Abstract
Low-temperature anaerobic digestion (LTAD) technology is underpinned by a diverse microbial community. The methanogenic archaea represent a key functional group in these consortia, undertaking CO2 reduction as well as acetate and methylated C1 metabolism with subsequent biogas (40 to 60% CH4 and 30 to 50% CO2) formation. However, the cold adaptation strategies, which allow methanogens to function efficiently in LTAD, remain unclear. Here, a pure-culture proteomic approach was employed to study the functional characteristics of Methanosarcina barkeri (optimum growth temperature, 37°C), which has been detected in LTAD bioreactors. Two experimental approaches were undertaken. The first approach aimed to characterize a low-temperature shock response (LTSR) of M. barkeri DSMZ 800T grown at 37°C with a temperature drop to 15°C, while the second experimental approach aimed to examine the low-temperature adaptation strategies (LTAS) of the same strain when it was grown at 15°C. The latter experiment employed cell viability and growth measurements (optical density at 600 nm [OD600]), which directly compared M. barkeri cells grown at 15°C with those grown at 37°C. During the LTSR experiment, a total of 127 proteins were detected in 37°C and 15°C samples, with 20 proteins differentially expressed with respect to temperature, while in the LTAS experiment 39% of proteins identified were differentially expressed between phases of growth. Functional categories included methanogenesis, cellular information processing, and chaperones. By applying a polyphasic approach (proteomics and growth studies), insights into the low-temperature adaptation capacity of this mesophilically characterized methanogen were obtained which suggest that the metabolically diverse Methanosarcinaceae could be functionally relevant for LTAD systems.
INTRODUCTION
Archaea are ubiquitous in low-temperature habitats such as polar marine waters (1, 2), alpine lakes (3), permafrost (4), and glacier ice (5). Methanogens represent the most characterized psychrophilic archaeal group (6). As such, low-temperature methanogenesis has been the focus of many studies, such as those focusing on measuring the methanogenic contribution to the global warming of cold areas (7) and also those using an astrobiological approach, where the ability of methanogens to survive under cold anoxic conditions has made them candidates as Earth analogues for extraterrestrial life (8).
Low-temperature methanogenic activity is also important from a biotechnological viewpoint, such as its application in low-temperature anaerobic digestion (LTAD) (9). Evidence of efficient LTAD treatment of wastewaters has been recorded in laboratory-scale trials which directly compared low-temperature bioreactor performance (chemical oxygen demand removal [COD] and biogas production) with traditional mesophilic configurations (10, 11). Experiments which comprised an initial mesophilic (37°C) bioreactor operation phase followed by a decrease to low-temperature conditions (≤15°C) have also been undertaken (12, 13). In these studies, low-temperature bioreactors achieved comparable performance levels to mesophilic systems after an initial period of “adaptation.” As a mesophilic inoculum was used to seed these bioreactors, a psychrotolerant capacity was deemed to be evident in the mixed microbial consortia underpinning these bioreactors. However, there still remains a significant knowledge gap relating to low-temperature methanogenic adaptation strategies, which require further elucidation for the optimization of LTAD systems.
There are three primary modes of methanogenic metabolism based on CO2 reduction, acetate decarboxylation, and methylotrophic activity (e.g., reduction of methylamines). Acetoclastic (acetate decarboxylation) methanogenesis has been recorded as being an important methanogenic pathway in low-temperature environments, including bioengineered systems (14). The order Methanosarcinales includes the only two known acetoclastic families, the Methanosaetaceae and the Methanosarcinaceae. The former have been historically categorized as strict acetoclastic methanogens although a recent genomic study highlighted the metabolic capacity for possible methyl group oxidation in three sequenced Methanosaetaceae strains (15). Nevertheless, this group has a minimum threshold concentration of 7 to 70 μM acetate (16) and has been documented to outcompete Methanosarcinaceae in environments where acetate concentrations are low (17, 18). However, in addition to acetate, the Methanosarcinaceae have the ability to utilize methylated compounds such as methanol and methylamines, with some species also able to use H2/CO2 as a carbon and energy source.
In many LTAD studies, Methanosaetaceae have been the predominant acetoclastic methanogenic group, with their abundance positively correlated to process efficiency (19–21) and granular sludge integrity (14). In contrast, low levels of Methanosarcinaceae have been found in well-functioning LTAD systems (14, 22). For example, in one study this group was detected below the quantification limit of the 16S rRNA gene assay, whereas Methanosaetaceae comprised 75% of total measured methanogenic 16S rRNA gene concentrations (23). However, a marked increase in Methanosarcinaceae signatures has been recorded during periods of bioreactor instability brought about through changes in operational parameters, e.g., temperature (14) and hydraulic retention time (24), with acetate accumulation apparent during this period. This would suggest that Methanosarcinaceae have the metabolic capacity to survive at low cell levels within a well-functioning LTAD system, where acetate concentrations are kept low. Thereafter, through a perturbation event this group can proliferate (acetate accumulation above utilization threshold). Thus, Methanosarcinaceae may have an important role in LTAD by ensuring a certain level of process stability during transient operation periods (11, 25).
The Methanosarcinaceae species Methanosarcina barkeri is a metabolically versatile methanogen which can grow on H2/CO2, methanol, various methylamines, and acetate as carbon and energy sources (26). This species has previously been the focus of a variety of studies, including growth experiments (27, 28), genomic investigations (29, 30), and enzymatic characterization (31, 32). In the present study, we employed a systematic, proteomics-based approach to characterize the low-temperature adaptive strategies of the methylotrophic M. barkeri strain DSMZ 800T, which was isolated from an anaerobic digester. Our hypothesis was that the organism would display an adaptive capacity to allow growth through the utilization of methanol and/or H2/CO2 under low-temperature conditions, thereby providing insights into the acetate-independent psychrotolerant growth and function of a Methanosarcina sp., which could be related to survival mechanisms employed by environmental strains in LTAD systems.
MATERIALS AND METHODS
Strain information, medium, and inoculum preparation.
The archaeal strain Methanosarcina barkeri DSMZ 800T (DSMZ, Braunschweig, Germany) was used throughout this study. Inoculum preparation was achieved using medium DMS 120 (pH 6.8) (33). After 40 ml of medium was added, each 60-ml vial was sealed with a butyl rubber bung and flushed with N2/CO2 (80:20, vol/vol) gas. Filter-sterilized reducing agents l-cysteine (2.5 mM) and Na2S · 9H2O (1.2 mM) were added after autoclaving as per DSMZ guidelines. In order to demonstrate that this strain was methylotrophic and to investigate substrate influence on growth, selected vials were supplemented with either methanol (156 mM; 50% [vol/vol] stock, filter sterilized), H2/CO2 (80:20, vol/vol), or methanol plus H2/CO2. Vials were inoculated with 2.5% (vol/vol) stock culture and incubated at 37°C on a shaker.
Batch culture setup.
When cells reached stationary phase (optical density at 600 nm [OD600] of ∼2), inoculum was added anaerobically to fresh vials (for each substrate mix) to an OD600 of ∼0.05 in 40 ml of medium. Separate vials were then incubated at 15°C and 37°C for comparative analysis on a shaker. OD600 measurements were taken at time intervals for each temperature, while separate vials for headspace methane measurements (percent) (34) were run in parallel. This experiment aimed to give direct insights into the capacity of M. barkeri to adapt to low temperatures and will therefore be referred to as the low-temperature adaptation strategy (LTAS) study from this point on. Molecular analysis was carried out on samples from cultures fed with methanol plus H2/CO2 (Table 1), and for this purpose, cells from 40 ml of culture were harvested by flash-freezing with liquid nitrogen followed by storage at −80°C. Samples were then thawed and centrifuged at 8,000 × g for 10 min at 4°C. The supernatant was discarded, and the recovered biomass pellet (approximately 1.5 g of wet weight per sample) was stored at −80°C until use.
