Approximately 70-mer oligonucleotide primers were designed to contain 50 bases that were specific to the start and end of the gene/operon of interest. Additional nucleotides were chemically added to the 3′ ends of both the forward (5′-GTGTAGGCTGGAGCTGCTTC-3′) and reverse (5′-CATATGAATATCCTCCTTA-3′) primers and served as sites for PCR amplification of the antibiotic resistance gene from the pKD3 or pKD4 plasmid. The primers used for the generation of mutants are listed in Table S2 in the supplemental material. The mutations were confirmed using the primers listed in Table S2 and were verified by sequencing. The mutations were then reintroduced into a wild-type Salmonella Typhimurium ST4/74 background via transduction with phage P22 HT105/1 int201 (33) using previously described protocols (34). Antibiotic resistance markers were removed using the pCP20 plasmid as described previously (32). The following mutations were constructed in ST4/74: ΔproP (strain CFS-0004), ΔproU (CFS-0005), and ΔosmU (CFS-0006). The ΔrpoE mutation was constructed previously in S. Typhimurium SL1344 (mutant strain JVS-1028) as described elsewhere (35). This deletion was subsequently transduced into ST4/74, yielding strain JH3630 (see Table S2 in the supplemental material).
Longer-term survival of deletion mutants.
Growth curves were generated for isogenic mutant strains, by using the method outlined above for the reference strain Salmonella Typhimurium ST4/74, to determine the point of ESP. ESP cultures of mutants were then dried onto stainless steel as outlined above and were examined for viability over a period of 6 weeks. Student t tests were carried out to determine if there were any statistically significant differences (cutoff P value, <0.05 by a two-tailed test) in desiccation survival between the wild-type strain ST4/74 and the deletion mutants.
Microarray data accession number.
Data from this study have been deposited in NCBI's Gene Expression Omnibus with GEO accession number GSE46763.
RESULTS
Experimental design, bacterial growth, and desiccation.
A static, room temperature (24°C), ca. 45% RH growth condition was chosen to represent growth during storage at room temperature and subsequent desiccation. The experimental design involved bacterial cells at early stationary phase (ESP) in order to limit variability between independent experiments (see Fig. S1 in the supplemental material). Following growth to ESP, bacterial cells were removed and were subsequently dried down onto stainless steel coupons. It has been stated previously that Salmonella is likely to enter a dormant state under reduced-moisture conditions, since it lacks the water necessary to support biological reactions (18). Furthermore, after extended periods of desiccation, important signals may be missed due to RNA degradation. A 4-h period of drying on the steel was chosen as the point at which the transcriptome was to be examined, since it was hypothesized that the mechanisms used by bacteria in the initial stages of desiccation may be critical for their long-term survival. Over this period, a cell loss of approximately 0.74 log CFU/ml was observed (data not shown). A further 0.72 log CFU/ml decrease in numbers, which could be attributed to osmotic shock, was observed after 30 min of rehydration (data not shown).
Identification of genes differentially expressed under desiccated conditions.
To examine the response of S. Typhimurium ST4/74 to desiccation, transcriptomic analysis was carried out on bacterial cells desiccated onto stainless steel for 4 h. These results were compared to those for a liquid-culture control with the same bacteria.
Desiccation stress (4 h after inoculation onto steel) caused differential expression of 266 genes (>5-fold change, with a P value of <0.05 [see Data set S1 in the supplemental material]). Of these, 79 genes were upregulated and 187 were downregulated. Some of the most highly upregulated genes expressed following desiccation are involved in the uptake of osmoprotectants, including proP and the proVWX operon. The OsmU ABC transporter system (the STM1491 to STM1494 genes, known as osmVWXY) was also upregulated under desiccation stress (36). This system is involved in the uptake of osmoprotectant molecules and is induced by osmotic stress using NaCl (36). Genes involved in trehalose biosynthesis (otsAB) were upregulated >11-fold over their level of expression in liquid culture. The global regulator rpoE and genes encoding sigma E-regulatory proteins (rseA and rseB) were upregulated 6- to 8-fold by desiccation stress.
An increase in the expression of genes involved in histidine biosynthesis (hisABCDGH) was observed, suggesting that this amino acid may play an important role in survival at low moisture. Genes involved in leucine biosynthesis, cysteine biosynthesis, and fatty acid metabolism were also upregulated under desiccation. A number of genes involved in the formation of Fe-S clusters (including iscA, iscS, and fdx), as well as the manganese superoxide dismutase gene sodA, which are induced under iron-limiting conditions (37, 38), were also upregulated. The nhaA gene, which encodes a Na+/H+ antiporter, was upregulated; this antiporter is associated with high-salinity environments and is used by bacteria to remove excess sodium from the cell (39).
The transcriptomic data showed that the majority of the differentially expressed genes were downregulated during desiccation (187 genes). These included genes involved in chemotaxis, including cheM, tcp, and tsr, and the flagellar genes fliS, fliT, and fliZ. Several other genes, involved in amino acid transport and metabolism, anaerobic metabolism, and carbohydrate transport and metabolism, were downregulated. The functions of more than half of the genes that showed >5-fold downregulation were unknown. Full gene lists are provided in Data set S1 in the supplemental material.
