Abstract
Accumulation of arsenic has potential health risks through consumption of food. Here, we inserted the arsenite [As(III)] S-adenosylmethionine methyltransferase (ArsM) gene into the chromosome of Pseudomonas putida KT2440. Recombinant bacteria methylate inorganic arsenic into less toxic organoarsenicals. This has the potential for bioremediation of environmental arsenic and reducing arsenic contamination in food.
TEXT
Arsenic is a class I human carcinogen that poses a health risk to humans. Arsenic exposure is linked to skin cancer, bladder cancer, diabetes, cardiovascular disease, and peripheral vascular disease (1, 2). The U.S. Environmental Protection Agency (EPA) ranks arsenic first on its Superfund List of Hazardous Substances (http://www.atsdr.cdc.gov/SPL/index.html).
Arsenic is released into the environment by geothermal activity, by dissolution of minerals, and by anthropogenic activities such industrial effluents, combustion of fossil fuels, and the use of arsenic-containing pesticides, herbicides, wood preservatives, and feed additives (3). As a result of the use of arsenic-contaminated irrigation water, arsenic accumulates in rice, the dietary staple for half the world's population (4). Arsenic methylation is a detoxification pathway (5, 6). Many organisms have genes that encode arsenite [As(III)] S-adenosylmethionine (SAM) methyltransferases (termed ArsM in microbes and AS3MT in higher organisms) that biotransform As(III) into methylated species, with volatile nontoxic trimethylarsine [TMAs(III)] (7) as the end product (5, 6, 8, 9). Pseudomonas putida is a Gram-negative bacterium found in water and soil, particularly in the rhizosphere at a relatively high population density (10). This microorganism has been studied extensively as a model for biodegradation of aromatic compounds such as naphthalene (11) and styrene (12, 13). Conventional remediation methods, such as soil excavation followed by coagulation filtration or ion exchange, are expensive, disruptive, and not widely used (14). Sphingomonas desiccabilis and Bacillus idriensis expressing arsM can remove arsenic from contaminated soil, but expression from a plasmid limits their utility (15). Pseudomonas species have the prospect of rhizoremediation of organic compounds (16) but have not been used for arsenic removal.
The objective of this study was to construct a strain of P. putida KT2440 with the potential for removal of arsenic from contaminated soil. We used the Chlamydomonas reinhardtii arsM gene encoding an ArsM orthologue (CrArsM). In vitro, purified CrArsM methylated As(III) to a variety of species (see Fig. S1A in the supplemental material). After 7 h, methylarsenite [MAs(III)] and dimethylarsenate [DMAs(V)] were produced in relatively equal amounts. After 14 h, the product was primarily DMAs(V), with lesser amounts of trimethylarsine oxide [TMAs(V)O] and no MAs(III). These results are consistent with sequential methylation steps to the mono-, di-, and trimethyl products. TMAs(III) gas could be detected on H2O2-impregnated filters by oxidation to TMAs(V)O (see Fig. S1B in the supplemental material). These results demonstrate that purified CrArsM catalyzes three sequential rounds of As(III) methylation and converts toxic inorganic arsenic to less toxic or nontoxic organic arsenicals.
The C. reinhardtii arsM gene behind the kanamycin promoter was integrated into the chromosome of P. putida KT2440, which does not have an arsM gene and does not methylate arsenic. Minitransposon delivery plasmid pBAM1 was used as a suicide vector to generate stable integrants that could express arsM constitutively. The arsM gene was cloned into pBAM1 (see Fig. S2 in the supplemental material) and subsequently transferred from Escherichia coli CC118λpir to P. putida KT2440 by tripartite conjugation with a helper strain. Wild-type P. putida has two chromosomal arsRBCH operons and can grow in the presence of 2 mM As(III), which provides a competitive advantage to P. putida in contaminated soil (10). This could be a crucial factor for sustaining growth of cells in the presence of indigenous bacterial populations (14). Cells of P. putida KT2440 expressing CrArsM were resistant to 7.5 to 10 mM As(III) in liquid basal salt M9 medium (Fig. 1). Biotransformation of arsenic by the cells was assayed with 25 μM As(III) or arsenate [As(V)] (Fig. 2). After 12 h, engineered P. putida biomethylated As(III) primarily to DMAs(V) and, to a lesser degree, methylarsenate [MAs(V)] (Fig. 2A). In a time-dependent fashion, the engineered cells produced dimethylarsine [DMAs(III)H] and TMAs(III) gases, identified by oxidizing them to DMAs(V) and TMAs(V)O with H2O2 (Fig. 2B). In addition, the product of the methylation reaction was quantified in cells of P. putida expressing CrArsM. After 48 h, the major product found in the culture medium was DMAs(V) (57% of total arsenic), with lesser amounts of MAs(V) (31%) and even less TMAs(V)O (8%) (Fig. 2C). The transgenic P. putida strain rapidly methylated As(V) (Fig. 2A, curve 4). There are two arsRCBH operons in the chromosome of P. putida, so it is reasonable to assume the chromosomally encoded ArsC reductase rapidly reduced As(V) to As(III), the substrate of CrArsM, allowing the soil bacterium to methylate both As(V) and As(III).