Table 1.
Overview of M. barkeri growth characteristics
| Parameter | Value of the parameter by expt and time (days)a |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LTAS at 37°C |
LTAS sampling at 15°C |
LTSR at: |
||||||||||||
| I |
II |
III |
IV |
37°C |
15°C |
|||||||||
| 0.8 | 1.8 | 3.7 | 7 | 1.8 | 3.7 | 7 | 11.8 | 17.2 | 24.2 | 29.2 | 37.2 | 3.5 | 7.2 | |
| OD600 | 0.53 | 0.56 | 2.13 | 1.35 | 0.05 | 0.31 | 0.25 | 0.23 | 0.21 | 0.24 | 0.35 | 0.76 | 1.92 | 0.84 |
| % CH4 | 1.8 | 19 | 46 | 43 | 1.2 | 1.9 | 2 | 3.7 | 6.7 | 15.6 | 25 | 46 | 58 | 46 |
H2/CO2 plus methanol was used as the substrate. LTAS, low-temperature adaptation strategies; LTSR, low-temperature shock response; I to IV, LTAS sampling points for iTRAQ analysis.
Continuous-culture setup.
Continuous cultures of M. barkeri were established in a modified 500-ml Schott bottle (fitted with a three-port lid; Fisher Scientific) with a working volume of 400 ± 100 ml, connected to both medium and waste reservoirs. Anaerobic conditions were maintained in the vessel by sealing with butyl stoppers, by using gas-impermeable tubing connected to the medium reservoir, and by regularly flushing with N2/CO2 (80:20, vol/vol) gas. The sterile medium was allowed to acclimatize at 37°C overnight without shaking prior to inoculation with 2.5% (vol/vol) culture. The flow rate from the reservoir was controlled by a peristaltic pump (Watson-Marlow 205U) and was monitored and adjusted to maintain a steady dilution rate of 2.0 h−1. The continuous-culture system was operated at 37°C, allowing a continuous culture to establish (3.5 days; OD600 of ∼1.9), after which point the temperature was dropped to 15°C. The system was further operated for 3.7 days (7.2 days of total running time). Samples for protein extraction (60 ml) were taken immediately before the temperature drop event and also at the end of the period of 15°C operation (Table 1). This experiment aimed to uncover the functional response of M. barkeri directly after a temperature drop to 15°C and will therefore be referred to as the low-temperature shock response (LTSR) experiment from this point on.
Cell viability measurements.
The viability of cells during the LTAS experiment was investigated using a Live/Dead BacLight kit (Invitrogen) (35) (Table 1). Anaerobic conditions were maintained in samples during incubation with BacLight probes by sealing samples with N2/CO2 (80:20, vol/vol) gas. A Nikon Eclipse E600 epifluorescence microscope (Carl Zeiss, Oberkochen, Germany) was used to assess cell viability. Live cells were indicated by green fluorescence (BS2 filter; excitation, 450 to 490 nm; long-pass [LP] emission filter, 515 nm) while dead cells were indicated by red fluorescence (CY3 filter; excitation, 546 ± 12 nm; LP emission, 590 nm). Ten images of a microscope field (×100 magnification) were taken for each sample and visualized using QCapture Pro software.
Protein extraction and quantification.
M. barkeri proteins were extracted from samples (Table 1) using a sonication method adapted from a previous LTAD proteomic study (36). Briefly, cell pellets were resuspended with 5 ml of noninterfering triethylammonium bicarbonate (TEAB) buffer. The cells were then disrupted by sonication (MSE Soniprep 150) at 40% amplitude for 30 s on ice. Fifteen pulses were applied with 30-s intervals to prevent thermal damage of cellular proteins. In order to remove any protein precipitates and cellular debris, lysates underwent centrifugation at 10,000 × g for 30 min at 4°C, and the resulting pellets were discarded. Protein precipitation from the supernatants was achieved through incubation with ice-cold acetone (3:1, vol/vol) at −20°C for 1 h, after which the samples underwent centrifugation at 10,000 × g for 15 min. The supernatants were discarded, and pellets were resuspended with 200 μl of TEAB buffer. Protein quantification was undertaken using a Calbiochem Non-Interfering Protein Assay kit (Merck KGaA, Darmstadt, Germany) by following the manufacturer's instructions.
iTRAQ labeling.
For each study (LTAS and LTSR), replicate protein extracts from specified time points were used in an eight-plex iTRAQ (isobaric tags for relative and absolute quantification) labeling experiment (Table 1). One hundred micrograms of protein extract in 20-μl samples was labeled with iTRAQ reagents according to the manufacturer's guidelines (ABSciex, Foster City, CA). Briefly, each sample was reduced and denatured, and cysteine residues were blocked before digestion with 10 μl of 25 μg/μl trypsin solution (1:20; Promega, Madison, WI) overnight at 37°C. The digested peptides were then labeled with iTRAQ tags as follows in the LTAS experiment: for duplicates of sample I, tags 113 and 114; for duplicates of sample II, tags 115 and 116; for duplicates of sample III, tags 117 and 118 tags; and for duplicates of sample IV, tags 119 and 121. For the LTSR experiment only four iTRAQ tags were required: for duplicate 37°C samples, tags 113 and 114; and for duplicate 15°C samples, tags 115 and 116. For each experimental setup, the samples were combined and subjected to peptide separation by cation exchange chromatography followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Cation exchange peptide separation and LC-MS/MS analysis.
After trypsin digestion and labeling, peptides from individual samples were combined and resuspended in 1 ml of cation exchange loading buffer (10 mM KH2PO4, pH 3.0, in 25% acetonitrile). The peptides were then separated by strong cation exchange chromatography on a PolySulfoethyl A column (PolyLC, Inc.) with an increasing salt gradient from 0 to 0.5 M over 30 min, and fractions were collected every 30 s. Fractions were pooled to give nine fractions of approximately equal peptide concentrations, as judged by inspection of the chromatogram, evaporated to dryness, and resuspended in 0.1% trifluoroacetic acid before desalting (PepClean C18 spin columns; ThermoScientific). Each fraction was further separated through reverse-phase chromatography directly before mass spectrometric analysis. This was achieved using an Eksigent nanoLC Ultra system coupled to a Nanoflex cHiPLC (ABSciex, Foster City, CA) equipped with a 200-μm by 0.5-mm ChromXP C18-CL 3-μm, 120-Å trap and 75-μm by 15-cm ChromXP C18-CL 3-μm, 120-Å column, using a gradient of increasing acetonitrile concentration containing 0.1% formic acid (15 to 40% acetonitrile in 40 min and 40 to 95% in a further 10 min, followed by 95% acetonitrile to clean the column). The eluent was sprayed into a TripleTOF 5600 mass spectrometer (ABSciex, Foster City, CA) and analyzed in information-dependent acquisition (IDA) mode, performing 0.25 s of MS followed by 2-s MS/MS analyses of the 20 most intense peaks seen by MS. These precursor masses were then excluded from analysis for the next 13 s. A rolling collision energy adjusted to 10 units higher than that normally used for peptides was employed to provide sufficient peptide fragmentation and generation of the iTRAQ reporter groups.
Two-dimensional gel electrophoresis (2-DGE).
Eight two-dimensional (2-D) gels were run in total, corresponding to two duplicate independent extractions and two technical replicates from 37°C and 15°C LTAS samples, by following the protocol described in a previous study (36) (Table 1). Gel images were processed and analyzed with PDQuest-Advanced software, version 8.0.1 (Bio-Rad). Spot counts were obtained using the spot detection wizard enabling the Gaussian model option as recommended by the manufacturer. Ratios of spot intensities were determined for each sample. Protein expression ratios greater than 1.5-fold were considered significant. The 1.5-fold cutoff point for significance was based on the geometric standard deviation of the entire set of proteins identified (at least two unique peptides at >99% confidence), which has been used in many proteomic studies as a conservative cutoff for biological significance (37, 38). Proteins of interest were excised from the gels and subjected to in-gel digestion prior to analysis using nanoflow liquid chromatography-electrospray ionization tandem mass spectrometry (nLC-ESI-MS/MS) (36).