Identification of genes differentially expressed following rehydration of previously desiccated cells.
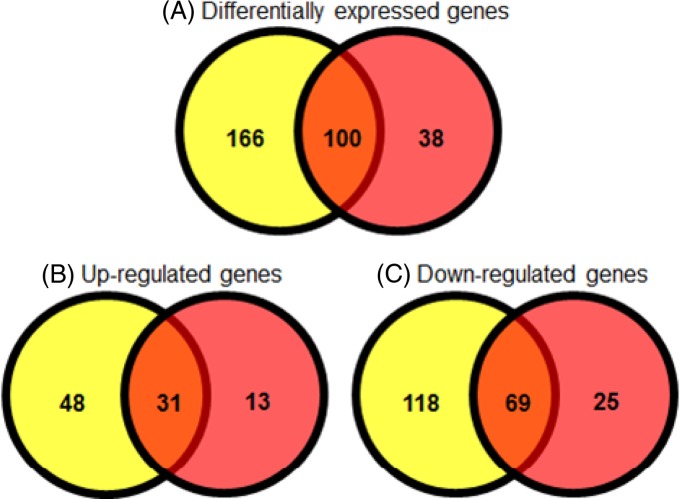
Food production facilities are commonly maintained as moisture-free environments to improve food safety. However, water may enter a facility via several routes, such as leaks in the pipe infrastructure, or during wet-cleaning of the facility. In an effort to mimic this scenario of moving from a desiccated to a hydrated condition, bacterial cells that had been dried onto stainless steel were rehydrated and were subjected to transcriptomic analysis. Comparison with the static planktonic culture revealed differential expression of 138 genes (>5-fold-change, with a P value of <0.05), of which 44 were upregulated while 94 were downregulated (see Data set S2 in the supplemental material). One hundred of these genes (72.4%) were also differentially expressed after desiccation (Fig. 1). The osmoprotectant transporter genes proP and proVWX remained highly expressed in cells after rehydration. Trehalose-biosynthetic genes (otsAB) and the osmU ABC transporter were no longer overexpressed postrehydration, suggesting that these genes are specifically required for survival in low-moisture environments. Phosphate transport genes (pstABCS) showed increased expression in rehydrated cells. As in cells in a desiccated state, the global regulator rpoE was upregulated following rehydration. A number of small RNAs were also upregulated, including ryeF and sTnc800, which were found solely after rehydration.
Fig 1.
Comparison of statistically significantly (P < 0.05) differentially expressed genes with >5-fold changes. Yellow circles represent desiccated samples, and red circles represent rehydrated samples. (A) Genes differentially expressed with >5-fold changes; (B) genes upregulated >5-fold; (C) genes downregulated >5-fold.
Several genes that were downregulated in desiccated cells also showed the same pattern of expression postrehydration; these included chemotaxis genes and those involved in anaerobic metabolism. Rehydration restored the expression levels of flagellar genes and the iron storage genes ftn and bfr. Other genes that were downregulated significantly following desiccation and returned to normal transcriptional levels upon rehydration included the cell division activator cedA and the RNase E regulator menG (24, 40).
Transcriptomic data were confirmed by quantitative real-time PCR (qRT-PCR) (Fig. 2). This showed statistically significant changes (P < 0.05) in the expression of genes tested in both desiccated and rehydrated samples, compared with the liquid-culture control. The same pattern of expression observed for the selected genes in the transcriptomic arrays was also obtained by qRT-PCR, thereby validating the data derived from microarrays.
Fig 2.
Validation of microarray data using qRT-PCR. The housekeeping gene gapA was used to normalize the data, and the fold change was calculated using the 2−ΔΔCT method.
Physiological systems required for survival of desiccation.
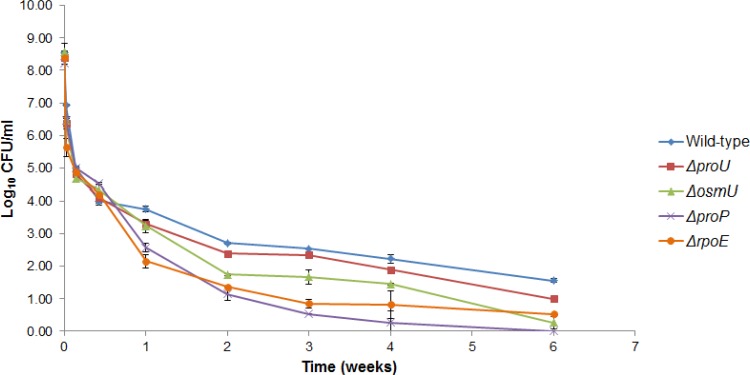
The realization that the proP, proU, and osmU genes were overexpressed in low-moisture environments led us to hypothesize that osmoprotectant transport systems could play a critical role when bacteria are dried. We tested this hypothesis by constructing a series of deletion mutants: the ΔproP, ΔproU, ΔosmU, and ΔrpoE mutants. All deletion mutants showed statistically significant (P < 0.05) decreases in viability from that for wild-type ST4/74 upon desiccation (Fig. 3). The greatest survival defect was seen for the S. Typhimurium ΔproP mutant, which became undetectable after 4 weeks of desiccation.