Fig 1.
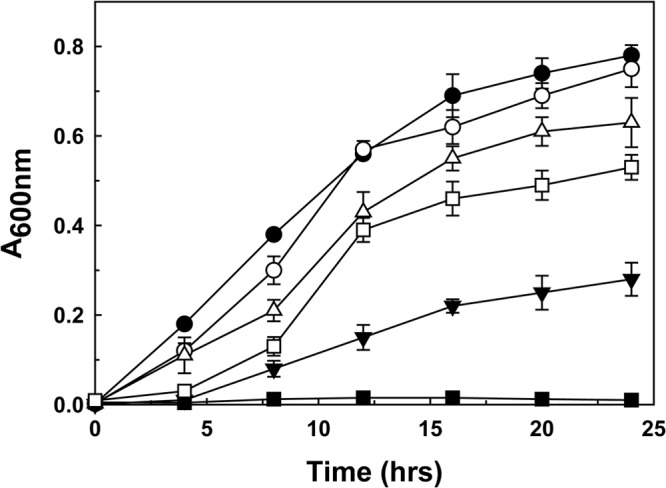
As(III) resistance in P. putida expressing arsM. Expression of arsM confers tolerance to As(III) at the indicated concentrations in M9 medium after overnight growth. Filled symbols, P. putida KT2440/pBAM1; open symbols, P. putida KT2440 with integrated arsM; circles, 2 mM As(III); inverted triangles, 7.5 mM As(III); squares, 10 mM As(III). Data are means ± standard error (SE) (n = 3 experiments).
Fig 2.
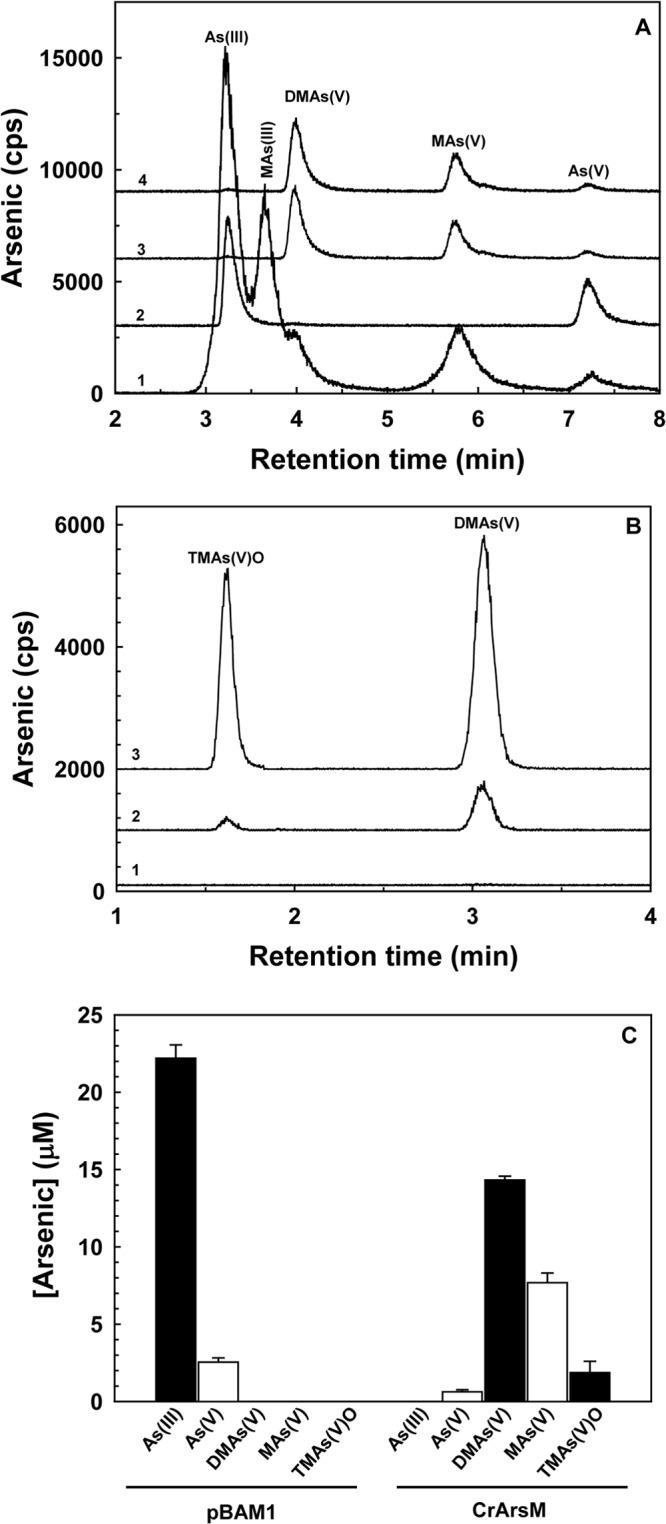
Biotransformation of As(III) in P. putida expressing arsM. (A) Cells of P. putida KT2440 bearing vector plasmid pBAM1 (control) or P. putida KT2440 with arsM stably integrated into the chromosome were grown overnight in Luria-Bertani medium with 25 μM As(III). Arsenical species in solution were separated and identified by anion-exchange high-performance liquid chromatography (HPLC) coupled to inductively coupled plasma mass spectroscopy (ICP-MS). Curve 1, standards; curve 2, P. putida cells bearing pBAM1 incubated with As(III); curve 3, P. putida with integrated arsM incubated with As(III); curve 4, P. putida with integrated arsM incubated with 25 μM As(V). (B) Volatilization of arsenic by P. putida with integrated arsM for 0 (curve 1), 12 h (curve 2), or 48 h (curve 3) was determined by trapping the gas on H2O2-impregnated filters, elution, and separation of species by HPLC–ICP-MS using an anion-exchange column. (C) After 48 h of growth with 25 μM As(III) in LB medium, arsenic species in solution produced by P. putida bearing pBAM1 (left) or with integrated arsM (right) were quantified by HPLC–ICP-MS. Data are means ± SE (n = 3 experiments).
Under comparable conditions, cells of E. coli BL21(DE3) expressing arsM produced mainly DMAs(V) (see Fig. S1A in the supplemental material), while P. putida expressing arsM produced significant amounts of MAs(V) (Fig. 2A and C). The monomethyl species is the precursor of the dimethyl species (17), indicating that the rate of methylation in E. coli is greater than that