Protein identification and quantification.
The MS/MS data recovered from iTRAQ and 2-DGE LC-MS/MS were analyzed as follows: MS/MS data for +2 to +5 charged precursor ions which exceeded 150 cps were processed using the Paragon search algorithm (39) within the ProteinPilot, version 4.1, software program (ABSciex, Foster City, CA) against an internal database comprising the M. barkeri Fusaro genome (DSM 804, with 3,616 proteins in the UniProtKB database) along with potential contaminants (UniProtKB/Swiss-Prot database, accessed 19 June 2012). The data were searched using the following parameters: a detected protein threshold (unused ProtScore) of >0.05, trypsin as the cleavage enzyme, one missed cleavage, iTRAQ modification of lysines and methyl methanethiosulfonate (MMTS) modification of cysteines as fixed modifications, and methionine oxidation selected as a variable modification. Peptide and protein lists were then exported from ProteinPilot to the ProteinPilot descriptive statistics template (PDST; ABSciex, Foster City, CA) for data management and additional analysis (e.g., Volcano and Quant false-discovery rate [FDR] worksheet). The FDR was calculated by ProteinPilot PDST using the embedded decoy search and was found to be <2% for all experiments.
RESULTS
Characteristics of M. barkeri grown at 37°C and 15°C.
The levels of growth of M. barkeri at 37°C and 15°C were compared through OD600 measurements with different substrate mixes (methanol plus H2/CO2, methanol alone, and H2/CO2 alone) (Fig. 1). The maximum specific growth rate (μmax) was found to be dependent on both temperature and substrate (Table 2). The OD600 measurements indicated a minimal lag phase for cultures grown at 37°C, regardless of substrate, with an increase in the OD600 correlating with an increase in percent CH4 production (Fig. 1). After a lag phase of ∼1 day, growth at 15°C exhibited a similar profile to that observed at 37°C. On day 3, however, a long stationary phase was recorded prior to a further steady increase in growth from day 17. Although the μmax was lower in the 15°C cultures, the cultures fed H2/CO2 plus methanol reached headspace percent CH4 levels similar to those of their mesophilic counterparts, peaking at 70% CH4 (±4.3% standard error [SE]; n = 3) on day 41 of the incubation compared with 64% (±6.7% SE; n = 3) on day 14 in 37°C vials (Fig. 1). The M. barkeri cultures with H2/CO2 as a sole substrate exhibited poor growth, and percent methane headspace values at both temperatures tested were lower than those of cultures fed with H2/CO2 plus methanol or with methanol only (Fig. 1). Although solubility of H2 increases with decreasing temperature, difficulties with substrate delivery through gas input has been recorded previously (40). Also, as this was a methanol-adapted strain of M. barkeri, it is plausible that the metabolic capacity to reduce CO2 using electrons from H2 was thermodynamically unfavorable under submesophilic conditions, as suggested by growth curve analysis (Fig. 1).
Fig 1.
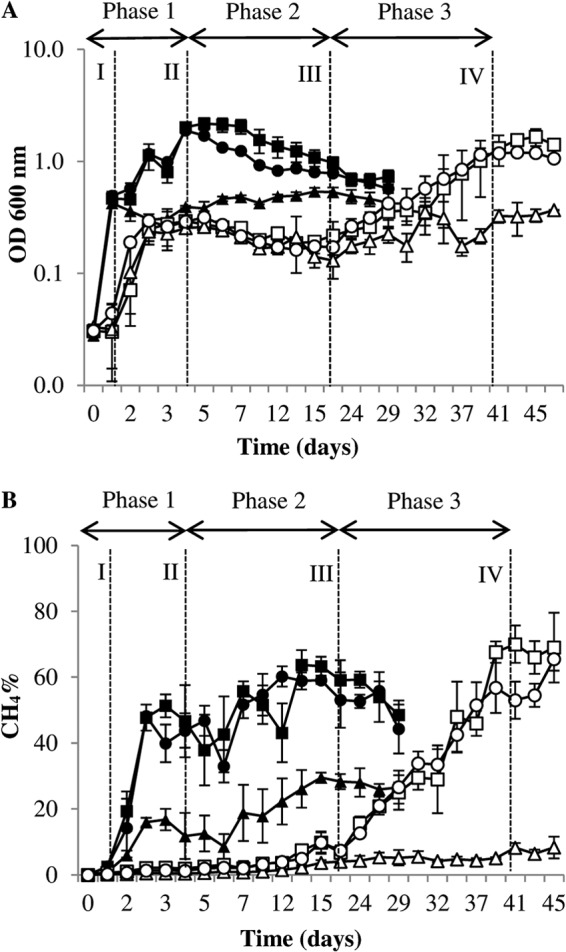
Growth profiles of M. barkeri incubated at 37°C and 15°C. Optical density (at 600 nm) measurements (A) and percent CH4 headspace measurements (B) were determined in tandem for M. barkeri grown at 37°C with H2/CO2 plus methanol ■, methanol alone ●, and H2/CO2 alone ▲ and at 15°C with H2/CO2 plus methanol □, methanol alone ○, and H2/CO2 alone △. Error bars indicate the standard deviations and are the result of at least three replicates. Data represent phase 1 (samples I and II), phase 2 (sample III), and phase 3 (sample IV) of the LTAS experiment.
Table 2.
Maximum specific growth rates of M. barkeri
| Substrate | Temp (°C) | μmax (day−1)a | % Decrease in μmax | Generation time (days) |
|---|---|---|---|---|
| H2/CO2 + methanol | 37 | 0.5 ± 0.03 | 0 | 1.4 |
| Methanol | 37 | 0.46 ± 0.06 | 8 | 1.5 |
| H2/CO2 | 37 | 0.11 ± 0.02 | 78 | 6.3 |
| H2/CO2 + methanol | 15 | 0.12 ± 0.07 | 0 | 6.2 |
| Methanol | 15 | 0.09 ± 0.03 | 25 | 10.7 |
| H2/CO2 | 15 | 0.03 ± 0.09 | 75 | 24.2 |
Values are means ± SD (n = 3). μmax, maximum specific growth rate (1/days).
Cell viability.
The viable cell fractions in 37°C samples increased from 41% (±6.5% SE; n = 10) at day 0.8 to 59% (±17.8% SE; n = 10) at day 1.8 (Fig. 2). The total cell count peaked after 3.7 days (OD600 of ∼2), but the viable cell fraction did not show any significant increase, with only 60% (±11.2% SE; n = 10) of total cells recorded as being viable at that point. At 15°C, however, a lower total cell count on day 7 was not reflective of the viable cell fraction, which was 79% (±13.8% SE; n = 10) (Fig. 2). This trend continued, as recorded at 17.2 days with 84% (±9.65% SE; n = 10) of cells being identified as viable. After this observed period of minimal growth but high viability, the total cell counts increased in 15°C samples, while the viable fraction decreased to 66% (±9.5% SE; n = 10) on day 37. The spatial arrangement of M. barkeri aggregates from both temperatures varied, with 37°C samples consisting of layered branching aggregates while the 15°C aggregates were more consistent in size, with clusters rarely exceeding 10 μm in diameter (Fig. 2).
Fig 2.