Fig 3.
The ProP, ProU, and OsmU osmoprotectant systems and the global regulator RpoE are required for optimal survival of desiccation. All deletion mutants showed significant decreases in viability from that for the wild type (P < 0.05). The ΔproP mutant showed the greatest survival defect.
DISCUSSION
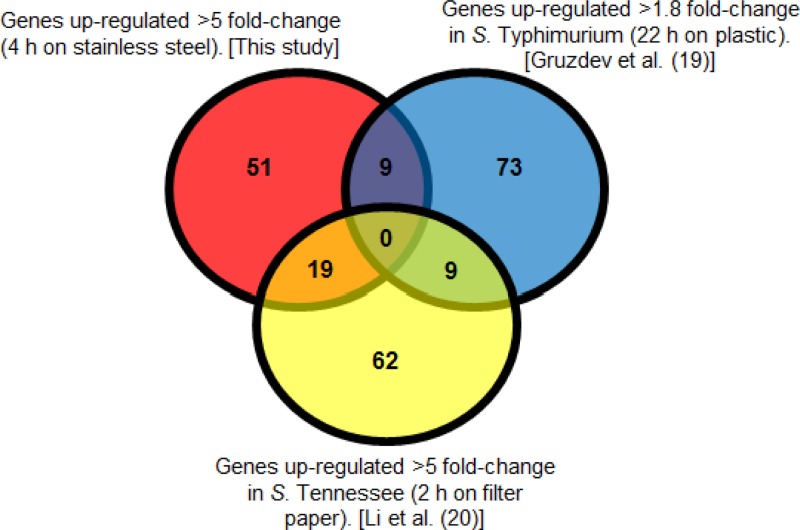
Salmonella strains have been the causative agents of several food-borne outbreaks due to low-moisture foods (7, 12, 15, 41, 42). Recently, two studies examining the transcriptomes of Salmonella species desiccated onto an abiotic surface have been published (19, 20). These are the first insights into transcriptional mechanisms contributing to survival under desiccation stress. A comparison of genes upregulated under desiccation stress from this study and those reports is shown in Fig. 4. From these comparative data, several differentially regulated genes are shared between these studies, but interestingly, the majority of upregulated genes identified are unique to each investigation. These discrepancies may arise from the different methodologies used in the different studies, including such factors as the surface used for desiccation, the length of the desiccation period, or the Salmonella serotype used. First, Gruzdev et al. examined the transcriptome after 22 h on a plastic surface (19). In that study, Salmonella suspensions from a stationary-phase culture were air dried on 90-mm petri dishes in a biosafety cabinet at 25°C for 22 h and were compared to a control culture held at the same temperature in a 50-ml tube (19). Therefore, signals that might have been present at an earlier point of desiccation (such as that examined in this study) might have been absent after 22 h due to the short life of RNA. Second, Li et al. examined Salmonella enterica serovar Tennessee after 2 h of desiccation on filter paper (20). Stationary-phase cultures were dried onto filter paper discs and were kept dry for 2 h, after which RNA was isolated and compared to that from the liquid control (20). The different serotype, time point, and surface examined may have contributed to the differences observed.
Fig 4.
Comparison of the genes identified as upregulated under desiccation by this study with those found upregulated by Gruzdev et al. (19) and by Li et al. (20).