in P. putida. The amount of CrArsM produced by cells of both species expressing arsM was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by quantitative Western blotting using an antibody to the six-histidine tag on CrArsM (see Fig. 4S in the supplemental material). More enzyme is produced in E. coli, most likely because more CrArsM is produced from a plasmid behind the strong T7 promoter than from the chromosomally carried gene in P. putida. Yet, lower expression in P. putida may be advantageous under environmental conditions, where overproduction of a heterologous protein can be detrimental to in situ performance (18). P. putida expressing arsM is 5-fold more resistant to arsenite than the wild-type strain, which gives it the ability to grow in arsenic-contaminated soils. Within 16 h, the cells transformed nearly all of the inorganic arsenic in the culture medium to the less toxic methylated species, including MAs(V), DMAs(V), TMAs(V)O, and eventually TMAs(III) gas, which further reduces the content of inorganic arsenic in soil and surface waters.
While this study provides a proof of concept, there is an opportunity to improve the process in P. putida, perhaps with a further engineered expression system that responds to arsenic in the medium (e.g., see reference 19). Thus, it is reasonable to propose that methylated arsenicals produced by engineered pseudomonads can ameliorate the effects of environmental contamination by inorganic arsenic. In addition, while higher plants do not have an arsM gene and do not methylate arsenic, rice transports and accumulates methylated arsenicals produced by paddy microorganisms (20). These organoarsenicals are not only less toxic than inorganic arsenic, but they are much more efficiently translocated from root to shoot and concentrated in the plant tissues. The methylated species can be taken up and concentrated in plants, creating possibilities for rhizo- and phytoremediation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R37 GM55425 to B.P.R.