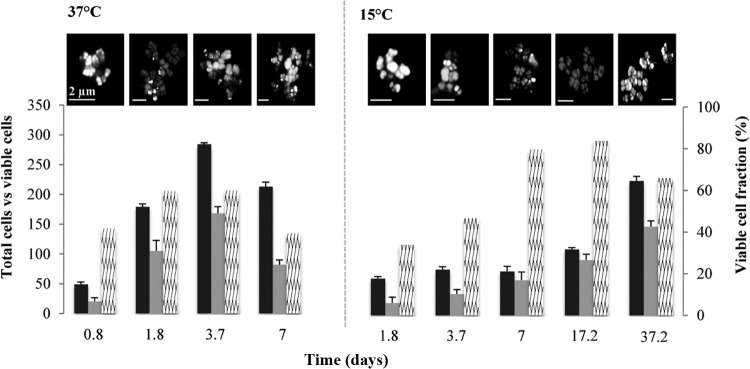
Cell viability of M. barkeri grown at 37°C and 15°C. Bar charts represent total cell counts (black bars), viable cell counts (gray bars), and the viable fraction (%) of total cell counts (hatched bars). Representative images from live/dead staining of M. barkeri taken from samples during the course of the growth curve incubations are shown above the graphs. Error bars indicate the standard deviations and are the result of at least three replicates.
Proteomics analysis.
In order to gain some insight into the low-temperature adaptation of M. barkeri, two iTRAQ experiments were performed, one involving cultures grown at 37°C and exposed to a temperature drop to 15°C (LTSR) and another experiment involving cultures grown at 15°C (LTAS). In the LTSR study, a total of 127 proteins (P ≤ 0.05) were detected in both 37°C and 15°C samples. From these, 20 proteins (16%) were found to be differentially expressed as a function of temperature by a 1.5-fold change or more (Table 3). Among these proteins, various functional categories were evident. Two proteins involved in CO2 methanogenesis were detected at higher levels at 37°C than at 15°C (Mbar_A1095 and Mbar_A1763) (Table 3). Methanogenesis from dimethylamine and methanol showed contrasting results, with a protein involved in the dimethylamine pathway (Mbar_A3605) identified at a higher level in the 37°C sample, while the corrinoid protein involved in the methanol pathway (Mbar_A3637) was upregulated at 15°C. Several information-processing enzymes were identified as being differentially expressed. These included an XRE domain protein (Mbar_A1033), which plays a key role in DNA binding (41), and an elongation factor 2 (EF-2) protein (Mbar_A3686), both of which were upregulated at 15°C (Table 3). In addition, a ribosomal protein (Mbar_A0614) was found to be expressed at a higher level at 15°C. Also, several stress proteins were upregulated at 37°C, including a heat shock protein (Mbar_A1543) and two subunits of the thermosome (Ths) protein (Mbar_A1084 and Mbar_A1201) (Table 3).
Table 3.
Differentially expressed proteins from iTRAQ experiments
| Gene function and locus | Proteinb | Sequence coverage (%) | Protein expression ratioa |
|||||
|---|---|---|---|---|---|---|---|---|
| LTSR (37°C/15°C) | P1 vs P2 |
P1 vs P3 |
P2 vs P3 (III/IV) | |||||
| I/III | II/III | I/IV | II/IV | |||||
| Methanogenesis genes | ||||||||
| CO2 specific | ||||||||
| Mbar_A0450 | Coenzyme F420 hydrogenase subunit G (FrhG) | 12 | 0.65 | 0.62 | 0.51 | 0.68 | NS | |
| Mono/di/trimethylamine specific | ||||||||
| Mbar_A0841 | Methylcobalamin:CoM methyltransferase (MtbA) | 8 | NS | 0.67 | 0.62 | 0.55 | 0.51 | NS |
| Mbar_A3605 | Dimethylamine methyltransferase (MtbB) | 8 | 3 | 0.68 | 0.63 | 0.56 | 0.42 | NS |
| Mbar_A1502 | Trimethylamine methyltransferase (MttB) | 7 | NS | 0.48 | 0.54 | 0.57 | 0.62 | 1.54 |
| Methanol specific | ||||||||
| Mbar_A1063 | Methanol corrinoid protein (MtaC) | 30 | NS | NS | NS | 0.45 | 0.44 | 0.64 |
| Mbar_A3637 | Methanol corrinoid protein (MtaC) | 12 | 0.3 | |||||
| Mbar_A1064 | Methanol:corroinoid methyltransferase (MtaB) | 17 | NS | NS | NS | 1.62 | 1.69 | 1.62 |
| Common | ||||||||
| Mbar_A1095 | F420-dependent H4MPT dehydrogenase (Mtd) | 34 | 3.3 | NS | NS | 0.67 | 0.62 | 0.62 |
| Mbar_A1763 | Formylmethanofuran dehydrogenase subunit B (FwdB) | 13 | 3.2 | |||||
| Mbar_A0893 | Methyl-coenzyme reductase subunit A (McrA) | 18 | NS | NS | NS | 0.51 | 0.57 | 0.56 |
| Mbar_A1182 | Methyl-coenzyme reductase subunit B (McrB) | 15 | NS | NS | NS | 0.65 | 0.55 | 0.43 |
| Mbar_A1256 | Tetrahydromethanopterin S-methyltransferase subunit G (MtrG) | 26 | 4.8 | NS | NS | NS | NS | NS |
| Cellular information processing genes | ||||||||
| Transcription | Putative nickel-responsive regulator 1 (NikR) | 29 | 2.7 | NS | NS | NS | 0.61 | 0.64 |
| Mbar_A2536 | DNA-directed RNA polymerase I, II and III, 7.3-kDa polypeptide | 37 | 0.56 | 0.52 | 0.47 | 0.45 | NS | |
| Translation | ||||||||
| Mbar_A3687 | 30S ribosomal protein S7P (RpsG) | 27 | 1.78 | 1.65 | 1.56 | NS | NS | |
| Mbar_A1149 | 30S ribosomal protein S17 (Rps17E) | 16 | NS | NS | 1.83 | 1.56 | 1.64 | |
| Mbar_A3231 | 50S ribosomal protein L21E (Rpl21) | 15 | 0.69 | 0.54 | 0.56 | 0.33 | NS | |
| Mbar_A0614 | 50S ribosomal protein L11P (RplA) | 16 | 0.3 | NS | NS | 0.68 | 0.64 | 0.65 |
| Mbar_A3173 | Aspartyl-tRNA synthetase (AspS) | 23 | NS | 1.66 | 1.99 | NS | NS | 0.45 |
| Mbar_A0454 | SSU ribosomal protein S19E (Rps19) | 4 | NS | 0.72 | 0.64 | 0.55 | 0.53 | NS |
| Mbar_A0623 | Isocitrate dehydrogenase (Idh1) | 4 | NS | NS | NS | 0.55 | 0.51 | NS |
| Mbar_A2048 | Leucyl-tRNA synthase (LeuS) | 17 | 2.6 | |||||
| Mbar_A3685 | Elongation factor 1 subunit A (EF1A) | 12 | NS | 0.78 | 0.67 | 0.69 | 0.63 | NS |
| Mbar_A3686 | Elongation factor 2 (EF-2) | 18 | 0.5 | NS | NS | 1.61 | 1.50 | 1.63 |
| DNA binding and repair | ||||||||
| Mbar_A1033 | Transcriptional regulator, XRE family | 8 | 0.3 | NS | NS | 1.59 | 1.52 | 1.54 |
| Mbar_A1182 | Nucleoid protein (MC1) | 22 | 0.63 | 0.53 | 0.49 | 0.35 | NS | |
| Mbar_A1584 | Archaeal histone (HmbA) | 30 | 0.62 | 0.56 | 0.50 | 0.54 | NS | |
| General metabolism genes | ||||||||
| Amino acid biosynthesis | ||||||||
| Mbar_A0248 | 3-Isopropylmalate dehydrogenase (AfkS) | 7 | 1.92 | 0.51 | 0.52 | 0.41 | 0.41 | NS |