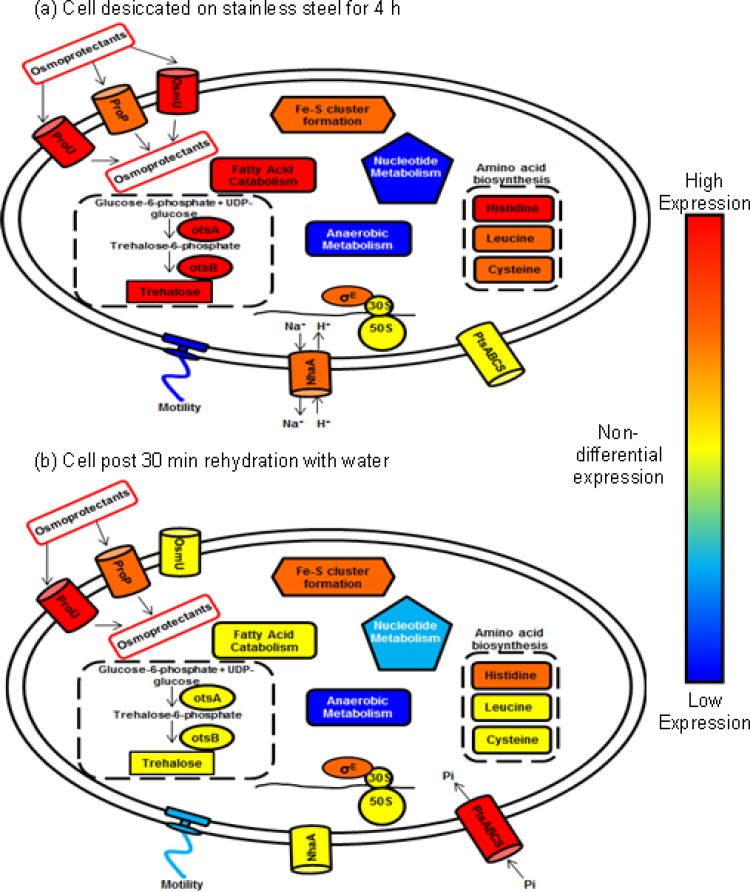
In this study, we investigated the response of a prototrophic Salmonella Typhimurium strain, ST4/74, to desiccation on industrial-grade stainless steel at the transcriptomic level. We identified key signals that were upregulated upon drying, and we subsequently confirmed their contributions to survival by using mutant studies. The effect of rehydrating dried cells with water was also compared, since water is often the vector by which bacteria can disseminate quickly and efficiently throughout the production environment (43). To our knowledge, this is the first study to examine, at a transcriptional level, the nature of the effect(s) of desiccation on an industrially relevant surface as well as the events that follow rehydration. Our findings are broadly summarized in a schematic shown in Fig. 5. The processes shown in Fig. 5a are related to signals to preserve the osmotic stability and homeostasis of the bacterial cell. Osmoprotectant influx via three transporters is increased in order to protect the cell from the deleterious effects of osmotic shock. The level of trehalose biosynthesis is significantly increased via the metabolism of glucose. Due to this energy requirement, fatty acid catabolism increases, producing components from which energy can be derived. The upregulation of Fe-S clusters was also noted, and this is likely linked to the increased demands for energy made by the terminal electron transport chain. Upregulation in the synthesis of some key amino acids, including histidine, leucine, and cysteine, was also observed. Motility-related gene expression was downregulated, a feature that would be consistent with the requirement for bacterial cells to shut down unnecessary energy demands. In contrast, Fig. 5b depicts changes occurring in the cell after rehydration. Osmoprotectant solute transport systems appear less active, and trehalose biosynthesis is no longer upregulated. Similarly, the need for fatty acid catabolism is diminished. Interestingly, an increase in the phosphate transport level occurred; this would support the thesis that the bacterial cell requires phosphate elements for the synthesis of key building blocks, such as nucleic acids necessary for growth. Levels of expression of motility-associated genes have also begun to return to normal. The effects of these changes are discussed in more detail below.
Fig 5.
Proposed model of cellular changes occurring during desiccation of a cell on steel for 4 h (a) and during rehydration of a desiccated cell with water (b). Each symbol represents a range of genes involved in particular physiological or regulatory processes. Blue indicates decreased expression; yellow indicates genes that were expressed at similar levels in the planktonic control bacteria; and red indicates increased expression.
When bacteria are exposed to desiccation stress, the water activity in these cells is lowered. To combat the loss of water, and to maintain survival, bacteria accumulate low-molecular-weight compatible solutes known as osmoprotectants (44). Examples of some known osmoprotectants include l-proline, betaine (N-trimethylglycine), choline, and trehalose (44). Three transporters of osmoprotectant solutes were recorded as upregulated 6.77- to 50.83-fold (see data set S1 in the supplemental material). These included proP, proU (proVWX), and osmU (osmVWXY; STM1491 to -94). The ProP system belongs to the major facilitator superfamily (MFS) of permeases, while both the ProU and OsmU systems are known ABC transporters (36, 45, 46). All three transporters have been shown to promote survival under NaCl stress in a broth culture system (36, 47). Unlike ProP and ProU, the OsmU transporter has a low affinity for l-proline and glycine betaine, and it is possible that OsmU recognizes an alternative, as yet uncharacterized substrate (36, 44, 48–50). Our study demonstrated that these genes are also required for survival on stainless steel, which is representative of a typical surface used in food production, and that the loss of any of the three transporters reduces the viability of S. Typhimurium ST4/74. It was noted that proP was critically important for survival. This observation may explain in part why the ST4/74 ΔproU and ΔosmU mutants remain viable for the duration of the survival assay, given that the ProP system continues to function. Interestingly, of these three systems, the proP-encoded transporter was the least upregulated (6.77-fold change), but it was nevertheless essential for persistence.
As mentioned above, Gruzdev et al. examined the transcriptome of S. Typhimurium SL1344 dehydrated on a plastic surface. That study did not identify any upregulated osmoprotectant transport systems (19). This discrepancy could possibly be accounted for by the different experimental protocol and controls used, since the transcriptome was measured at 22 h postdesiccation, and some of these earlier signals may have been absent/lost. Li et al. reported that both the ProU transport system and some of the genes of the OsmU transport system were upregulated when Salmonella cells were desiccated on paper discs, which supports our findings (20).