We thank Sabeeha Merchant, UCLA, for advice and suggestions for cloning the C. reinhardtii arsM gene.
Footnotes
Published ahead of print 3 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01133-13.
REFERENCES
- 1. Tchounwou PB, Centeno JA, Patlolla AK. 2004. Arsenic toxicity, mutagenesis, and carcinogenesis—a health risk assessment and management approach. Mol. Cell. Biochem. 255:47–55 [DOI] [PubMed] [Google Scholar]
- 2. Abernathy CO, Thomas DJ, Calderon RL. 2003. Health effects and risk assessment of arsenic. J. Nutr. 133:1536S–1538S [DOI] [PubMed] [Google Scholar]
- 3. Tchounwou PB, Patlolla AK, Centeno JA. 2003. Carcinogenic and systemic health effects associated with arsenic exposure—a critical review. Toxicol. Pathol. 31:575–588 [DOI] [PubMed] [Google Scholar]
- 4. Williams PN, Raab A, Feldmann J, Meharg AA. 2007. Market basket survey shows elevated levels of As in South Central U.S. processed rice compared to California: consequences for human dietary exposure. Environ. Sci. Technol. 41:2178–2183 [DOI] [PubMed] [Google Scholar]
- 5. Qin J, Lehr CR, Yuan C, Le XC, McDermott TR, Rosen BP. 2009. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc. Natl. Acad. Sci. U. S. A. 106:5213–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. 2006. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 103:2075–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cullen WR. 2005. The toxicity of trimethylarsine: an urban myth. J. Environ. Monit. 7:11–15 [DOI] [PubMed] [Google Scholar]
- 8. Ye J, Rensing C, Rosen BP, Zhu YG. 2012. Arsenic biomethylation by photosynthetic organisms. Trends Plant Sci. 17:155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yin XX, Chen J, Qin J, Sun GX, Rosen BP, Zhu YG. 2011. Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant Physiol. 156:1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canovas D, Cases I, de Lorenzo V. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5:1242–1256 [DOI] [PubMed] [Google Scholar]
- 11. Gomes NC, Kosheleva IA, Abraham WR, Smalla K. 2005. Effects of the inoculant strain Pseudomonas putida KT2442 (pNF142) and of naphthalene contamination on the soil bacterial community. FEMS Microbiol. Ecol. 54:21–33 [DOI] [PubMed] [Google Scholar]
- 12. Mooney A, Ward PG, O'Connor KE. 2006. Microbial degradation of styrene: biochemistry, molecular genetics, and perspectives for biotechnological applications. Appl. Microbiol. Biotechnol. 72:1–10 [DOI] [PubMed] [Google Scholar]
- 13. Ward PG, Goff M, Donner M, Kaminsky W, O'Connor KE. 2006. A two-step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ. Sci. Technol. 40:2433–2437 [DOI] [PubMed] [Google Scholar]
- 14. Wu CH, Wood TK, Mulchandani A, Chen W. 2006. Engineering plant-microbe symbiosis for rhizoremediation of heavy metals. Appl. Environ. Microbiol. 72:1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu S, Zhang F, Chen J, Sun G. Arsenic removal from contaminated soil via biovolatilization by genetically engineered bacteria under laboratory conditions. J. Environ. Sci. 23:1544–1550 [DOI] [PubMed] [Google Scholar]
- 16. Shim H, Chauhan S, Ryoo D, Bowers K, Thomas SM, Canada KA, Burken JG, Wood TK. 2000. Rhizosphere competitiveness of trichloroethylene-degrading, poplar-colonizing recombinant bacteria. Appl. Environ. Microbiol. 66:4673–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marapakala K, Qin J, Rosen BP. 2012. Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry 51:944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cases I, de Lorenzo V. 2005. Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int. Microbiol. 8:213–222 [PubMed] [Google Scholar]
- 19. Merulla D, Hatzimanikatis V, van der Meer JR. 15 January 2013. Tunable reporter signal production in feedback-uncoupled arsenic bioreporters. Microb. Biotechnol. [Epub ahead of print.] 10.1111/1751-7915.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lomax C, Liu WJ, Wu L, Xue K, Xiong J, Zhou J, McGrath SP, Meharg AA, Miller AJ, Zhao FJ. 2011. Methylated arsenic species in plants originate from soil microorganisms. New Phytol. 193:665–672 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