| Mbar_A0220 | Ketol-acid reductoisomerase (IlvC) | 9 | 1.78 | 0.54 | NS | 0.62 | 0.58 | NS |
| Mbar_A1431 | d-3-Phosphoglycerate dehydrogenase (PhgdH) | 37 | 2.17 | NS | NS | NS | NS | NS |
| Mbar_A1094 | Phosphoserine phosphatase (PspH) | 18 | 1.84 | NS | NS | NS | NS | NS |
| Mbar_A3623 | Tryptophan synthase alpha chain 1 (ThiC) | 12 | 3.19 | |||||
| Vitamin biosynthesis | ||||||||
| Mbar_A0597 | Phosphomethylpyrimidine synthase 1 (TrpA) | 10 | 1.66 | NS | 0.66 | NS | NS | NS |
| Mbar_A1056 | CobW protein | 4 | NS | 0.54 | NS | NS | NS | |
| Glycolysis and energy metabolism | ||||||||
| Mbar_A2189 | Glyceraldehyde-3-phosphate dehydrogenase (Gap2) | 6 | NS | NS | NS | 0.53 | 0.49 | NS |
| Mbar_A0392 | H+-transporting ATP synthase (NtpE) | 19 | NS | NS | NS | 0.47 | 0.44 | 0.66 |
| Electron transport | ||||||||
| Mbar_A1847 | Methanophenazine-reducing hydrogenase | 8 | NS | 0.61 | NS | 0.42 | 0.46 | NS |
| Mbar_A3651 | HesB protein | 13 | NS | 0.63 | 0.61 | 0.41 | 0.43 | 0.65 |
| Chaperones and stress proteins | ||||||||
| Mbar_A1084 | Themosome subunit A (Hsp60) | 22 | 2.67 | 0.62 | NS | 0.51 | 0.48 | 0.57 |
| Mbar_A1201 | Themosome subunit B (Hsp60) | 15 | 1.54 | NS | NS | 0.52 | 0.52 | 0.66 |
| Mbar_A3433 | Chaperone protein (DnaK) | 8 | NS | 0.56 | 0.55 | NS | NS | 1.56 |
| Mbar_A1543 | 60-kDa chaperone (GroEL) | 32 | 1.67 | |||||
| Mbar_A2503 | Proteasome subunit A (PrcA) | 12 | NS | 0.56 | 0.53 | 0.47 | 0.44 | NS |
| Mbar_A2249 | Peptidyl-prolyl isomerase (SlyD) | 20 | NS | 0.45 | 0.42 | NS | NS | 1.64 |
| Mbar_A0213 | Prefoldin subunit B (PrcA) | 21 | NS | 1.56 | NS | NS | NS | NS |
| Mbar_A0825 | Universal stress protein | 11 | 3.24 | NS | NS | 0.46 | 0.45 | 1.66 |
| Unknown-function genes | ||||||||
| Mbar_A0488 | Putative uncharacterized protein | 8 | NS | 0.52 | 0.64 | 0.53 | 0.61 | NS |
| Mbar_A1026 | Putative uncharacterized protein | 29 | NS | NS | 1.57 | 1.51 | 1.67 | |
| Mbar_A0376 | Putative uncharacterized protein | 17 | NS | NS | 0.57 | 0.51 | 0.64 | |
| Mbar_A1027 | Putative uncharacterized protein | 46 | 1.53 | 1.56 | 2.38 | 2.45 | 2.12 | |
| Mbar_A3219 | Putative uncharacterized protein | 15 | NS | NS | 0.41 | 0.43 | NS | |
For the LTSR experiment, the ratios represent values for growth at 37°C versus growth at 15°C (37°C/15°C). For the LTAS experiment, the ratios represent the values for the indicated sample(s) within phases 1 (P1, samples I and II), 2 (P2, sample III), and 3 (P3, sample IV). NS, no statistically significant difference. All samples were fed methanol plus H2/CO2.
CoM, coenzyme M; SSU, small subunit; H4MPT, tetrahydromethanopterin.
In conjunction with the M. barkeri growth profile at 15°C, the iTRAQ samples for the LTAS study were taken during the three phases observed: phase 1 (initial adaptation [samples I and II]), phase 2 (stationary phase [sample III]), and phase 3 (primary growth phase [sample IV]) (Fig. 1 and Table 1). In total, 104 proteins (P ≤ 0.05) were identified in all samples, with 41 (39%) found to be differentially expressed (1.5-fold difference) between two or more samples (Table 3).
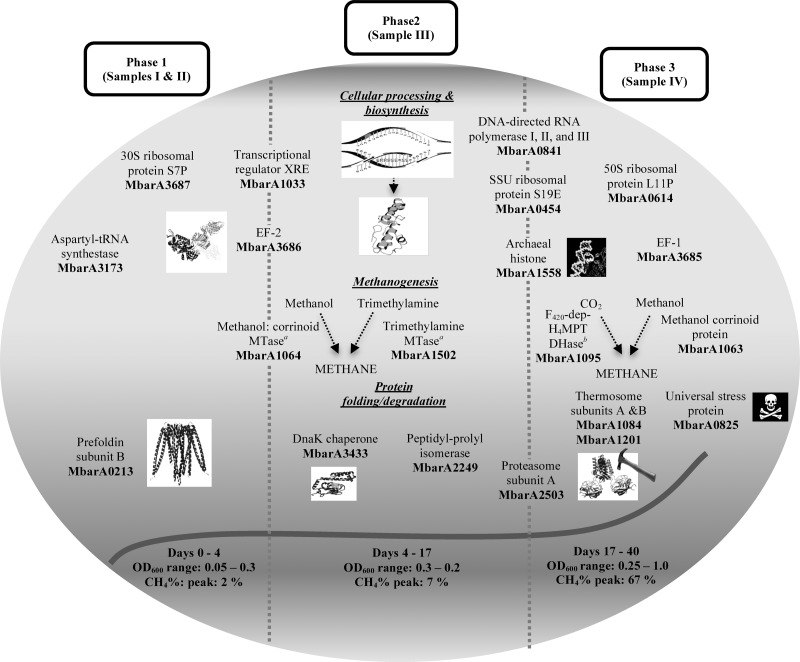
Figure 3 illustrates differentially expressed protein profiles for each of the phases investigated in the LTAS study. Proteins relating to CO2 and methanol methanogenesis as well as methylamine-specific proteins were found to be significantly expressed in one or more of these phases. A methanol metabolism protein (Mbar_A1063) was upregulated in phase 1 and phase 2 samples in comparison to the phase 3 sample. A trimethylamine protein (Mbar_A1502) was found to be expressed at a higher level in the phase 2 sample than in other samples. However, the majority of significantly expressed methanogenesis proteins were upregulated in the phase 3 sample (Fig. 3; Table 3).
Fig 3.
Differentially expressed proteins at specific phases of M. barkeri growth at 15°C are shown. Proteins located on broken lines represent a conserved result between particular phases. MTase, methyltransferase; SSU, small subunit; F420-dep-H4MPT DHase, F420-dependent methylenetetrahydromethanopterin dehydrogenase.
Four proteins relating to transcription and translation were upregulated in phase 2 and phase 3 samples compared with phase 1 samples (Table 3). These included a protein involved in transcriptional RNA synthesis (Mbar_A2536) and a protein biosynthesis catalyst (Mbar_A3685). However, the EF-2 protein (Mbar_A3686) as well as a transcriptional regulator (Mbar_A1033) was found to be upregulated in phase 1 and phase 2 samples compared with the phase 3 sample level (Table 3). This result was consistent with the LTSR study in which both of these proteins were detected at higher levels at 15°C, suggesting that they might be important for M. barkeri during initial stages of low-temperature adaptation. Two other DNA binding and repair proteins were found to be upregulated in phase 2 and phase 3 samples compared to phase 1 samples. These included a DNA regulator (Mbar_A1584) involved in transcription (42). General metabolism proteins identified in the LTAS study were found to be involved in diverse pathways, including amino acid and vitamin biosynthesis, glycolysis, and electron transport (Table 3). Six of these proteins, which included an oxidoreductase protein (Mbar_A2189) essential in glycolysis and biosynthesis of secondary metabolites, were expressed at higher levels in the phase 3 sample than in the phase 1 samples.