Our data highlight the importance of both proP and proU in Salmonella Typhimurium ST4/74 following rehydration in water. Although the osmU operon (STM1491 to -94) was highly upregulated under low-moisture conditions, it appeared to be redundant after rehydration. Deletion of osmU resulted in a marked reduction in long-term viability from that of the wild-type strain, indicating the functional importance of osmU in desiccation survival (Fig. 3). OsmU has been reported to alleviate growth inhibition in a ΔproP ΔproU double mutant; however, deletion of osmU alone can increase resistance to high salt levels and virulence due to an increase in the production of trehalose (36, 51). It is possible, therefore, that the regulation of osmU and the regulation of the otsAB genes, involved in trehalose production, are somehow linked, in a manner as yet undefined. While trehalose-biosynthetic genes were upregulated under desiccation stress, a situation similar to that for osmU, they returned to basal levels following rehydration in water. The induction of the cytoplasmic trehalase (encoded by treF) under desiccation stress suggests that bacteria catabolize trehalose to produce glucose, an efficient source of metabolic energy (52). Trehalose production in desiccated Salmonella cells has been measured previously (20). The results from that study showed a significant increase in trehalose production in an S. Tennessee isolate that was more tolerant to desiccation than an S. Typhimurium LT2 control (20). This finding further confirms the role of the otsAB genes in desiccation tolerance.
Models of osmoregulation in enteric bacteria suggest that in order to initiate the transport of osmoprotectants, an increase in K+ transport is required to stimulate the transcription of the genes required, either directly or via the formation of potassium glutamate (53–56). Balaji et al. reported that an alternative series of events, involving the upregulation of both proU and proP prior to the activation of a potassium ion transport channel, such as kdpFABC, may occur in Salmonella Typhimurium (56). The data from our study would appear to support this hypothesis, since no genes involved in potassium transport were found to be upregulated above 5-fold after 4 h. In contrast, however, Gruzdev et al. reported that the genes most highly upregulated after 22 h of desiccation were those of the kdpFABC operon, which would coincide with this alternative activation model (19).
A study of Escherichia coli showed that the overproduction of histidine biosynthesis gene products can lead to osmosensitivity (57). Conversely, upregulation of histidine biosynthesis was observed under desiccation stress and also, to some extent, after rehydration with water. Similar increased expression was reported by Gruzdev et al. after the dehydration of Salmonella Typhimurium on a plastic surface (19). These observations suggest that increasing the amount of proteins that contain histidine could provide the bacteria with some form of stabilization or protection under low-moisture conditions. Gruzdev et al. further explored this observation by deleting hisD to eliminate histidine biosynthesis; however, the tolerance of the deletion mutant to dehydration was comparable to that of the auxotrophic reference strain (19). Furthermore, the choice of the model bacterium in the latter study may be somewhat unfortunate, and the results consequently difficult to interpret, given that this particular reference strain is a histidine auxotroph. Further investigation into the role of this amino acid in desiccation tolerance may be warranted.
In our study, a gene encoding an alternative sigma factor, rpoE, was found to be upregulated both under desiccation stress and after rehydration. This sigma factor, σE, plays a key role in the regulation of genes involved in cell envelope stress, together with genes that respond to heat, starvation, osmotic shock with NaCl, and desiccation (18, 19, 35, 58–60). σE clearly plays a key role when cells are exposed to desiccation stress on stainless steel coupons, since the long-term survival of a ΔrpoE deletion mutant of S. Typhimurium was significantly reduced from that of the wild type (P < 0.05) (Fig. 3).
Previous studies have indicated that the formation of filaments and the production of curli fimbriae can aid the survival and long-term persistence of Salmonella species and E. coli under low-moisture conditions (16, 17, 61–64). However, no genes for the biosynthesis of curli fimbriae (csgDEFG or csgBAC) or cellulose (bcsABZC) were upregulated following 4 h of desiccation in our study. Moreover, none of these genes were upregulated in the studies reported by Gruzdev et al. and Li et al. (19, 20). This observation may indicate that while the formation of filaments may be important for survival in a low-moisture broth-based system, this may not be the case when bacteria are dried onto an abiotic food contact surface. White et al. demonstrated that bacteria lacking the ability to produce two main components found in a Salmonella biofilm, cellulose and curli fimbriae, exhibit a phenotype of susceptibility to desiccation (17). Since none of these genes were found to be upregulated under desiccation, it is possible that isolates capable of forming these structures prior to drying have a greater ability to attach to surfaces but that curli fimbriae and cellulose are not required once drying has occurred.
When cells were rehydrated with water after initially being desiccated, the expression of several of these genes, including proP, proU, and rpoE, as well as a number of Fe-S cluster-related genes (iscA, iscS, fdx and nifU), is required. An inorganic phosphate ABC transporter encoded by pstSCAB was observed to be upregulated only when dried cells were rehydrated with water. Phosphate is essential for the synthesis of nucleic acids and phospholipids and for phosphorylation-based signaling pathways within the bacterial cell (25). The induction of this transporter may suggest that these bacteria have entered the early stages of a lag phase, as they attempt to scavenge a central building block of the cell (25). We speculate that this occurrence may prime bacteria for growth if and when nutrients again became available.