Proteins involved in oxidative stress, protein folding, and proteolysis were also represented in the LTAS data set (Table 3). Two thermosome proteins (Mbar_A1084 and Mbar_A1201) were upregulated in the phase 3 sample compared with phase 1 sample levels, and this was also the case for a universal stress protein (Mbar_A0825) (Fig. 3). Also, a peptidyl prolyl isomerase (Mbar_A2249) was upregulated in the phase 2 sample compared to levels in other samples. Although little is known regarding chaperone-like activity of the peptidyl prolyl isomerase, a study on Methanococcoides burtonii reported the upregulation of this protein when the organism was grown at 4°C compared to 26°C (43), therefore suggesting that this protein may play an important role in low-temperature adaptation of M. barkeri. Finally, a DnaK heat shock protein (Mbar_A3433) was found to be upregulated in phase 2, and a proteolysis protein (Mbar_A2503) was found to be upregulated in the phase 3 sample compared to phase 1 samples (Fig. 3 and Table 3).
In order to complement the iTRAQ results, 2-DGE was undertaken on M. barkeri samples grown at 37°C and 15°C (LTAS study). These samples were taken at time points with similar OD600 and percent CH4 levels for each temperature (Table 1). An average of 179 (standard deviation [SD], 18.4; n = 6) reproducible protein spots were detected on 2-D gels, from which 79 (44%) were found to be upregulated at 37°C while 39 (22%) proteins were upregulated at 15°C, and 61 (34%) were conserved for each temperature. Two proteins representing differential expression for each temperature were excised and identified using nLC-ESI-MS/MS. The universal stress protein (Mbar_A0825) was found to be upregulated (up 4.5-fold) at 37°C, while a proteasome (Mbar_A2503) was expressed at a higher level in the 15°C sample (up 2.9-fold) (Fig. 4). In the LTSR iTRAQ experiment, the universal stress protein was upregulated at 37°C while the proteasome was upregulated at 15°C, thus showing consistency between the two proteomic techniques applied (Table 3). A protein responsible for methyl-coenzyme M reduction (Mbar_A0893) was conserved for each temperature, which was in agreement with the LTSR iTRAQ experiment.
Fig 4.
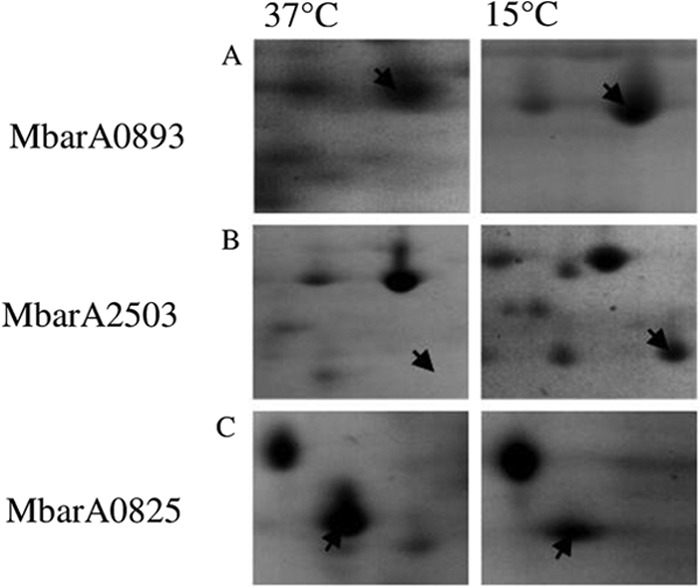
2-DGE gel sections comprising proteins extracted from M. barkeri grown at 37°C and 15°C. (A) Methyl-coenzyme M reductase subunit G protein (Mbar_A0893) conserved for each temperature (expression level at 37°C/expression level at 15°C, up 1.24-fold). (B) Proteasome subunit A (Mbar_A2503) upregulated at 15°C (expression level at 15°C/expression level at 37°C, up 2.87-fold). (C) Universal stress protein (Mbar_A0825) upregulated at 37°C (expression level at 37°C/expression level at 15°C, up 3.81-fold).
DISCUSSION
The low-temperature adaptation of M. barkeri was investigated through two experimental approaches: (i) low-temperature shock response (LTSR) and (ii) low-temperature adaptation strategy (LTAS). In the LTSR experiment, we applied an iTRAQ method to characterize the protein expression profile of M. barkeri before and after a temperature drop from 37°C to 15°C. In the LTAS experiment, M. barkeri growth at 15°C was characterized through proteomic samples taken at specific time points to coincide with a particular phase of growth. Also, cell viability measurements were recorded in the LTAS experiment, which included measurements at the proteomics sampling time points, in order to give a concise overview of the submesophilic growth capacity of M. barkeri. For the LTSR experiment, a total of 20 (13%) proteins were found to be upregulated at either 37°C or 15°C, while in the LTAS experiment there were 43 (41%) proteins found to be differentially expressed during incubation at 15°C (Table 3). Overall, this study provided new insights into the low-temperature adaptation capacity of M. barkeri.
Methanogenesis from methanol was found to be significant for M. barkeri at 15°C, with corrinoid protein upregulation (Mbar_A3637) at 2.8-fold at this low temperature compared with 37°C (LTSR). During growth at 15°C (LTAS), two methanol metabolism proteins (Mbar_A1063 and Mbar_A1064) were found to be upregulated at different time points. Taken together, these results indicate the ability of M. barkeri to efficiently utilize methanol under submesophilic conditions. Two proteins involved in H2/CO2 metabolism proteins were found to be upregulated at 37°C (Mbar_A1095, up 3.3-fold; Mbar_A1763, up 3.2-fold), which might suggest a preference for H2/CO2 at mesophilic temperatures. The pathway involved in trimethylamine and dimethylamine metabolism was found to be active in M. barkeri during growth at 15°C. Interestingly, a methyltransferase protein (Mbar_A1502), which catalyzes trimethylamine reduction to methyl-coenzyme M, was upregulated during the stationary phase compared to other stages of growth (Table 3). Thus, the ability to undertake methylamine methanogenesis at 15°C may confer an advantage for M. barkeri survival in a low-temperature environment.
Proteins involved in translation were detected in both iTRAQ experiments. The EF-2 protein (Mbar_A3686) was upregulated at 15°C compared to 37°C (LTSR), and, furthermore, it was found to be upregulated in the initial phases of growth at 15°C (LTAS) (phase 1 and phase 2 samples compared to phase 3 sample) (Fig. 3). Therefore, this protein may play a role in the initial stages of low-temperature adaptation in M. barkeri. The EF-2 protein has been the focus in many studies relating to Methanococcoides burtonii, a psychrotolerant methanogen isolated from Ace Lake, Antarctica (44–46). It has been suggested that the low stability of EF-2 (e.g., fewer salt bridges) facilitates high enzymatic activity during growth of M. burtonii at low temperatures. Therefore, it is conceivable that EF-2 may also confer an advantage in M. barkeri during submesophilic growth. The importance of low-temperature DNA binding and repair was evident through the upregulation of the XRE transcriptional regulator (Mbar_A1033) as well as two other DNA binding proteins found at 15°C.
Cellular chaperone proteins were predominant at 37°C. A chaperonin protein (Mbar_A1543) was upregulated at 37°C (LTSR), which relates to a previous study where a corresponding RNA transcript was upregulated under heat stress conditions (47). In addition to this, a thermosome (Ths) protein was found to be upregulated at 37°C (LTSR) (Table 3). The Ths protein was also found to be upregulated in the latter phase of growth at 15°C (LTAS, phase 3) (Fig. 3), suggesting an important functional role under submesophilic exponential growth conditions. A DnaK chaperone (Mbar_A3433), which was found to be upregulated during stationary-phase growth at 15°C (LTAS, phase 2) (Fig. 3), has been reported to play an important role in cell survival under cell stress conditions (48), particularly heat stress (49). The recombinant expression of DnaK from the psychrophilic Shewanella sp. in Escherichia coli was found to confer the ability to grow at 15°C to an E. coli mutant strain (50). Together with a peptidyl propyl isomerase (Mbar_A2249), these proteins are important for low-temperature adaptation in M. barkeri and require further examination to characterize their respective roles in more detail.
Cell viability measurements recorded during growth of M. barkeri at 15°C (LTAS) gave insights into the capacity of this methanogen to survive under submesophilic conditions. This was highlighted with M. barkeri cells showing 83% (±9.65% SE; n = 10) cell viability after 17 days of incubation at 15°C. It is at this point (end of phase 2) of incubation that viable M. barkeri cells (only cultures with methanol) exhibited complete adaption to cold conditions as shortly after this sampling point the primary growth phase was recorded, corresponding to OD600 and percent CH4 readings reaching comparable levels to those of the 37°C samples (exponential phase). Together with proteomic data, these findings contribute to a better understanding of Methanosarcina survival in low-temperature environments, e.g., EF-2-mediated protein synthesis and chaperone activity (DnaK/proteasome system) facilitating psychrophilic adaptation.
It is conceivable that Methanosarcina spp. utilize substrates other than acetate in well-functioning LTAS applications, which in turn may offer a favorable advantage through bypassing direct substrate competition. If an alternative metabolic route was accessible in LTAD, such as C1 metabolism, this might promote the survival of Methanosarcina spp. (possibly in a long lag phase observed in phase 2 of the LTAS experiment). Acetate accumulation brought about through a perturbation event, such as after a temperature drop, may then provide Methanosarcina with enough substrate to accumulate above the threshold value (>1 mM), which was evident in previous low-temperature (14) and high-temperature (51) anaerobic digestion studies. Targeted functional approaches are required to uncover the metabolic and adaptation strategies of Methanosarcina spp. in a mixed community context. The application of a polyphasic experimental design would be ideal, combining, for example, DNA quantification and proteomics methods with isotope labeling studies (e.g., separate incubations with 14C-labeled methanol, H2/CO2, and acetate). Together, these approaches may shine some light on the functional role of this methanogen in LTAD systems.
In a process context, procuring a psychrophilic seed biomass in future LTAD applications may not be required for studies characterizing the psychrotolerant capabilities of methanogens and other microbial groups underpinning mesophilic seed biomass. In addition to this, bioreactor design and operation can be directed away from a performance-related paradigm, where differences between successful and unsuccessful bioreactor trials are still poorly understood. Instead, community-based approaches may be undertaken, e.g., system-based mathematical modeling.
ACKNOWLEDGMENTS
We are grateful to colleagues in the Microbial Ecology Lab and in Microbiology at the National University of Ireland, Galway, for all the help given in preparation of the manuscript.
We thank the Wellcome Trust for funding mass spectrometry equipment at the University of St. Andrews. This research was financially supported by Science Foundation Ireland (grants RFP 08/RFP/EOB1343 and 06/CP/E006).
Footnotes
Published ahead of print 3 May 2013
REFERENCES
- 1. Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM. 2003. Abundance and distribution of planktonic archaea and bacteria in the waters west of the Antarctic peninsula. Limnol. Oceanogr. 48:1893–1902 [Google Scholar]
- 2. Galand PE, Lovejoy C, Vincent WF. 2006. Remarkably diverse and contrasting archaeal communities in a large arctic river and the coastal Arctic Ocean. Aquat. Microb. Ecol. 44:115–126 [Google Scholar]
- 3. Pernthaler J, Glockner FO, Unterholzner S, Alfreider A, Psenner R, Amann R. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wagner D, Kobabe S, Pfeiffer EM, Hubberten HW. 2003. Microbial controls on methane fluxes from a polygonal tundra of the Lena Delta, Siberia. Permafrost Periglac. Process. 14:173–185 [Google Scholar]
- 5. Battin TJ, Willie A, Sattler B, Psenner R. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cavicchioli R. 2006. Cold adapted archaea. Nat. Rev. Microbiol. 4:331–343 [DOI] [PubMed] [Google Scholar]
- 7. Hoj L, Olsen RA, Torsvik VL. 2008. Effects of temperature on the diversity and community structure of known methanogenic groups and other archaea in high Arctic peat. ISME J. 2:37–48 [DOI] [PubMed] [Google Scholar]
- 8. Morozova D, Wagner D. 2007. Highly resistant methanogenic archaea from Siberian permafrost as candidates for the possible life on Mars. Int. J. Astrobiol. 6:59–87 [Google Scholar]
- 9. Collins G, Enright AM, Scully C, Mahony T, O'Flaherty V. 2006. Application and biomolecular monitoring of psychrophilic anaerobic digestion. Water Sci. Technol. 54:41–47 [DOI] [PubMed] [Google Scholar]
- 10. Connaughton S, Collins G, O'Flaherty V. 2006. Psychrophilic and mesophilic anaerobic digestion of brewery effluent: a comparative study. Water Res. 40:2503–2510 [DOI] [PubMed] [Google Scholar]
- 11. O'Reilly J, Lee C, Collins G, Chinalia FA, Mahony T, O'Flaherty V. 2009. Quantitative and qualitative analysis of methanogenic communities in mesophilically and psychrophilically cultivated anaerobic granular biofilms. Water Res. 43:3365–3374 [DOI] [PubMed] [Google Scholar]
- 12. McHugh S, Carton M, Collins G, O'Flaherty V. 2004. Reactor performance and microbial community dynamics during anaerobic biological treatment of wastewater at 16–37°C. FEMS Microbiol. Ecol. 48:369–378 [DOI] [PubMed] [Google Scholar]
- 13. McHugh S, Collins G, O'Flaherty V. 2006. Long-term, high-rate anaerobic biological treatment of whey wastewaters at psychrophilic temperatures. Bioresour. Technol. 97:1669–1678 [DOI] [PubMed] [Google Scholar]
- 14. McKeown R, Scully C, Enright AM, Chinalia FA, Lee C, Mahony T, Collins G, O'Flaherty V. 2009. Psychrophilic methanogenic community development during long-term cultivation of anaerobic granular biofilms. ISME. J. 3:1231–1242 [DOI] [PubMed] [Google Scholar]
- 15. Zhu J, Zheng H, Guomin A, Zhang G, Liu D, Xiaoli L, Dong X. 2012. The genome characteristics and predicted function of methyl-group oxidation pathway in the obligate aceticlastic methanogens, Methanosaeta spp. PLoS One 7:e36756. 10.1371/journal.pone.0036756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mori K, Lino T, Suzuki Yamaguchi K-IK, Kamagata Y. 2012. Aceticlastic and NaCl-requiring methanogen “Methanosaeta pelagica” sp. nov., isolated from marine tidal flat sediment. Appl. Environ. Microbiol. 78:3416–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fey A, Conrad R. 2000. Effect of temperature on carbon and electron flow and on the archaeal community in methanogenic rice field soil. Appl. Environ. Microbiol. 66:4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Griffin ME, McMahon KD, Mackie RI, Raskin L. 1998. Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol. Bioeng. 57:342–355 [DOI] [PubMed] [Google Scholar]