Desiccation stress caused a marked reduction in a number of genes involved in motility and chemotaxis. Similar results have been observed previously (20). This inhibition was partially maintained postrehydration, suggesting that the cells are expending energy elsewhere. However, the expression of some flagellum-encoding genes did return to normal levels. It is possible that motility would be induced at a later stage to aid in dissemination throughout the aqueous environment.
Conclusion.
The mechanism(s) used by Salmonella to promote its survival under low- and intermediate-moisture conditions is poorly understood. Our study investigated the response of Salmonella Typhimurium ST4/74 to desiccation and subsequent rehydration at a transcriptomic level. Several features were identified that contributed to desiccation survival, including the importance of the ProP, ProU, and OsmU osmoprotectant transporters. In addition, other cellular responses were observed, including the increase in trehalose production and histidine biosynthesis and the upregulation of an alternative sigma factor encoded by rpoE. While this study provides early information on the initial response of Salmonella Typhimurium to desiccation, further characterization to elucidate the importance of other upregulated systems is warranted so as to gain a more holistic understanding of the process(es) involved in desiccation tolerance. This investigation constitutes the first in-depth study of the processes occurring within a previously dried cell upon the reintroduction of moisture. The findings of our study may aid manufacturers of low-moisture foods in the design of effective control strategies by, for example, using the insight into desiccation stress to optimize cleaning and sanitation regimes. Such approaches could be aimed at the elimination of desiccated Salmonella cells from the production environment, thereby improving food safety and protecting public health.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Irish Research Council in conjunction with Unilever's Safety & Environmental Assurance Centre.
Footnotes
Published ahead of print 10 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00515-13.
REFERENCES
- 1.Podolak R, Enache E, Stone W, Black DG, Elliott PH. 2010. Sources and risk factors for contamination, survival, persistence, and heat resistance of Salmonella in low-moisture foods. J. Food Prot. 73:1919–1936 [DOI] [PubMed] [Google Scholar]
- 2.Hiramatsu R, Matsumoto M, Sakae K, Miyazaki Y. 2005. Ability of Shiga toxin-producing Escherichia coli and Salmonella spp. to survive in a desiccation model system and in dry foods. Appl. Environ. Microbiol. 71:6657–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotzekidou P. 1998. Microbial stability and fate of Salmonella Enteritidis in halva, a low-moisture confection. J. Food Prot. 61:181–185 [DOI] [PubMed] [Google Scholar]
- 4.Nummer B, Smith J. 2012. Survival of Salmonella in a high sugar, low water-activity, peanut butter flavored candy fondant. Food Control 27:184–187 [Google Scholar]
- 5.Gruzdev N, Pinto R. 2012. Persistence of Salmonella enterica during dehydration and subsequent cold storage. Food Microbiol. 32:415–422 [DOI] [PubMed] [Google Scholar]
- 6.Breuer T. 1999. CDC investigations: the May 1998 outbreak of Salmonella Agona linked to cereal. Cereal Foods World 44:185–186 [Google Scholar]
- 7.Rowe B, Hutchinson DN, Gilbert RJ, Hales BH, Begg NT, Dawkins HC, Jacob M, Rae FA, Jepson M. 1987. Salmonella Ealing infections associated with consumption of infant dried milk. Lancet ii:900–903 [DOI] [PubMed] [Google Scholar]
- 8.Kapperud G, Gustavsen S, Hellesnes I, Hansen AH, Lassen J, Hirn J, Jahkola M, Montenegro MA, Helmuth R. 1990. Outbreak of Salmonella typhimurium infection traced to contaminated chocolate and caused by a strain lacking the 60-megadalton virulence plasmid. J. Clin. Microbiol. 28:2597–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis DA, Paramathasan R, White DG, Neil LS, Tanner AC, Hill SD, Bruce JC, Stuart JM, Ridley AM, Threlfall EJ. 1996. Marshmallows cause an outbreak of infection with Salmonella enteritidis phage type 4. Commun. Dis. Rep. CDR Rev. 6:R183–R186 [PubMed] [Google Scholar]
- 10.Rushdy AA, Stuart JM, Ward LR, Bruce J, Threlfall EJ, Punia P, Bailey JR. 1998. National outbreak of Salmonella Senftenberg associated with infant food. Epidemiol. Infect. 120:125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werber D, Dreesman J, Feil F, van Treeck U, Fell G, Ethelberg S, Hauri AM, Roggentin P, Prager R, Fisher IS, Behnke SC, Bartelt E, Weise E, Ellis A, Siitonen A, Andersson Y, Tschäpe H, Kramer MH, Ammon A. 2005. International outbreak of Salmonella Oranienburg due to German chocolate. BMC Infect. Dis. 5:7. 10.1186/1471-2334-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC 2007. Multistate outbreak of Salmonella Tennessee infections associated with peanut butter—United States, 2006–2007. MMWR Morb. Mortal. Wkly. Rep. 56:521–524 [PubMed] [Google Scholar]
- 13.Gruzdev N, Pinto R, Sela S. 2011. Effect of desiccation on tolerance of Salmonella enterica to multiple stresses. Appl. Environ. Microbiol. 77:1667–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spector MP, Kenyon WJ. 2012. Resistance and survival strategies of Salmonella enterica to environmental stresses. Food Res. Int. 45:455–481 [Google Scholar]
- 15.Beuchat LR, Komitopoulou E, Beckers H, Betts RP, Bourdichon F, Fanning S, Joosten HM, Ter Kuile BH. 2013. Low water activity foods: increased concern as vehicles of foodborne pathogens. J. Food Prot. 76:150–172 [DOI] [PubMed] [Google Scholar]
- 16.Mattick KL, Jorgensen F, Legan JD, Cole MB, Porter J, Lappin-Scott HM, Humphrey TJ. 2000. Survival and filamentation of Salmonella enterica serovar Enteritidis PT4 and Salmonella enterica serovar Typhimurium DT104 at low water activity. Appl. Environ. Microbiol. 66:1274–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White AP, Gibson DL, Kim W, Kay WW, Surette MG. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X, Li Z, Zhang W. 2012. Transcriptome sequencing of Salmonella enterica serovar Enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol. 30:311–315 [DOI] [PubMed] [Google Scholar]
- 19.Gruzdev N, McClelland M, Porwollik S, Ofaim S, Pinto R, Saldinger-Sela S. 2012. Global transcriptional analysis of dehydrated Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 78:7866–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Bhaskara A, Megalis C, Tortorello ML. 2012. Transcriptomic analysis of Salmonella desiccation resistance. Foodborne Pathog. Dis. 9:1143–1151 [DOI] [PubMed] [Google Scholar]
- 21.Rankin J, Taylor R. 1966. The estimation of doses of Salmonella Typhimurium suitable for the experimental production of disease in calves. Vet. Rec. 78:706–707 [DOI] [PubMed] [Google Scholar]
- 22.Richardson EJ, Limaye B, Inamdar H, Datta A, Manjari KS, Pullinger GD, Thomson NR, Joshi RR, Watson M, Stevens MP. 2011. Genome sequences of Salmonella enterica serovar Typhimurium, Choleraesuis, Dublin, and Gallinarum strains of well-defined virulence in food-producing animals. J. Bacteriol. 193:3162–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoiseth SK, Stocker B. 1981. Aromatic-dependent Salmonella Typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239 [DOI] [PubMed] [Google Scholar]
- 24.Kröger C, Dillon SC, Cameron ADS, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hébrard M, Händler K. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. U. S. A. 109:E1277–E1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, Cameron ADS, Alston M, Stringer MF, Betts RP, Baranyi J, Peck MW, Hinton JCD. 2012. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J. Bacteriol. 194:686–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhir VK, Dodd C. 1995. Susceptibility of suspended and surface-attached Salmonella Enteritidis to biocides and elevated temperatures. Appl. Environ. Microbiol. 61:1731–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vatanyoopaisarn S, Nazli A, Dodd CE, Rees CE, Waites WM. 2000. Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl. Environ. Microbiol. 66:860–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang YH, Speed T. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579–588 [DOI] [PubMed] [Google Scholar]
- 29.Thompson A, Rowley G, Alston M, Danino V, Hinton JCD. 2006. Salmonella transcriptomics: relating regulons, stimulons and regulatory networks to the process of infection. Curr. Opin. Microbiol. 9:109–116 [DOI] [PubMed] [Google Scholar]
- 30.Ygberg SE, Clements MO, Rytkönen A, Thompson A, Holden DW, Hinton JCD, Rhen M. 2006. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 74:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 32.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmieger H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75–88 [DOI] [PubMed] [Google Scholar]
- 34.Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 35.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JCD, Vogel J. 2006. σE dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62:1674–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frossard SM, Khan AA, Warrick EC, Gately JM, Hanson AD, Oldham ML, Sanders DA, Csonka LN. 2012. Identification of a third osmoprotectant transport system, the OsmU system, in Salmonella enterica. J. Bacteriol. 194:3861–3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsolis RM, Bäumler AJ, Heffron F. 1995. Role of Salmonella Typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect. Immun. 63:1739–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247–281 [DOI] [PubMed] [Google Scholar]
- 39.Padan E, Gerchman Y, Rimon A, Rothman A, Dover N, Carmel-Harel O. 1999. The molecular mechanism of regulation of the NhaA Na+/H+ antiporter of Escherichia coli, a key transporter in the adaptation to Na+ and H+. Novartis Found. Symp. 221:183–199. [DOI] [PubMed] [Google Scholar]
- 40.Katayama T, Takata M, Sekimizu K. 1997. CedA is a novel Escherichia coli protein that activates the cell division inhibited by chromosomal DNA over-replication. Mol. Microbiol. 26:687–697 [DOI] [PubMed] [Google Scholar]