- 19. Collins G, Mahony T, O'Flaherty V. 2006. Stability and reproducibility of low-temperature anaerobic biological wastewater treatment. FEMS Microbiol. Ecol. 55:449–458 [DOI] [PubMed] [Google Scholar]
- 20. Diaz EE, Stams AJ, Amils R, Sanz JL. 2006. Phenotypic properties and microbial diversity of methanogenic granules from a full-scale upflow anaerobic sludge bed reactor treating brewery wastewater. Appl. Environ. Microbiol. 72:4942–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y, Xu H, Show K, Tay J. 2002. Anaerobic granulation technology for wastewater treatment. J. Microbiol. Biotechnol. 18:99–113 [Google Scholar]
- 22. Siggins A, Enright AM, O'Flaherty V. 2011. Low-temperature (7°C) anaerobic treatment of a trichloroethylene-contaminated wastewater: microbial community development. Water Res. 45:4035–4046 [DOI] [PubMed] [Google Scholar]
- 23. Siggins A, Enright AM, O'Flaherty V. 2011. Methanogenic community development in anaerobic granular bioreactors treating trichloroethylene (TCE)-contaminated wastewater at 37°C and 15°C. Water Res. 45:2452–2462 [DOI] [PubMed] [Google Scholar]
- 24. O'Reilly J, Lee C, Chinalia FA, Collins G, Mahony T, O'Flaherty V. 2010. Microbial community dynamics associated with biomass granulation in low-temperature (15 °C) anaerobic wastewater treatment bioreactors. Bioresour. Technol. 101:6336–6344 [DOI] [PubMed] [Google Scholar]
- 25. McKeown RM, Scully C, Mahony T, Collins G, O'Flaherty V. 2009. Long-term (1243 days), low-temperature (4–15 °C), anaerobic biotreatment of acidified wastewaters: bioprocess performance and physiological characteristics. Water Res. 43:1611–1620 [DOI] [PubMed] [Google Scholar]
- 26. Bock AK, Schonheit P. 1995. Growth of Methanosarcina barkeri (Fusaro) under nonmethanogenic conditions by the fermentation of pyruvate to acetate: ATP synthesis via the mechanism of substrate level phosphorylation. J. Bacteriol. 177:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mazumder TK, Nishio N, Fukuzaki S, Nagai S. 1986. Effect of sulfur-containing compounds on growth of Methanosarcina barkeri in defined medium. Appl. Environ. Microbiol. 52:617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muller V, Blaut M, Gottschalk G. 1986. Utilization of methanol plus hydrogen by Methanosarcina barkeri for methanogenesis and growth. Appl. Environ. Microbiol. 52:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feist AM, Scholten JCM, Palsson BO, Brockman FJ, Ideker T. 2006. Modelling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2:2006.0004. 10.1038/msb4100046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WM, Saunders E, Tapia R, Sowers KR. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188:7922–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buchenau B, Thauer RK. 2004. Tetrahydrofolate-specific enzymes in Methanosarcina barkeri and growth dependence of this methanogenic archaeon on folic acid or p-aminobenzoic acid. Arch. Microbiol. 182:313–325 [DOI] [PubMed] [Google Scholar]
- 32. Goenrich M, Duin EC, Mahlert F, Thauer RK. 2005. Temperature dependence of methyl-coenzyme M reductase activity and of the formation of the methyl-coenzyme M reductase red2 state induced by coenzyme B. J. Biol. Inorg. Chem. 10:333–342 [DOI] [PubMed] [Google Scholar]
- 33. Nomura T, Nagao T, Yoshihara A, Tokumoto H, Konishi Y. 2006. Selective immobilization of aceticlastic methanogens to support material. J. Soc. Powder Technol. Jpn. 43:653–659 (In Japanese.) [Google Scholar]
- 34. Clesceri LS, Greenberg AE, Eaton AD. (ed). 1998. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, DC [Google Scholar]
- 35. Hao X, Wang Q, Zhang X, Cao Y, van Loosdrecht MCM. 2009. Experimental evaluation of decrease in bacterial activity due to cell death and activity decay in activated sludge. Water Res. 43:3604–3612 [DOI] [PubMed] [Google Scholar]
- 36. Abram F, Enright AM, O'Reilly J, Botting CH, Collins G, O'Flaherty V. 2011. A metaproteomic approach gives functional insights into anaerobic digestion. J. Appl. Microbiol. 110:1550–1560 [DOI] [PubMed] [Google Scholar]
- 37. Gozal D, Jortani S, Ayelet BS, Kheirandish-Gozal L, Bhattacharjee R, Kim J, Capdevilla S. 2009. Two-dimensional differential in-gel electrophoresis proteomic approaches reveal urine candidate biomarkers in pediatric obstructive sleep apnea. Am. J. Respir. Care Med. 180:1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang G, Spellman DS, Skolink EY, Neubert TA. 2006. Quantitative phosphotyrosine proteomics of EphB2 signaling by stable isotope labeling with amino acids in cell culture (SILAC). J. Proteome Res. 5:581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shilova IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6:1638–1655 [DOI] [PubMed] [Google Scholar]
- 40. Rittmann BE. 2007. The membrane biofilm reactor is a versatile platform for water and wastewater treatment. Environ. Eng. Res. 12:157–175 [Google Scholar]
- 41. Veit K, Ehlers C, Ehrereich A, Salmon K, Hovey R, Gunsalus RP, Deppenmeier U, Schmitz RA. 2006. Global transcriptional analysis of Methanosarcina mazei strain Gö1 under different nitrogen availabilities. Mol. Genet. Genomics 276:41–55 [DOI] [PubMed] [Google Scholar]
- 42. Weidenbach K, Ehlers C, Kock J, Ehrenreich A, Schmitz RA. 2008. Insights into the NrpR regulon in Methanosarcina mazei Gö1. Arch. Microbiol. 190:319–332 [DOI] [PubMed] [Google Scholar]
- 43. Goodchild A, Raftery M, Saunders NF, Guilhaus M, Cavicchioli R. 2005. Cold adaptation of the Antarctic archaeon, Methanococcoides burtonii assessed by proteomics using ICAT. J. Proteome Res. 4:473–480 [DOI] [PubMed] [Google Scholar]
- 44. Gu J, Hilser VJ. 2009. Sequence-based analysis of protein energy landscapes reveals nonuniform thermal adaptation within the proteome. Mol. Biol. Evol. 26:2217–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Siddiqui KS, Cavicchioli R, Thomas T. 2002. Thermodynamic activation properties of elongation factor 2 (EF-2) proteins from psychrotolerant and thermophilic Archaea. Extremophiles 6:143–150 [DOI] [PubMed] [Google Scholar]
- 46. Thomas T, Cavicchioli R. 2000. Effect of temperature on stability and activity of elongation factor 2 proteins from Antarctic and thermophilic methanogens. J. Bacteriol. 182:1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang W, Culley DE, Brockman FL. 2006. DNA microarray analysis of anaerobic Methanosarcina barkeri reveals responses to heat shock and air exposure. J. Ind. Microbiol. Biotechnol. 33:784–790 [DOI] [PubMed] [Google Scholar]
- 48. Clarens M, Macario AJ, Conway de Marcario E. 1995. The archaeal dnaK-dnaJ gene cluster: organisation and expression in the methanogen Methanosarcina mazei. J. Mol. Biol. 250:191–201 [DOI] [PubMed] [Google Scholar]
- 49. De Biase A, Marcario AJ, Conway de Marcario E. 2002. Effect of heat stress on promoter binding by transcription factors in the cytosol of the archaeon Methanosarcina mazeii. Gene 282:189–197 [DOI] [PubMed] [Google Scholar]
- 50. Yoshimune K, Galkin A, Kulalova L, Yoshimura T, Esaki N. 2005. Cold-active DnaK of an Antarctic psychrotroph Shewanella sp. Ac10 supporting the growth of dnaK-null mutant of Escherichia coli at cold temperatures. Extremophiles 9:145–150 [DOI] [PubMed] [Google Scholar]
- 51. Hori T, Haruta S, Ueno Y, Ishii M, Igarashi Y. 2006. Dynamic transition of a methanogenic population in response to the concentration of volatile fatty acids in a thermophilic anaerobic digester. Appl. Environ. Microbiol. 72:1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]