- 41.Brouard C, Espié E, Weill FX, Kérouanton A, Brisabois A, Forgue AM, Vaillant V, de Valk H. 2007. Two consecutive large outbreaks of Salmonella enterica serotype Agona infections in infants linked to the consumption of powdered infant formula. Pediatr. Infect. Dis. J. 26:148–152 [DOI] [PubMed] [Google Scholar]
- 42.Isaacs S, Aramini J, Ciebin B, Farrar J, Ahmed R, Middleton D, Chandran A, Harris L, Howes M, Chan E. 2005. An international outbreak of salmonellosis associated with raw almonds contaminated with a rare phage type of Salmonella Enteritidis. J. Food Prot. 68:191–198 [DOI] [PubMed] [Google Scholar]
- 43.Beuchat L, Komitopoulou E, Betts R, Beckers H, Bourdichon F, Joosten H, Fanning S, ter Kuile B. 2011. Persistence and survival of pathogens in dry foods and dry food processing environments. ILSI Europe Report Series; ILSI Europe, Brussels, Belgium [Google Scholar]
- 44.Csonka LN. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacMillan SV, Alexander DA, Culham DE, Kunte HJ, Marshall EV, Rochon D, Wood JM. 1999. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim. Biophys. Acta 1420:30–44 [DOI] [PubMed] [Google Scholar]
- 46.Stirling D, Hulton C, Waddell L, Park S, Stewart G, Booth I, Higgins C. 1989. Molecular characterization of the proU loci of Salmonella Typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol. Microbiol. 3:1025–1038 [DOI] [PubMed] [Google Scholar]
- 47.Dunlap VJ, Csonka LN. 1985. Osmotic regulation of l-proline transport in Salmonella Typhimurium. J. Bacteriol. 163:296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Csonka LN. 1982. A third l-proline permease in Salmonella Typhimurium which functions in media of elevated osmotic strength. J. Bacteriol. 151:1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cairney J, Booth I, Higgins C. 1985. Osmoregulation of gene expression in Salmonella Typhimurium: proU encodes an osmotically induced betaine transport system. J. Bacteriol. 164:1224–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cairney J, Booth I, Higgins C. 1985. Salmonella Typhimurium proP gene encodes a transport system for the osmoprotectant betaine. J. Bacteriol. 164:1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilonieta M, Nagy TA, Jorgensen DR, Detweiler CS. 2012. A glycine betaine importer limits Salmonella stress resistance and tissue colonization by reducing trehalose production. Mol. Microbiol. 84:296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horlacher R, Uhland K, Klein W, Ehrmann M, Boos W. 1996. Characterization of a cytoplasmic trehalase of Escherichia coli. J. Bacteriol. 178:6250–6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee SJ, Gralla JD. 2004. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell 14:153–162 [DOI] [PubMed] [Google Scholar]
- 54.Booth I, Higgins C. 1990. Enteric bacteria and osmotic stress: intracellular potassium glutamate as a secondary signal of osmotic stress? FEMS Microbiol. Lett. 75:239–246 [DOI] [PubMed] [Google Scholar]
- 55.Epstein W. 1986. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Lett. 39:73–78 [Google Scholar]
- 56.Balaji B, O'Connor K, Lucas JR, Anderson JM, Csonka LN. 2005. Timing of induction of osmotically controlled genes in Salmonella enterica serovar Typhimurium, determined with quantitative real-time reverse transcription-PCR. Appl. Environ. Microbiol. 71:8273–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray ML, Hartman PE. 1972. Overproduction of hisH and hisF gene products leads to inhibition of cell division in Salmonella. Can. J. Microbiol. 18:671–681 [DOI] [PubMed] [Google Scholar]
- 58.McMeechan A, Roberts M, Cogan TA, Jørgensen F, Stevenson A, Lewis C, Rowley G, Humphrey TJ. 2007. Role of the alternative sigma factors σE and σS in survival of Salmonella enterica serovar Typhimurium during starvation, refrigeration and osmotic shock. Microbiology 153:263–269 [DOI] [PubMed] [Google Scholar]
- 59.Alba BM, Gross CA. 2004. Regulation of the Escherichia coli σE dependent envelope stress response. Mol. Microbiol. 52:613–619 [DOI] [PubMed] [Google Scholar]
- 60.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4:e2. 10.1371/journal.pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kieboom J, Kusumaningrum HD, Tempelaars MH, Hazeleger WC, Abee T, Beumer RR. 2006. Survival, elongation, and elevated tolerance of Salmonella enterica serovar Enteritidis at reduced water activity. J. Food Prot. 69:2681–2686 [DOI] [PubMed] [Google Scholar]
- 62.Shaw MK. 1968. Formation of filaments and synthesis of macromolecules at temperatures below the minimum for growth of Escherichia coli. J. Bacteriol. 95:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vestby L, Møretrø T, Ballance S, Langsrud S, Nesse L. 2009. Survival potential of wild type cellulose deficient Salmonella from the feed industry. BMC Vet. Res. 5:43. 10.1186/1746-6148-5-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stackhouse RR, Faith NG, Kaspar CW, Czuprynski CJ, Wong AC. 2012. Survival and virulence of Salmonella enterica serovar Enteritidis filaments induced by reduced water activity. Appl. Environ. Microbiol. 78:2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.